Abstract
ComX and CSF are Bacillus subtilis extracellular signaling peptides. Many different strains of B. subtilis do not communicate due to strain-specific variation of ComX. We demonstrate that CSF is a species-specific signaling molecule that partially compensates for the lack of ComX-mediated communication between different strains of B. subtilis.
Quorum sensing is the ability of cells to sense and respond to a high cell density. The gram-positive bacterium Bacillus subtilis may use as many as five secreted peptide signals to mediate quorum sensing and activate the transcription factor ComA (4, 8, 18). ComA is part of a two-component regulatory system and is activated by phosphorylation via the ComP histidine-protein kinase (11, 30). ComX pheromone is an extracellular signaling peptide that activates the phosphorylation of ComP (20, 27, 28). In addition to ComX, the CSF, PhrF, PhrK, and possibly PhrH peptides also stimulate ComA activity by first being transported into the cell by an oligopeptide permease (4, 19). In the cytoplasm, they bind to their cognate Rap protein to antagonize the inhibitory activity of the Rap proteins on ComA (8, 11). CSF is also able to inhibit ComA-controlled gene expression when present at high concentrations through an incompletely defined mechanism (19).
Knowing that five extracellular signaling peptides stimulate ComA activity raises the question of whether the different signaling peptides provide different information for B. subtilis. Genes activated by ComA include those involved in extracellular and membrane functions and regulators of development, including genetic competence and sporulation (10). At least two of these peptides, ComX and CSF, by virtue of their secretion into the growth medium, activate these processes in response to high cell density (5, 17, 20, 26). ComX also appears to signal whether there are other B. subtilis organisms of the same strain present in the environment. Genetic polymorphisms were found for different strains of B. subtilis and its close relative, Bacillus mojavensis, in genes involved in producing and responding to the ComX pheromone, comQXP (1, 2, 28, 29). Each strain produces a ComX that specifically activates ComA-controlled gene expression in that strain; however, in some cases, a limited set of pheromones from other strains partially activate or inhibit ComA-controlled gene expression (2, 28). Since ComX is a strain-specific signaling molecule, this raised the question of whether other ComA-activating signal peptides exhibited strain-specific variation. Here, we show that CSF is a conserved signaling peptide produced by B. subtilis and other closely related species and that CSF can partially compensate for the lack of ComX pheromone communication between strains.
Conservation of rapC-phrC from Bacillus strains producing different ComX pheromones.
To determine the CSF pherotypes of B. subtilis and B. mojavensis strains, we sequenced the rapC-phrC operon, encoding CSF and its cytoplasmic receptor RapC, from six strains previously shown to have different comQXP sequences (28) (Table 1). Chromosomal DNA was isolated from B. subtilis strains RO-OO-2, RO-FF-1, and RS-B-1 and B. mojavensis strains RO-B-2, RO-H-1, and RO-C-2 by using a standard protocol (9). The rapC-phrC locus was then PCR amplified using Taq polymerase (Qiagen) and primers that corresponded to the sequence of the B. subtilis 168 strain. For each strain, three independent PCR products were sequenced from two independent chromosomal DNA preparations.
TABLE 1.
Strains used in this study
| Species and straina | Relevant genotypeb | Source or constructionc |
|---|---|---|
| B. subtilis JH642 derivatives | ||
| B. subtilis BAL188 (JRL293) | amyE::(srfA-lacZΩ1974 cat) | 27 |
| B. subtilis BAL201 (AG1520) | ΔcomQ::spc | 20 |
| B. subtilis BAL218 (JH642) | trpC2 pheA1 | 22 |
| B. subtilis BAL941 | ΔphrC::tet | 16 |
| B. subtilis BAL1141 | thrC::(xylAp-xylR erm) | pRDC19→BAL218 |
| B. subtilis BAL1385 | thrC::(comQXPJH642erm) | pBL58→BAL218 |
| B. subtilis BAL1386 | thrC::(comQXPRS-B-1erm) | pBL220→BAL218 |
| B. subtilis BAL1391 | thrC::(comQXPJH642erm) ΔcomQ::spc | BAL201→BAL1385 |
| B. subtilis BAL1392 | thrC::(comQXPRS-B-1erm) ΔcomQ::spc | BAL201→BAL1386 |
| B. subtilis BAL1397 | thrC::(xylAp-xylR erm) ΔcomQ::spc | BAL201→BAL1141 |
| B. subtilis BAL1416 | amyE::(srfA-lacZΩ1974 cat) ΔphrC::tet | BAL941→BAL188 |
| B. subtilis BAL1419 | thrC::(comQXPRS-B-1erm) ΔcomQ::spc ΔphrC::tet | BAL941→BAL1392 |
| Non-B. subtilis JH642 derivatives | ||
| B. mojavensis RO-B-2 | Wild type | 23 |
| B. mojavensis RO-C-2 | Wild type | 23 |
| B. subtilis RO-FF-1 | Wild type | 23 |
| B. mojavensis RO-H-1 | Wild type | 23 |
| B. subtilis RO-OO-2 | Wild type | 23 |
| B. subtilis RS-B-1 | Wild type | 23 |
| B. subtilis BAL1399 | As RO-OO-2, except ΔphrC::tet | BAL941→RO-OO-2 |
| B. subtilis BAL1400 | As RS-B-1, except ΔphrC::tet | BAL941→RS-B-1 |
| B. subtilis BAL1404 | As RO-FF-1, except ΔphrC::tet | BAL941→RO-FF-1 |
Names in parentheses are original laboratory strain names.
All JH642 derivatives contain trpC2 and pheA1 mutations.
The direction of the strain construction is indicated as DNA→recipient strain.
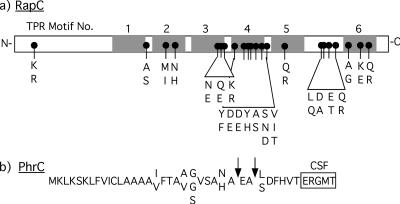
Sequencing data showed that the rapC-phrC regions were greater than 95% and 97% identical between strains of B. subtilis and B. mojavensis, respectively. The rapC-phrC region showed the greatest difference between strains of B. subtilis and B. mojavensis, with a range of 89 to 90% identity. In contrast, the hypervariable region of comQXP from these strains showed ∼70% identity (2, 28). None of the differences were predicted either to affect the function of rapC-phrC (Fig. 1) or to change the region corresponding to the ComA-binding box in the promoter region of rapC, which suggests that this promoter would be similarly regulated in these strains. Most of the amino acid differences in RapC and PhrC across the strains were conservative and would not be predicted to affect the function of these proteins. In particular, the sequence of the mature CSF peptide was conserved among all of the strains (Fig. 1B), and all of the PhrC protein variants were predicted by the SignalP 3.0 program to have a functional signal sequence and signal peptidase cleavage sites (7). These data suggest that for strains that produce different ComX pheromones, they all produce identical CSF peptides.
FIG. 1.
Locations of amino acid differences in the RapC and PhrC proteins of B. subtilis and B. mojavensis. (a) Black dots show the locations of amino acid differences. Below these dots are the amino acids found at these positions. The positions of these substitutions are shown relative to the known structural feature of RapC. (b) Linear amino acid sequence of PhrC. Where there are amino acid differences, two or three amino acids are listed. The amino acid shown above the linear sequence is that found in B. subtilis strains. The amino acids shown below the linear sequence are those found in B. mojavensis strains. Arrows indicate putative signal peptidase cleavage sites as determined by SignalP 3.0 (7). The sequence of the mature CSF peptide is boxed.
B. subtilis strains secrete a functional CSF peptide.
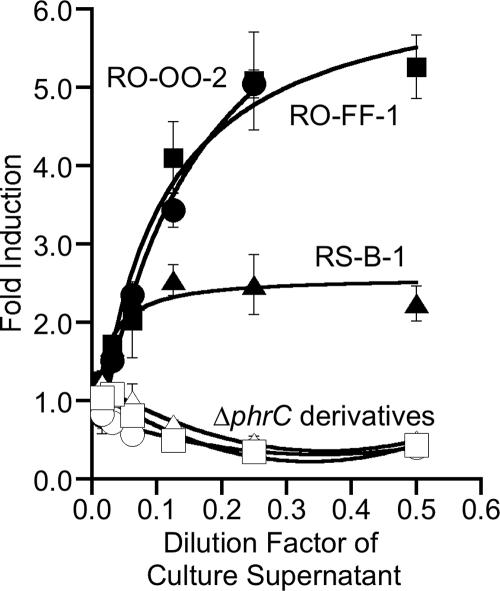
To confirm that different B. subtilis strains secrete a functional CSF peptide, we asked whether three wild B. subtilis strains could secrete a factor that would induce ComA-controlled gene expression in the laboratory strain of B. subtilis. As the wild strains do not produce a form of ComX pheromone which the laboratory strain can sense and respond to, any ComA-activating factor secreted by the wild strains should be CSF or another Phr peptide. B. subtilis strains RO-OO-2, RO-FF-1, and RS-B-1 were grown under standard conditions for the production of CSF by the laboratory strain (16, 17, 26), and then cell-free culture supernatants were harvested. These culture supernatants were tested for CSF activity using a standard CSF detection assay (16, 17, 26) in which dilutions of the culture supernatants were incubated with cells from the reporter strain BAL188 (srfA-lacZ) and then assayed for β-galactosidase specific activity (21). The srfA-lacZ fusion in the reporter strain is a fusion of the promoter region of the ComA-activated srfA gene to the gene for β-galactosidase (27). Culture supernatants from strains RO-OO-2, RO-FF-1, and RS-B-1 induced the expression of srfA-lacZ (Fig. 2). It is curious that the culture supernatant from strain RS-B-1 maximally induced srfA-lacZ expression 2.5-fold, whereas the other tested strains maximally induced srfA-lacZ expression 5.0-fold. The reason for this difference is most likely due to ComX of strain RS-B-1 (ComXRS-B-1) inhibiting ComA-controlled gene expression by the laboratory strain, as ComXRS-B-1 is very similar in sequence to ComXRO-H-1, which has been shown to inhibit ComA-controlled gene expression by the laboratory strain (2). Furthermore, the slopes of the dose-response curves of the different strains are identical and differ only in the induction level at which they plateau, suggesting that that these strains produce similar levels of the stimulating factor, either CSF or another Phr peptide.
FIG. 2.
Wild-type isolates of B. subtilis secrete CSF. Culture supernatants from B. subtilis wild isolates and their ΔphrC derivatives were tested for the ability to induce ComA-controlled gene expression in the BAL188 (JH642 srfA-lacZ) reporter strain. The increase in induction was calculated as the level of β-galactosidase specific activity for cells treated with culture supernatant divided by the level of activity for untreated cells. Plotted is the average increase in induction from three independent experiments with the standard error of the mean. Filled and open symbols correspond to results for culture supernatants from strains that either have the gene for CSF or do not (ΔphrC), respectively. Conditioned medium was from the following B. subtilis strains: RO-OO-2 (circles), RO-FF-1 (squares), and RS-B-1 (triangles).
To demonstrate that the factor in the culture supernatants that was able to induce srfA-lacZ expression in the laboratory strain was CSF, a ΔphrC mutation was introduced into the RO-OO-2, RO-FF-1, and RS-B-1 strains. Genetically competent cells of these strains were prepared by growing the strains in a defined minimal medium (14) to an optical density at 600 nm (OD600) of 2 to 3; these cells were then transformed with chromosomal DNA from BAL941 (ΔphrC::tet), using standard protocols (12). Culture supernatants from these ΔphrC derivatives were assayed, as described above, and were found to be unable to induce srfA-lacZ expression (Fig. 2). These data indicate that phrC, and by extension CSF, was required for the inducing activity produced by the wild B. subtilis strains.
Effect of CSF on the quorum response of cultures of mixed-ComX-pherotype cells.
To determine the role of CSF in the context of a mixed community producing different ComX pheromones, we created cultures in which 10% of the cells were of a “reporter strain” that produced and responded to ComXJH642 and had a srfA-lacZ fusion. The remaining 90% of cells were of a “neighbor strain” that produced ComXJH642, ComXRS-B-1, or no ComX pheromone. To create isogenic neighbor strains, either the comQXP region from JH642 or RS-B-1 or a heterologous gene was introduced into the chromosome at the thrC locus. The comQXP locus was PCR amplified, from 200 bp upstream of comQ to 11 bp downstream of comP, and then cloned into the BamHI and EcoRI sites of the thrC integration vector pRDC19 (3). The resulting plasmids, pBL58 [thrC::(comQXPJH642 erm)], pBL220 [thrC::(comQXPRS-B-1 erm)], and pRDC19 [thrC::(xylAp-xylR erm)], were each transformed into BAL218, and transformants in which thrC was replaced with the indicated alleles were selected. To prevent ComX pheromone production from the native locus, the ΔcomQ::spc allele was backcrossed into these strains.
The reporter and neighbor strains were grown separately in defined minimal medium with shaking at 37°C to an OD600 of ∼0.1, at which point the reporter strain and one of the neighbor strains were mixed. β-Galactosidase specific activity was measured during growth of the mixed culture, and the percentage of the culture that was comprised by each of the two strains was measured at the end of growth as the number of CFU per ml on the appropriate selective agar plates.
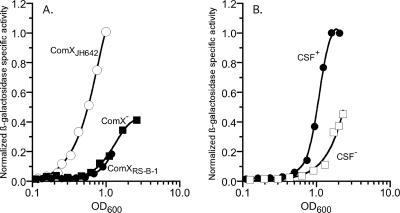
When the reporter strain was grown in the presence of a neighbor strain producing ComXJH642, the expression of srfA-lacZ was induced at an OD600 of ∼0.2 (Fig. 3A). In contrast, srfA-lacZ expression was induced two cell doublings later when the reporter strain was grown in the presence of a strain that either did not produce ComX (n = 3; P < 0.01) or produced ComXRS-B-1 (n = 3; P < 0.05) (Fig. 3A). These data confirm that the laboratory strain, JH642, does not respond to the ComX pheromone from the RS-B-1 strain. They further indicate that the inability to sense the presence of cells from other strains of B. subtilis via the ComX pheromone delays the expression of ComA-controlled genes when cells are grown in a mixed community.
FIG. 3.
CSF partially compensates for the lack of ComX pheromone communication and leads to the induction of ComA-controlled gene expression at a lower cell density. Mixed cultures in which 10% of the cells were of a reporter strain that had a srfA-lacZ fusion and 90% were of a neighbor strain producing a different complement of extracellular signaling peptides were grown. β-Galactosidase specific activity measurements were normalized by dividing the level of β-galactosidase specific activity for each point by the highest level of β-galactosidase specific activity obtained for the group of strains assayed in one experiment. The graphs shown are representative of three independent experiments. (A) For all three cultures, all cells produced CSF, and BAL188 (srfA-lacZ ComXJH642) was the reporter strain. The neighbor strain either produced ComXJH642 (BAL1391) (open circles), ComXRS-B-1 (BAL139) (filled circles), or no ComX pheromone (ComX−; BAL1397) (filled squares). (B) BAL1416 (srfA-lacZ ΔphrC ComXJH642) does not produce CSF and was the reporter strain. The neighbor strain produced ComXRS-B-1 and was either phenotypically CSF+ (BAL1392) (filled circles) or CSF− (BAL1419) (open squares).
We next determined what role CSF had in activating ComA-controlled gene expression in a mixed culture that does not communicate by ComX pheromone. To accomplish this, we determined the effect of the presence and absence of CSF in the mixed-culture assay described above. Strains lacking CSF were created by backcrossing the ΔphrC mutation into the reporter strain (BAL188) and the neighbor strain expressing ComXRS-B-1 (BAL1419) (Table 1). Two mixed cultures were then set up, in which the reporter strain, which cannot produce CSF (BAL1416), was grown with either a neighbor strain that expressed comQXPRS-B-1 and CSF (BAL1392) or a neighbor strain that expressed comQXPRS-B-1 but not CSF (BAL1419) (Fig. 3B). For the culture in which CSF was not produced, srfA-lacZ expression was induced at 0.5 cell doublings later than it was induced in the culture that produced CSF (n = 3; P < 0.02). These data indicate that CSF mediates communication between different ComX pherotype strains and leads to the induction of ComA-controlled gene expression at a lower cell density than can be achieved without CSF. CSF only partially compensated for the lack of ComX pheromone communication, as ComA-controlled gene expression was delayed when strains of different ComX pherotypes were grown together, even in the presence of CSF.
Conclusions and implications.
The reason that B. subtilis produces multiple signaling molecules that converge to regulate the quorum response is unclear. In light of the strain specificity exhibited by ComX pheromone, we set out to determine whether the CSF signaling molecule was also strain specific. Our data indicate that CSF is not; it is a conserved signaling molecule produced by strains of B. subtilis and the closely related species B. mojavensis. Furthermore, CSF can mediate communication between strains that are unable to communicate via ComX. This is the first evidence that a bacterium that exhibits strain level specificity for a secreted signaling molecule produces an additional signaling molecule to allow communication between strains. Strain-specific variation has also been observed for the extracellular signaling peptides that induce genetic competence in Streptococcus pneumoniae (31) and virulence gene expression in Staphylococcus aureus (15) and Bacillus cereus (24), and it will be interesting to learn whether these bacteria also produce a species-specific signaling molecule, as does B. subtilis.
It is unknown why bacteria secrete multiple signaling molecules that differ in the ability to communicate the relatedness of the surrounding bacteria. In addition to the fact that B. subtilis produces strain- and species-specific signals, Vibrio harveyi produces species- and genus-specific signals, as well as a universal signal (6, 13, 32). The fact that bacteria as distantly related as V. harveyi and B. subtilis exhibit this phenomenon suggests that this may be a common property of signaling molecules produced by many bacterial species.
To understand why B. subtilis produces both strain- and species-specific signaling molecules, it will be necessary to discern the selective pressure that has led to the strain level of variation in ComX pheromone. Since ComX pheromone is required for the development of genetic competence (i.e., the natural ability to take up DNA), one consequence of ComX pheromone variation is sexual isolation from different strains (25). Therefore, it has been proposed that by B. subtilis cells responding only to ComX pheromone from genetically identical clones, these cells avoid the uptake of DNA from closely related, but not genetically identical, strains that could carry harmful genetic elements (1). If sexual isolation is the fitness-enhancing factor that has selected for the diversification of the ComX pheromone, then what advantage does communication via CSF provide for strains of different ComX pherotypes? Perhaps B. subtilis has to strike a balance between sexual isolation and cooperation for other processes activated by ComX and CSF through the ComA transcription factor (e.g., surfactin or secreted pectate lyase production) (10). And so, although B. subtilis possibly excludes harmful genetic variation via the lack of ComX communication, it can still gain the benefits of cooperation by signaling via the conserved molecule CSF. If CSF accumulates to sufficient levels to be sensed by B. subtilis cells, which is dependent on the accumulation of ComX itself (17), the likelihood of the existence of a high density of cooperative cells may be sufficiently great to make induction of the quorum response possibly beneficial. Clearly, there are many experiments yet to be done to understand the potential benefits offered to a bacterium that can communicate at both the strain and species levels.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the obtained sequences are as follows: AY664811 (B. subtilis RO-OO-2), AY667584 (B. subtilis RO-FF-1), AY667585 (B. subtilis RS-B-1), AY667586 (B. mojavensis RO-B-2), AY672642 (B. mojavensis RO-H-1), and AY672643 (B. mojavensis RO-C-2).
Acknowledgments
We thank Denise Guzman for her assistance in sequencing the rapC-phrC loci and Fredrick Cohn (Wesleyan University) for the B. subtilis and B. mojavensis strains sequenced in this study.
This work was supported by NIH Public Health Service grant AI48616 to B.A.L. M.P. was supported in part by USPHS National Research Service Award GM07185.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Ansaldi, M., and D. Dubnau. 2004. Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J. Bacteriol. 18615-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaldi, M., D. Marolt, T. Stebe, I. Mandic-Mulec, and D. Dubnau. 2002. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol. Microbiol. 441561-1573. [DOI] [PubMed] [Google Scholar]
- 3.Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M.-L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16851-856. [DOI] [PubMed] [Google Scholar]
- 4.Auchtung, J. M., C. A. Lee, and A. D. Grossman. 2006. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol. 1885273-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon Schneider, K., T. M. Palmer, and A. D. Grossman. 2002. Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1794043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 8.Bongiorni, C., S. Ishikawa, S. Stephenson, N. Ogasawara, and M. Perego. 2005. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J. Bacteriol. 1874353-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bron, S. 1990. Plasmids, p. 75-174. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, England.
- 10.Comella, N., and A. D. Grossman. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 571159-1174. [DOI] [PubMed] [Google Scholar]
- 11.Core, L., and M. Perego. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 491509-1522. [DOI] [PubMed] [Google Scholar]
- 12.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 13.Henke, J. M., and B. L. Bassler. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 1866902-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 1714121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 2762027-2030. [DOI] [PubMed] [Google Scholar]
- 16.Lanigan-Gerdes, S., A. N. Dooley, K. F. Faull, and B. A. Lazazzera. 2007. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis. Mol. Microbiol. 651321-1333. [DOI] [PubMed] [Google Scholar]
- 17.Lazazzera, B. A., I. G. Kurtser, R. S. McQuade, and A. D. Grossman. 1999. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J. Bacteriol. 1815193-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazazzera, B. A., T. Palmer, J. Quisel, and A. D. Grossman. 1999. Cell density control of gene expression and development in Bacillus subtilis, p. 27-46. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, DC.
- 19.Lazazzera, B. A., J. M. Solomon, and A. D. Grossman. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89917-925. [DOI] [PubMed] [Google Scholar]
- 20.Magnuson, R., J. Solomon, and A. D. Grossman. 1994. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77207-216. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 22.Perego, M., and J. A. Hoch. 1988. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J. Bacteriol. 1702560-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts, M. S., and F. M. Cohan. 1995. Recombination and migration rates in natural populations of Bacillus subtilis and Bacillus mojavensis. Evolution 491081-1094. [DOI] [PubMed] [Google Scholar]
- 24.Slamti, L., and D. Lereclus. 2005. Specificity and polymorphism of the PlcR-PapR quorum-sensing system in the Bacillus cereus group. J. Bacteriol. 1871182-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon, J. M., and A. D. Grossman. 1996. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 12150-155. [DOI] [PubMed] [Google Scholar]
- 26.Solomon, J. M., B. A. Lazazzera, and A. D. Grossman. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 102014-2024. [DOI] [PubMed] [Google Scholar]
- 27.Solomon, J. M., R. Magnuson, A. Srivastava, and A. D. Grossman. 1995. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 9547-558. [DOI] [PubMed] [Google Scholar]
- 28.Tortosa, P., L. Logsdon, B. Kraigher, Y. Itoh, I. Mandic-Mulec, and D. Dubnau. 2001. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J. Bacteriol. 183451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran, L. S., T. Nagai, and Y. Itoh. 2000. Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 371159-1171. [DOI] [PubMed] [Google Scholar]
- 30.Weinrauch, Y., R. Penchev, E. Dubnau, I. Smith, and D. Dubnau. 1990. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 4860-872. [DOI] [PubMed] [Google Scholar]
- 31.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 1813144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6191-197. [DOI] [PubMed] [Google Scholar]