Abstract
The gram-negative enteric bacterium Proteus mirabilis is a frequent cause of urinary tract infections in individuals with long-term indwelling catheters or with complicated urinary tracts (e.g., due to spinal cord injury or anatomic abnormality). P. mirabilis bacteriuria may lead to acute pyelonephritis, fever, and bacteremia. Most notoriously, this pathogen uses urease to catalyze the formation of kidney and bladder stones or to encrust or obstruct indwelling urinary catheters. Here we report the complete genome sequence of P. mirabilis HI4320, a representative strain cultured in our laboratory from the urine of a nursing home patient with a long-term (≥30 days) indwelling urinary catheter. The genome is 4.063 Mb long and has a G+C content of 38.88%. There is a single plasmid consisting of 36,289 nucleotides. Annotation of the genome identified 3,685 coding sequences and seven rRNA loci. Analysis of the sequence confirmed the presence of previously identified virulence determinants, as well as a contiguous 54-kb flagellar regulon and 17 types of fimbriae. Genes encoding a potential type III secretion system were identified on a low-G+C-content genomic island containing 24 intact genes that appear to encode all components necessary to assemble a type III secretion system needle complex. In addition, the P. mirabilis HI4320 genome possesses four tandem copies of the zapE metalloprotease gene, genes encoding six putative autotransporters, an extension of the atf fimbrial operon to six genes, including an mrpJ homolog, and genes encoding at least five iron uptake mechanisms, two potential type IV secretion systems, and 16 two-component regulators.
Proteus mirabilis is not a common cause of urinary tract infections (UTI) in normal hosts (65). Surveys of uncomplicated cystitis or acute pyelonephritis show that P. mirabilis causes only a few percent of cases. Even in patients with recurrent UTI, the incidence of infection by this organism is only a few percentage points higher. However, this organism infects a very high proportion of patients with complicated urinary tracts, that is, urinary tracts with functional or anatomic abnormalities or with chronic instrumentation. In these patients, not only does this bacterium cause cystitis and acute pyelonephritis (20-22, 65, 73), but the production of urinary stones, a hallmark of infection with this organism (23), further compromises the already complicated urinary tract. P. mirabilis has sporadically been reported to be a causative agent of bacteremia and nosocomial infections (59); additional evidence suggests that this species is associated with rheumatoid arthritis (62).
P. mirabilis is a highly motile gram-negative bacterium (52). The ability of P. mirabilis to differentiate from short vegetative swimmer cells to an elongated, highly flagellated swarmer form, originally noted by Hauser in 1885, is a distinctive feature of this organism (63). P. mirabilis is well known to microbiologists as the species that forms a bull's-eye pattern on an agar plate as it swarms over the entire surface of the medium, although the swarm morphotype is rarely found in the urinary tract (28).
On the other hand, adherence is also a key virulence property of P. mirabilis. This organism attaches to and swarms across the surface of urinary catheters to gain a foothold in the urinary tract (32). Once within the bladder, bacteria counteract the flushing motion of urine flow by attaching to epithelial cells (29). Adherence factors may also play a role in the ability of this organism to colonize the kidneys or cross the renal barrier into the bloodstream. Five fimbriae have been described to date for P. mirabilis: mannose-resistant Proteus-like (MR/P) fimbriae (1, 6), uroepithelial cell adhesin (UCA) or nonagglutinating fimbriae (NAF) (17, 79), P. mirabilis fimbriae (PMF) (1, 48), ambient-temperature fimbriae (ATF) (46), and P. mirabilis P-like pili (PMP) (11). MR/P fimbriae, UCA, and PMF have been shown to play a role in UTI pathogenesis in virulence studies (6, 48, 79). Among these, the MR/P fimbriae have been best studied. During infection, these phase-variable fimbriae are selected for and contribute significantly to the fitness of P. mirabilis (6, 81).
Clinically, P. mirabilis is perhaps best known for its ability to form stones in the bladder and kidney (40), as well as its ability to form crystalline biofilms on the outer surface and in the lumen of indwelling urinary catheters (57). Urolithiasis (stone formation), a major clinical problem in patients infected with P. mirabilis, is caused by the induction of urease, which hydrolyzes urea to ammonia, causing the local pH to rise and subsequent precipitation of magnesium ammonium phosphate (struvite) and calcium phosphate (apatite) crystals (23, 56, 57). The stones resulting from aggregation of such crystals complicate infection for three reasons. First, P. mirabilis caught within the interstices of the forming stones is difficult to clear with only antibiotics. Second, a stone serves as a nidus for non-P. mirabilis bacteria to establish UTI, which also are difficult to eradicate. Third, a stone can obstruct urine flow; pelvic and renal stones are often associated with pyonephrosis and/or chronic pyelonephritis.
We and other laboratories have previously identified virulence determinants based on prominent phenotypes such as hemolysis, adherence, and motility (5, 6, 15, 46, 47, 54, 55, 71). Genetic screens including signature-tagged mutagenesis have uncovered additional virulence properties with more subtle in vitro phenotypes (13, 82). The complete nucleotide sequence now defines each coding sequence (CDS) in the bacterium and opens the door for a more thorough analysis of its strategy for pathogenesis. Here we describe the 4.063-Mb genome of P. mirabilis HI4320 that was isolated from the urine of a nursing home patient with a long-term (≥30 days) indwelling urinary catheter (31, 78). This strain is considered a low-passage clinical isolate. The sequence reveals that P. mirabilis possesses many features of a true pathogen and has refined the ability to switch from multiple adherence mechanisms to a highly motile state.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
P. mirabilis strain HI4320, a prototypical isolate that is representative of the species (54), was isolated at a concentration of ≥105 CFU/ml from the urine of an elderly female nursing home patient with a long-term (≥30 days) indwelling catheter (78). This scenario is representative of where P. mirabilis is most commonly isolated in the United States, the United Kingdom, and indeed worldwide. This strain has been the subject of numerous virulence studies with the CBA mouse model of ascending UTI, including a signature-tagged mutagenesis study in which over 3,000 mutants were screened for attenuation of virulence (13, 82).
Genomic DNA preparation.
P. mirabilis was cultured in 100 ml Luria broth overnight at 37°C with aeration. The culture was split into 10 10-ml portions for DNA isolation. Genomic DNA from strain HI4320 was isolated using Qiagen Genomic-tips with the genomic DNA buffer set (Qiagen) according to the manufacturer's instructions. Purified DNA was pooled and quantified by spectrophotometry; an A260/A280 ratio of 2 was found, confirming the purity of the DNA sample.
Cloning and sequencing.
Genome assembly was obtained from 60,288 paired end sequences (giving eightfold coverage) derived from seven genomic shotgun libraries (four in pUC19 with insert sizes of 2.6 to 3.0, 3.0 to 3.3, 3.3 to 4.0, and 2.0 to 4.0 kb; two in pMAQ1b_SmaI with insert sizes of 4.0 to 5.0 and 5.0 to 6.0 kb; and one in pBACehr with insert sizes of 25 to 40 kb) using dye terminator chemistry with ABI3700 automated sequencers.
Sequence analysis, annotation, and comparative genomics.
The sequence was assembled, finished, and annotated as described previously (69), using Artemis (66) to collate data and facilitate annotation. The DNA and predicted protein sequences of P. mirabilis were compared to those of the sequenced Escherichia coli genomes using the Artemis Comparison Tool (ACT) (14). Orthologous gene sets were calculated by using reciprocal best-match FASTA comparisons with subsequent manual curation. Pseudogenes had one or more mutations that prevented translation; each of the inactivating mutations was checked against the original sequencing data. Horizontally acquired DNA was identified by anomalies in G+C content and GC deviation combined with the presence of mobility genes, such as genes encoding integrases and transposases. Horizontally acquired DNA was also identified by the Alien Hunter algorithm, which implements the interpolated variable order motifs theory and predicts compositionally deviating regions (74), and by anomalies in the G+C content and GC deviation. Additional analysis of the mrp and mrp′ fimbrial operons and flagellar regulon was conducted using Lasergene GeneQuest software (v.7.0.0; DNASTAR).
RNA extraction and Northern blotting.
P. mirabilis HI4320 was cultured to mid-exponential phase (optical density at 600 nm, 0.7). RNA was isolated by using the method of Maroncle et al. (45). Briefly, 2 ml of a bacterial culture was collected by centrifugation (6,000 × g, 8 min, 4°C). The bacterial pellet was suspended in 10 mM morpholinepropanesulfonic acid (MOPS) (pH 7.3), 10 mM KCl, and 5 mM MgCl2. An equal volume of a solution containing 20 mM MOPS (pH 7.5), 40 mM EDTA, 200 mM NaCl, and 1% sodium dodecyl sulfate was added, and the mixture was boiled for 2 min. Lysates were extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1). Then 0.05 volume of 1 M sodium acetate was added, followed by 0.7 volume of 100% ethanol. The RNA was allowed to precipitate for 20 min at −20°C and collected by centrifugation (9,500 × g, 15 min, 4°C). The pellet was washed once with 70% ethanol, allowed to air dry, and resuspended in RNase-free water. Contaminating DNA was removed with DNase (Ambion).
Probes corresponding to 5S rRNA, the 5′ and 3′ ends of both 16S and 23S rRNA, and an insertion sequence within some copies of 23S rRNA were constructed by PCR amplifying the chromosomal regions corresponding to rRNA genes using oligonucleotide primers shown in Table S1 in the supplemental material. These PCR amplicons were ligated into pCR2.1 (Invitrogen). E. coli Top10 was electroporated with the ligation mixture, and plasmids with inserts cloned in the orientation opposite to that of the T7 promoter were selected. Plasmids were linearized with SpeI to facilitate transcription. The T7 promoter was used to transcribe probes for Northern blotting using a DIG Northern starter kit (Roche) according to the manufacturer's instructions. Total RNA from HI4320 was separated on a gel and hybridized with labeled probes according to the DIG Northern starter kit protocol.
Nucleotide sequence accession numbers.
The sequence and annotation of the P. mirabilis chromosome and plasmid have been deposited in the EMBL database under accession numbers AM942759 and AM942760, respectively.
RESULTS
General features of the P. mirabilis genome.
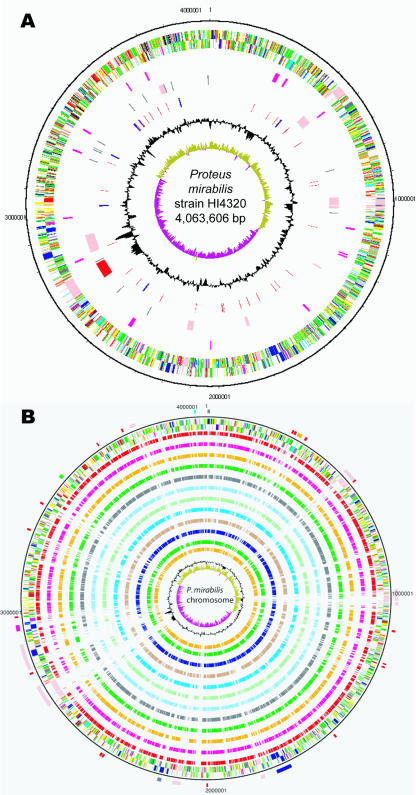
The P. mirabilis genome consists of a 4.063-Mb chromosome and a 36,289-nucleotide (nt) plasmid (pHI4320) containing 3,685 and 55 CDSs, respectively. The origin of replication was assigned based on the GC deviation of the genome and was found to be between mioC (PMI3054) and gidA (PMI3055). The P. mirabilis chromosome is considerably smaller than the chromosomes of other Enterobacteriaceae (average, 4.6 Mb), including uropathogenic strains of E. coli (CFT073, 5.23 Mb; UTI89, 5.21Mb; 536, 4.94 Mb). General features of the genome are shown in Table 1, and schematic representations of the P. mirabilis chromosome and plasmid are shown in Fig. 1 and 2, respectively.
TABLE 1.
General features of the P. mirabilis genome
| Genetic element | Size (bp) | G+C content (%) | No. of coding sequences | Coding density | Avg gene size (bp) | No. of insertion elements | No. of rRNA operons | No. of tRNA genes | No. of noncoding RNA genes | No. of pseudogenes |
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | 4,063,606 | 38.9 | 3,693 | 0.90 | 941 | 15 | 7 | 83 | 18 | 24 |
| Plasmid | 36,289 | 36.21 | 53 | 1.46 | 606 | 0 | 0 | 0 | 0 | 0 |
FIG. 1.
Circular representations of the genome of P. mirabilis. (A) The circles indicate (from the outside in) the positions of the following genes: circles 1 and 2, all genes (transcribed clockwise and counterclockwise); circle 3, fimbrial operons; circle 4, mobile elements (pink, prophages; red, integrative and conjugative elements; black, insertion elements); circle 5, RNA genes (blue, rRNA genes; red, tRNA genes); circle 6, G+C content (plotted using a 10-kb window); circle 7, GC deviation ([G − C]/[G + C] plotted using a 10-kb window; khaki, values more than 1; purple, values less than 1). (B) Comparison of P. mirabilis with other enteric genomes. The outer scale shows the positions (in bp). The circles indicate (from the outside in) the positions of the following genes. Circles 1 and 2 show all genes transcribed clockwise and counterclockwise, respectively (for an explanation of the colors, see below). Circles 3 to 14 show the positions of P. mirabilis genes that have orthologs (as determined by reciprocal FASTA) in E. coli UTI89, E. coli CFT073, E. coli 536, E. coli APEC O1, E. coli O157:H7 Sakai, E. coli E24377A, E. coli K-12, P. luminescens, K. pneumoniae, S. enterica serovar Typhimurium, Y. enterocolitica, and Y. pseudotuberculosis, respectively. Circle 15 shows a plot of the G+C content (plotted using a 10-kb window), and circle 16 shows a plot of GC deviation ([G − C]/[G + C] plotted using a 10-kb window; khaki, values more than 1; purple, values less than 1). The positions of the following important regions (mentioned in the text) are indicated on the outermost circle: putative toxins (black), fimbrial operons (red), Zap proteases (orange), flagella (dark blue), type III secretion system (purple), O antigen biosynthesis (green), urease (light blue), and mobile elements (pink). Colors in both panels for genes in circles 1 and 2 indicate the following: dark blue, cell processes, adaptation, or pathogenicity; black, energy metabolism; red, information transfer; dark green, surface associated; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central or intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical proteins; brown, pseudogenes; pink, mobile elements; gray, miscellaneous.
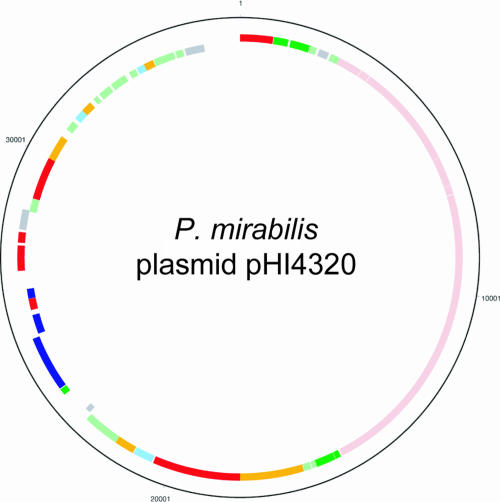
FIG. 2.
Circular representation of the P. mirabilis HI4320 plasmid. Circles 1 and 2 (from the outside in) show all 55 CDSs transcribed clockwise and counterclockwise, respectively. For an explanation of the colors, see the legend to Fig. 1. Of particular interest, CDSs indicated by pink encode a type IV conjugal transfer pilus, and CDSs indicated by blue encode a proticin and immunity system.
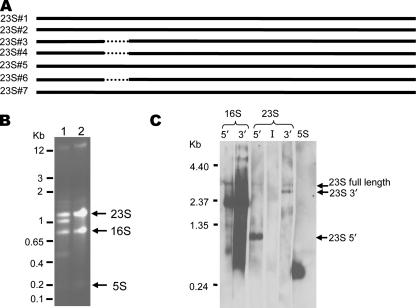
One of the most striking features of the HI4320 genome is related to the seven complete rRNA operons; six of them are 16S-23S-5S operons, and one is a 16S-23S-5S-5S operon. Four of the seven 23S rRNA genes have an intervening 173-nt sequence between nt 542 and 543 (Fig. 3A). The insertion appears to be removed and degraded during processing, resulting in splitting of the 23S rRNA into 542- and 2,359-nt fragments. Both the intact 23S rRNA from the three uninterrupted operons and the processed 542- and 2,359-nt 23S rRNA fragments are visible when total RNA from P. mirabilis HI4320 is separated on an agarose gel (Fig. 3B, lane 1). Northern blot analysis of P. mirabilis rRNA confirmed that the extra two rRNA species compared to an E. coli RNA control are the processed 23S rRNA fragments (Fig. 3C, lanes 23S-5′ and 23S-3′). Furthermore, a probe specific for the insertion sequence does not hybridize with any of the rRNA species, confirming the rapid excision and degradation of this fragment (Fig. 3C, lane 23S-I). Insertion sequences in P. mirabilis 23S rRNA have been reported previously (51). The insertion in the 23S rRNA of strain HI4320 appears to correspond with one of the previously reported insertion sites in P. mirabilis (helix 25) (51).
FIG. 3.
(A) Alignment of the seven 23S rRNA gene copies in HI4320. The dotted lines indicate where an insertion is present in four of seven copies of the 23S rRNA gene. Insertions are present in rRNA gene sequences 1, 2, 5, and 7. (B) RNA was isolated from P. mirabilis HI4320 and uropathogenic E. coli strain CFT073 and resolved on a 1.5% agarose gel. Lane 1, HI4320 RNA; lane 2, CFT073 RNA. The positions of double-stranded DNA molecular weight markers are indicated on the left. (C) Probes were designed to recognize the beginning and end of the 16S rRNA gene; the beginning, insertion sequence, and end of the 23S rRNA gene; and the 5S rRNA gene. These probes were used in a Northern blot analysis of HI4320 RNA. The intervening sequence of the 23S rRNA gene is believed to be excised and degraded (51), leaving behind the 5′ and 3′ fragments, which appear as the second and fourth bands in the agarose gel in panel B.
P. mirabilis mobile/extrachromosomal genome.
In addition to the 4-Mb chromosome, P. mirabilis HI4320 was also found to carry a plasmid. Not all P. mirabilis strains carry this plasmid or other plasmids. The P. mirabilis HI4320 plasmid, designated pHI4320, is highly related to R6K from E. coli (34) and encodes the π protein (encoded by PMIP01) and R6K replication proteins (encoded by PMIP37 to PMIP39). Interestingly, two independent transposon insertions in one of the plasmid CDSs (PMIP09) that encodes a conjugal transfer protein were previously found to alter the virulence of P. mirabilis (13). Since it is clear that this CDS is not involved in virulence per se, it is possible that the plasmid carries other genes that might contribute to virulence. The in silico analysis of the plasmid sequence did not reveal any obvious or known virulence factors. However, the plasmid carries CDSs encoding a bacteriocin and its immunity system (PMIP30 and PMIP31), which may confer a competitive advantage to the bacterium in the urinary tract.
The genome sequence also revealed that other potentially laterally acquired elements have also contributed to the genome evolution of this strain. These elements include both prophage and genomic islands, some of which have an anomalous G+C content or GC skew compared to the surrounding DNA. The genome contains three apparently complete prophages (PMI0456 to PMI0530, PMI0892 to PMI0947, and PMI1917 to PMI2007) and three degenerate prophages (PMI0953 to PMI0967, PMI1710 to PMI1727, and PMI3468 to PMI3493). In addition, the genome has a conjugative transposon (PMI2423 to PMI2491) similar to the ICE R391 from Providencia rettgeri (12) and another large genomic island (PMI2549 to PMI2642) which encodes an adhesin (PMI2575), as well as siderophore biosynthesis and transport genes (nrpSUTABG).
Motility and chemotaxis.
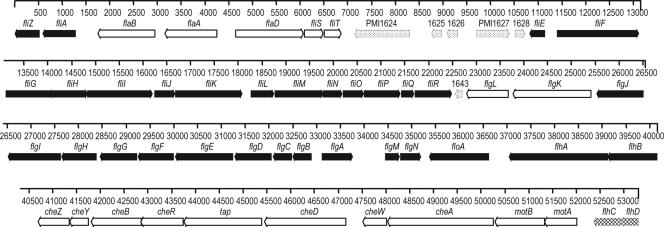
Interestingly, all flagellum-related genes are located within a single 53.3-kb locus (Fig. 4). The chemotaxis genes cheA, cheW, cheD, tap, cheR, cheB, cheY, and cheZ are also located within this locus. The localization of all flagellar genes within one contiguous locus is unusual, if not unique, among bacterial species. This locus also includes six CDSs (PMI1624 to PMI1628 and PMI1643) which encode proteins with unknown functions; however, it is worth noting that one of these CDSs (PMI1627) encodes a protein that is weakly similar (26% amino acid sequence identity) to the major fimbrial subunit (FloA) of CS18 fimbriae in E. coli (26). The role played by these genes in motility, if any, remains to be determined. Six other genes outside the flagellar locus encode methyl-accepting chemotaxis proteins (PMI1180, PMI2380, PMI2381, PMI2671, PMI2808, and PMI2809), and one aerotaxis gene, aer (PMI2813), was identified. Genes encoding several previously identified regulators of P. mirabilis flagellar motility were also located in the chromosome, including umoA (PMI3115), umoB (PMI3018), umoC (PMI1939), umoD (PMI0876) (19), rssBA (PMI1696 and PMI1697) (35), wosA (PMI0608) (25), and the rcsBCD regulatory system (PMI1729 to PMI1731) composed of genes encoding the sensor kinase RcsC, the phosphotransfer intermediate protein RcsD (YojN) (72) (also described as rsbA [42]), and the response regulator RcsB (16).
FIG. 4.
All genes that encode flagellar components are located in a 54-kb contiguous region of the HI4320 genome. There are 50 genes that are known to be involved in flagellar structure, regulation, or chemotaxis in this locus. In addition, there are five hypothetical or unknown genes between fliT and fliE (indicated by stippled arrows). It is unusual among the Enterobacteriaceae to find all the flagellar genes in one locus of the chromosome. Class 1 genes are indicated by cross-hatched arrows, class 2 genes are indicated by filled arrows, and class 3 genes are indicated by open arrows.
Fimbriae and pili.
Five fimbriae have been previously identified and studied to varying degrees: MR/P fimbriae (1, 6), UCA (NAF) (17, 79), PMF (MR/K) (1, 5), ATF (46), and PMP (11). Analysis of the HI4320 genome sequence revealed 12 additional previously undescribed chaperone-usher fimbrial operons, bringing the total to 17 potential fimbriae encoded by P. mirabilis (Table 2). Included in this list is an apparent duplication of the mrp operon immediately adjacent to mrpI (Fig. 5). The duplicated mrp operon, designated mrp′, does not appear to have an invertible element in its promoter (38), suggesting that expression of this operon is not responsive to the MrpI recombinase. Interestingly, the previously characterized atf fimbrial operon (47) was described as having only three genes (atfABC); the sequence revealed an additional three genes in the operon (atfABCDEJ). This places P. mirabilis among the bacteria encoding the most distinct types of fimbriae. Ten of these fimbrial gene clusters encode homologs of MrpJ, a repressor of motility (M. M. Pearson and H. L. T. Mobley, submitted for publication).
TABLE 2.
Chaperone-usher fimbrial operons
| Genes | Designation |
|---|---|
| PMI0254-PMI0261 | mrp′ (66.7% identical at the nucleotide level to mrp operon) |
| PMI0262-PMI0271 | mrp (mannose-resistant/Proteus-like fimbria) |
| PMI0296-PMI0304 | Fimbria 3 |
| PMI0532-PMI0536 | uca (uroepithelial cell adhesin); also called naf (nonagglutinating fimbria) |
| PMI1060-PMI1067 | Fimbria 5 |
| PMI1185-PMI1190 | Fimbria 6 |
| PMI1193-PMI1197 | Fimbria 7 |
| PMI1464-PMI1470 | Fimbria 8 |
| PMI1877-PMI1881 | pmf (P. mirabilis fimbria); also called MR/K |
| PMI2207-PMI2214 | Fimbria 10 |
| PMI2216-PMI2224 | pmp (P. mirabilis P-like pili) |
| PMI2533-PMI2539 | Fimbria 12 |
| PMI2728-PMI2733 | atf (ambient temperature fimbria) |
| PMI2997-PMI3003 | Fimbria 14 |
| PMI3086-PMI3093 | Fimbria 15 |
| PMI3348-PMI3352 | Fimbria 16 |
| PMI3435-PMI3440 | Fimbria 17 |
FIG. 5.
The well-studied mrp fimbrial operon, mrpABCDEFGHJ, is required for full virulence in the CBA/J mouse model of ascending UTI (6). The mrpI gene encodes a recombinase that controls the invertible promoter element for the mrp operon (38). Sequence analysis of the completed HI4320 genome revealed a duplication of the mrp operon immediately upstream of mrpI. This mrp operon, currently designated mrp′ (open arrows), is missing the mrpB gene and the mrpI recombinase but otherwise is highly similar to mrp (filled arrows). The percentages of similarity of the predicted MR/P′ protein sequences to the MR/P protein sequences are indicated above the mrp′ genes.
In addition to the 17 fimbrial operons, there are 13 putative orphan fimbrial genes that are not part of a complete operon. PMI2622 to PMI2624 encode a fimbrial usher, a chaperone containing a frameshift and two internal stop codons, and a structural protein; this region likely comprises a cryptic fimbrial operon. The other genes (PMI1033, PMI1627, PMI1812, PMI1920, PMI1922, PMI1924, PMI1929, PMI2154, and PMI3023) are isolated from other fimbrial genes but could act in trans to further increase the variable complement of fimbriae (33). Notably, PMI1627 is located within the flagellar locus.
There are several genes which are potentially involved in the assembly of type IV pili, such as genes encoding a putative prepilin (PMI2315), a prepilin peptidase-dependent protein (PMI2314), and a prepilin peptidase (PMI0342), suggesting that P. mirabilis may produce one or more type IV pili.
Nonfimbrial adhesins and toxins.
P. mirabilis HI4320 possesses several genes that may encode adhesins; PMI1359 and PMI2950 are similar (46 and 31% amino acid identity, respectively) to the ail (attachment and invasion locus) gene of Yersinia enterocolitica (50).
Six potential autotransporter (type V secretion system) proteins are encoded by P. mirabilis HI4320. Three of these proteins (encoded by PMI0844, PMI2126, and PMI2341) have the canonical autotransporter C terminus (pfam03797) and also possess serine protease motifs and thus may serve as toxins. The other three putative autotransporters (encoded by PMI2122, PMI2174, and PMI2575) are members of the trimeric autotransporter family (C terminus with the pfam03895 motif) (18) and appear to be adhesins.
In addition to the adhesins, the genome carries several CDSs that potentially encode toxins. PMI0004 and PMI2043 encode products with homology to the cytotoxin RtxA (43). Two contiguous CDSs, PMI1747 and PMI1748, encode a binary toxin similar to the apoptotic toxin XaxAB from the insect pathogen Xenorhabdus nematophila (75). PMI0023 encodes a putative intimin/invasin. Three contiguous CDSs, PMI2028 to PMI2030, encode a type I secretion system that may be involved in the secretion of some of these toxins and adhesins, particularly the two RtxA paralogs.
Genes encoding all the components necessary to form a type III secretion system were identified (PMI2681 to PMI2704). This type III secretion system, which does not appear to play a role in the urinary tract, is described in more detail elsewhere (61).
The genome also encodes two two-partner secretion systems (PMI0592 and PMI0593; PMI2056 and PMI2057). The PMI0592-PMI0593 system is similar to the cdiABI locus that appears to encode a contact-dependent inhibition system. E. coli strains that express this system are able to halt the growth of other E. coli strains that lack cdiABI in a nonlethal manner (4). The second two-partner secretion system (PMI2056 and PMI2057) was previously identified as containing the hemolysin genes hpmBA (71).
Other virulence factors.
The previously characterized immunoglobulin A-degrading metalloprotease (zap) locus zapEABCD (PMI0276 to PMI0281) (76) was found to have three additional copies of zapE at the beginning of the locus (PMI0282 to PMI0285). The role of these zapE paralogs in Zap metalloprotease function has not been determined yet.
LPS and capsule.
Although at least 74 O serogroups have been identified for Proteus species (80), the structure of P. mirabilis HI4320 lipopolysaccharide (LPS) has not been elucidated yet. Table S2 in the supplemental material lists genes predicted to be involved in LPS and enterobacterial common antigen biosynthesis.
To date, the P. mirabilis capsular polysaccharide (CPS) has been defined for only one strain, ATCC 49565; in this case, the CPS was formed from the same structure as the LPS O chain (10). Thus, it is likely that genes listed in Table S2 in the supplemental material also contribute to P. mirabilis CPS formation (PMI3187 to PMI3197). Additionally, the glycosyl transferase gene cpsF (PMI3190) has been shown to contribute to P. mirabilis CPS (24). Five genes implicated in the regulation of capsule synthesis have been located in HI4320: rcsA (PMI1680), rcsB (PMI1730), rcsC (PMI1731), rcsD (yojN/rsbA) (PMI1729), and rcsF (PMI2262).
Iron acquisition.
The genome encodes a large number of proteins that are potentially involved in scavenging a wide range of iron substrates. At least eight iron uptake loci were annotated in the P. mirabilis genome, including afuCBA (PMI0188 to PMI0190), sitDCBA (PMI1024 to PMI1027), the B12 transporter locus btuCD (PMI1041 to PMI1042), the hemin uptake system locus hmuR1R2STUV (PMI1425 to PMI1430), the ferric siderophore receptor locus ireA (PMI1945), and the ferrous iron transport locus feoAB (PMI2920 to PMI2921). A possible heme-binding system is encoded by PMI3120 and PMI3121, and a putative iron receptor is located adjacent to a fecR homolog located at PMI3706-PMI3707. There are also three other iron-related ABC transporters (encoded by PMI0229 to PMI0238, PMI2675 to PMI2677, and PMI2957 to PMI2960) and 10 additional TonB-dependent receptors (encoded by PMI0233, PMI0363, PMI0409, PMI0842, PMI1548, PMI2596, PMI2618, PMI2619, PMI2680, and PMI3246).
Intracellular stress.
Three superoxide dismutases are encoded by the following P. mirabilis genes: sodA (PMI3036) (Mn superoxide dismutase), sodB (PMI1397) (Fe superoxide dismutase), and sodC (PMI1333) (CuZn superoxide dismutase). Genes encoding catalase (katA; PMI1740), soxRS (PMI2705 and PMI1597), and oxyR (PMI3241) were also annotated in the chromosome.
Drug resistance.
P. mirabilis HI4320 is known to be tetracycline resistant and displays intermediate resistance to chloramphenicol. Genes conferring these resistance traits were located in the genome; tetAJ (PMI2399) encodes a tetracycline resistance protein, and cat (PMI0728) encodes chloramphenicol acetyltransferase. Other potential resistance genes located in the chromosome include a tellurite resistance operon (ter; PMI2383 to PMI2393), the bicyclomycin (sulfonamide) resistance gene bcr (PMI0829), a catB3-like putative acetyltransferase gene (PMI1693), and the kasugamycin resistance gene ksgA (PMI2334). Genes involved in multidrug efflux or CDSs that predict possible antimicrobial resistance are listed in Table S3 in the supplemental material.
Metabolism.
P. mirabilis has intact pathways for glycolysis, the tricarboxylic acid cycle, gluconeogenesis, and the pentose phosphate shunt. However, several redundant enzymes that are present in these pathways in E. coli are missing in P. mirabilis, including fumAB, tktB, talA, pfkB, and gpmI.
Arguably, the most important aspect of P. mirabilis metabolism is its ability to degrade urea. The urea-inducible urease gene cluster PMI3681 to PMI3688 (ureRDABCEFG) encodes a multimeric nickel-metalloenzyme that hydrolyzes the abundant substrate in urine; this enzyme is required for virulence and urolithiasis (stone formation) and has been well characterized (27, 30).
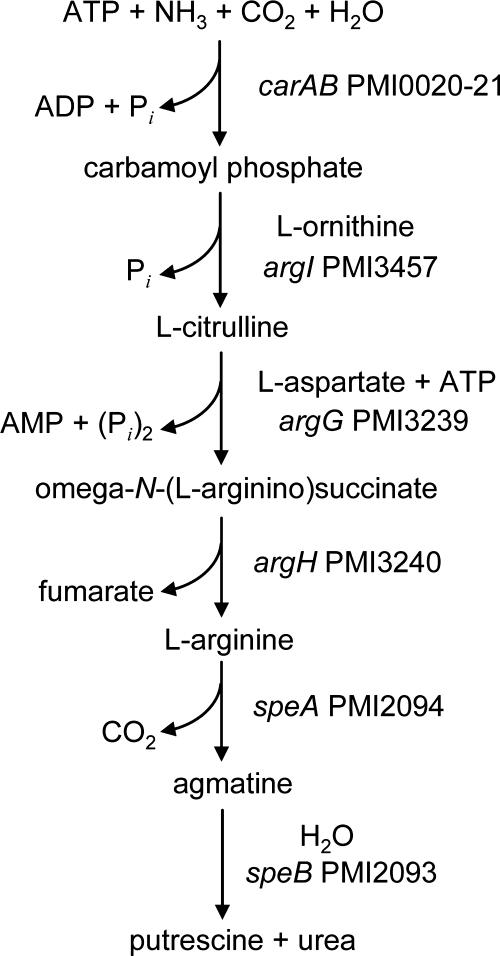
The polyamine putrescine has been previously shown to act as an extracellular signal required for swarming in P. mirabilis (70). We identified two different likely pathways for the production of putrescine by P. mirabilis. The first pathway involves a single enzyme, ornithine decarboxylase (SpeF) (encoded by PMI0307), which decarboxylates ornithine to form putrescine. Interestingly, both import of the substrate (ornithine) and export of the product (putrescine) are mediated by a single protein, a putrescine-ornithine antiporter (encoded by PMI0306 [potE]). The second putrescine biosynthesis pathway includes several steps, as shown in Fig. 6.
FIG. 6.
Predicted generation of putrescine and urea from ammonia. P. mirabilis HI4320 possesses seven genes potentially encoding a pathway that converts ammonia, ATP, carbon dioxide, and water into putrescine and urea in six steps.
It is intriguing that P. mirabilis can potentially synthesize urea (Fig. 6) and hydrolyze urea using its urease. It may be that the synthesis of urea is useful when the bacterium is outside the urinary tract and lacks this substrate. Another interesting possibility can be raised: in the presence of excess urea, one could speculate that urea is consumed in the reverse reactions to synthesize ATP for energy stores. If this pathway is indeed reversible, this could also explain why P. mirabilis is rarely found as swarmer cells in the urinary tract (28); excess urea could lead to consumption of putrescine, which contributes to swarming.
Regulation.
Sixteen potential two-component regulator systems were identified and are summarized in Table S4 in the supplemental material. There appears to be a surprising lack of genes encoding synthesis or degradation proteins of the messenger cyclic di-GMP. Only one gene (PMI3101) was found that encodes a putative signaling protein with a GGDEF domain (used in cyclic di-GMP synthesis). No genes encoding EAL or HD-GYP domains, associated with the degradation of cyclic di-GMP (64, 67), were found.
Genes encoding at least seven sigma factor subunits for RNA polymerase have been identified in the genome. The primary sigma factor RpoD (σ70, σA) is encoded by PMI2372. Genes encoding two sigma factors similar to those involved in heat shock and cell envelope stress were identified: rpoH (PMI3608) encoding the heat shock sigma factor σH or σ32 and rpoE (PMI1894) encoding σE or σ24. An rpoN gene, encoding a σ54 sigma factor involved in nitrogen regulation, is encoded by PMI3648. The rpoF (fliA, PMI1618) gene encodes σF (σ28), the class 2 activator in the flagellar cascade (Fig. 4). A FecR homolog is encoded by PMI3708. Finally, the stress and stationary-phase sigma factor RpoS is encoded by PMI2236.
We also identified 18 small noncoding RNA genes that are likely involved in several regulatory functions. Chief among these are two copies of ryhB, which encodes a 90-nt RNA that down-regulates iron storage and iron-using proteins under iron-limiting conditions (49).
Comparative genomics.
The genome of P. mirabilis was compared (by reciprocal FASTA) to the genomes of 12 members of the Enterobacteriaceae, including seven E. coli strains (UTI89, CFT073, 536, APEC O1, O157:H7 Sakai, E24377A, and K-12), Photorhabdus luminescens, Klebsiella pneumoniae, Salmonella enterica serovar Typhimurium, Y. enterocolitica, and Yersinia pseudotuberculosis (Fig. 1B). This comparison identified 784 CDSs that are unique to P. mirabilis compared to the 12 enteric genomes mentioned above (see Table S5 in the supplemental material). Interestingly, a large number of these P. mirabilis-specific CDSs encode virulence-related functions, such as fimbriae, adhesins, autotransporters, O antigen, and proteases (see Table S5 in the supplemental material).
Comparative genomic analysis also revealed 1,747 P. mirabilis CDSs that are shared with the three sequenced genomes of uropathogenic E. coli, the genomes of strains 536, UTI89, and CFT073, including basic metabolic pathways. In addition, of the 131 genes that were previously identified as uropathogenic E. coli specific (present in 10 uropathogenic E. coli strains but not in three commensal strains) (44), 25 are present in P. mirabilis (Table 3). Eleven of these genes are related to iron acquisition, a critical virulence trait of uropathogens. Also, the genes encoding an uncharacterized ABC transporter (PMI1480 to PMI1484) and two regulatory systems (ibrBA [PMI0984 and PMI0985] [68] and PMI2963) were found both on the uropathogenic E. coli-specific list and in P. mirabilis HI4320.
TABLE 3.
P. mirabilis CDSs that are present in the set of 131 uropathogenic E. coli-specific genesa
| Gene no. | Designation | Annotation | CFT073 open reading frame |
|---|---|---|---|
| PMI0229 | ABC transporter, permease protein (FecCD transport family) | c2516 | |
| PMI0984 | ibrB | Putative immunoglobulin-binding regulator | c2484 |
| PMI0985 | ibrA | Putative immunoglobulin-binding regulator | c2485 |
| PMI1233 | Putative dihydrodipicolinate synthase | c0761 | |
| PMI1426 | hmuR2 | Hemin receptor | c4308 (chuA) |
| PMI1427 | hmuS | Hemin transport protein | c4307 (chuS) |
| PMI1428 | hmuT | Hemin-binding periplasmic protein | c4313 (chuT) |
| PMI1429 | hmuU | Hemin transport system permease protein HmuU | c4317 (chuU) |
| PMI1480 | ABC transporter, substrate-binding protein | c5081 | |
| PMI1481 | ABC transporter, permease protein | c5080 | |
| PMI1482 | ABC transporter, permease protein | c5079 | |
| PMI1483 | ABC transporter, ATP-binding protein | c5078 | |
| PMI1484 | ABC transporter, ATP-binding protein | c5077 | |
| PMI2145 | gatY | Putative tagatose-1,6-bisphosphate aldolase | c4018 |
| PMI2597 | MFS family transporter | c2420 | |
| PMI2601 | nrpU | Putative siderophore biosynthetic protein | c2430 |
| PMI2603 | nrpA | Putative siderophore ABC transporter, ATP-binding/permease protein | c2422 |
| PMI2604 | nrpB | Putative siderophore ABC transporter, ATP-binding/permease protein | c2421 |
| PMI2676 | Putative iron compound ABC transporter, permease | c1651 (fecD) | |
| PMI2677 | Putative iron compound ABC transporter, ATP-binding protein | c1650 | |
| PMI2679 | modD | Putative pyrophosphorylase | c1647 (modD) |
| PMI2935 | Putative membrane protein | c4774 | |
| PMI2937 | Putative exported protein | c4776 | |
| PMI2963 | Probable transcriptional regulator | c1810 | |
| PMI3232 | Putative amidohydrolase/metallopeptidase | c4924 |
See reference 44.
Whole-genome alignment (using the ACT software) revealed low levels of synteny and orthology between P. mirabilis and the enteric bacteria mentioned above (an example of an alignment of the genomes of P. mirabilis, E. coli UTI89, and Y. enterocolitica is shown in Fig. S1 in the supplemental material; similar profiles were observed when the whole genomes of P. mirabilis and other enteric bacteria were compared). Thus, P. mirabilis appears to be distantly related to the other sequenced enteric species.
DISCUSSION
P. mirabilis, a gram-negative enteric bacterium, is the first bacterial agent of complicated UTI or catheter-associated UTI to be sequenced. This opportunistic pathogen is well equipped to colonize a compromised urinary tract. It can use urea as a nitrogen source, and this nitrogenous waste compound is plentiful in the urinary tract. P. mirabilis has extraordinary potential for adherence to host tissue. To our knowledge, this bacterium encodes the highest number of fimbriae of any organism sequenced thus far. However, it can nimbly shift between adherent and motile phenotypes, indicating the presence of a sophisticated regulatory network that controls the modulation between alternate lifestyles. Fimbriae are encoded at loci around the chromosome, while all flagellar genes are contiguous. This bacterium has numerous virulence factors that have been demonstrated experimentally. It is interesting, however, that single gene mutations can fully attenuate the bacterium (13, 82).
Along with Providencia and Morganella spp., P. mirabilis produces urease. The synthesis of this multimeric nickel-metalloenzyme is urea inducible. Since urine contains approximately 0.4 M urea (23), urease is always synthesized (31). The products of urea hydrolysis, ammonia and carbon dioxide, alkalanize the urinary tract, resulting in a pH ranging from 8 to 9, which often results in precipitation of normally soluble ions and thus the formation of bladder and kidney stones. Beyond urolithiasis, the environment created by P. mirabilis colonization requires that surface-localized bacterial processes (e.g., enzymatic catalysis, fimbrial adhesin-receptor interaction, iron acquisition, and cytotoxin activity) perform optimally at an alkaline pH. For example, the surface-localized Proteus toxic agglutinin (encoded by PMI2341) has a pH optimum for the subtilase-like protease of 9.0, making this molecule well suited to proteolyze its targets in a P. mirabilis-colonized urinary tract (2). Certainly, in the life cycle of P. mirabilis environments that lack urea are encountered. It is therefore interesting that there is a pathway by which urea can be synthesized de novo (Fig. 6) from ammonia and carbon dioxide.
Virulence determinants that mediate colonization have been identified in numerous studies (6, 13, 30, 53, 82); however, the genome sequence revealed previously unknown candidates that one would predict also contribute to the pathogenesis of infections. For example, several fimbriae have been shown to contribute to P. mirabilis virulence. Prior to the sequencing of the P. mirabilis genome, only five chaperone-usher fimbriae had been identified in this organism (MR/P fimbriae, UCA, PMF, ATF, and PMP). However, P. mirabilis potentially can produce 17 unique fimbriae, which is more fimbriae than any other bacterial species sequenced to date encodes. Notably, Salmonella enterica serovar Typhi has a 4.81-Mb genome (60), which is nearly 800 kb larger than the P. mirabilis HI4320 genome, yet it has only 12 fimbrial operons. Thus, the density of fimbrial operons in the relatively small P. mirabilis HI4320 genome indicates the likely importance of these fimbriae for this organism. Determining the expression profiles of these fimbriae may reveal new roles in virulence or may provide clues to the life cycle of P. mirabilis in other niches, such as the soil or the gastrointestinal tract.
P. mirabilis differentiates between the vegetative fimbriated swimmer cell and the hyperflagellated swarmer cell (8). The discovery that all the flagellar genes of P. mirabilis are at a single contiguous chromosomal locus, which is unusual in bacteria, presents interesting possibilities for regulation of flagellar expression. The close proximity of all flagellar genes could be key to the ability of P. mirabilis to rapidly switch from swimmer cell to swarmer cell.
Several studies have documented coordinated expression of virulence genes with the swarming phenomenon (3, 9, 41, 76). In particular, the MR/P and NAF (UCA) fimbrial operons are downregulated during swarming (36). Conversely, fimbrial expression may repress flagellar motility; overexpression of MrpJ, whose gene is cotranscribed with the mrp fimbrial operon, represses flagellin expression and motility (39). At least 14 other paralogs of MrpJ are present in this strain, and 10 are associated with fimbrial operons. These MrpJ paralogs are being assessed to determine their ability to repress motility when fimbriae are expressed (Pearson and Mobley, submitted). The unique organization of the P. mirabilis flagellar regulon, coupled with the surprisingly high number of fimbrial operons, suggests that the decision to stick or swim is central to the lifestyle of this opportunistic pathogen. As both motility and adherence contribute to the ability of P. mirabilis to cause disease, further research is necessary to determine how this organism switches from a swarming state to an adherent state and back again.
With the exception of multiple fimbriae and iron acquisition systems, P. mirabilis may have fewer redundant pathways for virulence than other genera in the Enterobacteriaceae. This species is more easily attenuated by a single mutation than other uropathogenic species, such as E. coli. In signature-tagged mutagenesis studies of E. coli, for example, it was difficult to isolate transposon mutants that were attenuated by more than 10-fold in the CBA mouse model of ascending UTI (7). On the other hand, signature-tagged mutagenesis studies of P. mirabilis using identical reagents yielded more than 30 transposon mutants that were attenuated ≥10,000-fold (13, 82).
As the aging population continues to expand, more individuals will be at risk for P. mirabilis UTI (78). In the United States, the proportion of the population that is ≥65 years old is projected to increase from 12.4% (30 million) in 2000 to 19.6% (71 million) in 2030 (http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5206a2.htm). In 1997, the United States had the highest health care spending per person who was ≥65 years old ($12,100) (http://fesportal.fes.de/pls/portal30/docs/folder/ipg/ipg1_2002/artjacobzone.htm); nursing home and home health care expenditures doubled from 1990 to 2001, reaching approximately $132 billion (37). The absolute number of persons who are ≥65 years old residing in 18,000 nursing homes in the United States is currently estimated to be 1.5 million to 1.9 million (58). In these facilities urinary incontinence, a very frequent complication, is treated with long-term (≥30 days) urinary catheterization. Nearly 100% of these patients become bacteriuric (78), often leading to fever and bacteremia and sometimes to death (77). P. mirabilis and the related species Providencia stuartii and Morganella morganii account for more than one-half of such infections (57, 78). With the new understanding derived from the complete genome sequence of P. mirabilis, novel antibiotic targets and identification of protective antigens for use in vaccines may be forthcoming and thus useful for controlling these infections.
Supplementary Material
Acknowledgments
We thank Carrie Poore for genomic DNA isolation. We acknowledge the support of the Wellcome Trust Sanger Institute core sequencing and informatics groups.
This work was funded by the Wellcome Trust (Sanger Institute) and Public Health Service grant AI059722 (to H.L.T.M.). M.M.P. was supported in part by Molecular Mechanisms of Microbial Pathogenesis training grant T32 AI7528 and by National Research Service award F32 AI068324.
Footnotes
Published ahead of print on 28 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adegbola, R. A., D. C. Old, and B. W. Senior. 1983. The adhesins and fimbriae of Proteus mirabilis strains associated with high and low affinity for the urinary tract. J. Med. Microbiol. 16427-431. [DOI] [PubMed] [Google Scholar]
- 2.Alamuri, P., and H. L. T. Mobley. A novel autotransporter of uropathogenic Proteus mirabilis is both a cytotoxin and an agglutinin. Mol. Microbiol., in press. [DOI] [PubMed]
- 3.Allison, C., H. C. Lai, and C. Hughes. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 61583-1591. [DOI] [PubMed] [Google Scholar]
- 4.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 3091245-1248. [DOI] [PubMed] [Google Scholar]
- 5.Bahrani, F. K., S. Cook, R. A. Hull, G. Massad, and H. L. Mobley. 1993. Proteus mirabilis fimbriae: N-terminal amino acid sequence of a major fimbrial subunit and nucleotide sequences of the genes from two strains. Infect. Immun. 61884-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahrani, F. K., G. Massad, C. V. Lockatell, D. E. Johnson, R. G. Russell, J. W. Warren, and H. L. Mobley. 1994. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 623363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 451079-1093. [DOI] [PubMed] [Google Scholar]
- 8.Belas, R. 1996. Proteus mirabilis swarmer cell differentiation and urinary tract infection, p. 271-298. In H. L. Mobley and J. W. Warren (ed.), Urinary tract infections: molecular pathogenesis and clinical management. ASM Press, Washington, DC.
- 9.Belas, R., and R. Suvanasuthi. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 1876789-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beynon, L. M., A. J. Dumanski, R. J. McLean, L. L. MacLean, J. C. Richards, and M. B. Perry. 1992. Capsule structure of Proteus mirabilis (ATCC 49565). J. Bacteriol. 1742172-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bijlsma, I. G., L. van Dijk, J. G. Kusters, and W. Gaastra. 1995. Nucleotide sequences of two fimbrial major subunit genes, pmpA and ucaA, from canine-uropathogenic Proteus mirabilis strains. Microbiology 1411349-1357. [DOI] [PubMed] [Google Scholar]
- 12.Böltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 1845158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burall, L. S., J. M. Harro, X. Li, C. V. Lockatell, S. D. Himpsl, J. R. Hebel, D. E. Johnson, and H. L. Mobley. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 722922-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 213422-3423. [DOI] [PubMed] [Google Scholar]
- 15.Chippendale, G. R., J. W. Warren, A. L. Trifillis, and H. L. Mobley. 1994. Internalization of Proteus mirabilis by human renal epithelial cells. Infect. Immun. 623115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemmer, K. M., and P. N. Rather. 2007. Regulation of flhDC expression in Proteus mirabilis. Res. Microbiol. 158295-302. [DOI] [PubMed] [Google Scholar]
- 17.Cook, S. W., N. Mody, J. Valle, and R. Hull. 1995. Molecular cloning of Proteus mirabilis uroepithelial cell adherence (uca) genes. Infect. Immun. 632082-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 19.Dufour, A., R. B. Furness, and C. Hughes. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol. Microbiol. 29741-751. [DOI] [PubMed] [Google Scholar]
- 20.Eriksson, S., J. Zbornik, H. Dahnsjo, P. Erlanson, O. Kahlmeter, H. Fritz, and C. A. Bauer. 1986. The combination of pivampicillin and pivmecillinam versus pivampicillin alone in the treatment of acute pyelonephritis. Scand. J. Infect. Dis. 18431-438. [DOI] [PubMed] [Google Scholar]
- 21.File, T. M., Jr., J. S. Tan, S. J. Salstrom, and L. Johnson. 1985. Timentin versus piperacillin in the therapy of serious urinary tract infections. Am. J. Med. 7991-95. [DOI] [PubMed] [Google Scholar]
- 22.Gentry, L. O., B. A. Wood, M. D. Martin, and J. Smythe. 1980. Cefamandole alone and combined with gentamicin or tobramycin in the treatment of acute pyelonephritis. Scand. J. Infect. Dis. Suppl. 2596-100. [PubMed] [Google Scholar]
- 23.Griffith, D. P., D. M. Musher, and C. Itin. 1976. Urease. The primary cause of infection-induced urinary stones. Investig. Urol. 13346-350. [PubMed] [Google Scholar]
- 24.Gygi, D., M. M. Rahman, H. C. Lai, R. Carlson, J. Guard-Petter, and C. Hughes. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 171167-1175. [DOI] [PubMed] [Google Scholar]
- 25.Hatt, J. K., and P. N. Rather. 2008. Characterization of a novel gene, wosA, regulating FlhDC expression in Proteus mirabilis. J. Bacteriol. 1901946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honarvar, S., B. K. Choi, and D. M. Schifferli. 2003. Phase variation of the 987P-like CS18 fimbriae of human enterotoxigenic Escherichia coli is regulated by site-specific recombinases. Mol. Microbiol. 48157-171. [DOI] [PubMed] [Google Scholar]
- 27.Island, M. D., and H. L. T. Mobley. 1995. Proteus mirabilis urease: operon fusion and linker insertion analysis of ure gene organization, regulation, and function. J. Bacteriol. 1775653-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansen, A. M., C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2003. Visualization of Proteus mirabilis morphotypes in the urinary tract: the elongated swarmer cell is rarely observed in ascending urinary tract infection. Infect. Immun. 713607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansen, A. M., V. Lockatell, D. E. Johnson, and H. L. Mobley. 2004. Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect. Immun. 727294-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, B. D., C. V. Lockatell, D. E. Johnson, J. W. Warren, and H. L. Mobley. 1990. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 581120-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, B. D., and H. L. Mobley. 1988. Proteus mirabilis urease: genetic organization, regulation, and expression of structural genes. J. Bacteriol. 1703342-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, B. V., E. Mahenthiralingam, N. A. Sabbuba, and D. J. Stickler. 2005. Role of swarming in the formation of crystalline Proteus mirabilis biofilms on urinary catheters. J. Med. Microbiol. 54807-813. [DOI] [PubMed] [Google Scholar]
- 33.Klemm, P., B. J. Jørgensen, B. Kreft, and G. Christiansen. 1995. The export systems of type 1 and F1C fimbriae are interchangeable but work in parental pairs. J. Bacteriol. 177621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontomichalou, P., M. Mitani, and R. C. Clowes. 1970. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under either relaxed or stringent control. J. Bacteriol. 10434-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, H. C., P. C. Soo, J. R. Wei, W. C. Yi, S. J. Liaw, Y. T. Horng, S. M. Lin, S. W. Ho, S. Swift, and P. Williams. 2005. The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J. Bacteriol. 1873407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Latta, R. K., A. Grondin, H. C. Jarrell, G. R. Nicholls, and L. R. Bérubé. 1999. Differential expression of nonagglutinating fimbriae and MR/P pili in swarming colonies of Proteus mirabilis. J. Bacteriol. 1813220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levit, K., C. Smith, C. Cowan, H. Lazenby, A. Sensenig, and A. Catlin. 2003. Trends in U.S. health care spending, 2001. Health Aff. (Millwood) 22154-164. [DOI] [PubMed] [Google Scholar]
- 38.Li, X., C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2002. Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol. Microbiol. 45865-874. [DOI] [PubMed] [Google Scholar]
- 39.Li, X., D. A. Rasko, C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2001. Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 204854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, X., H. Zhao, C. V. Lockatell, C. B. Drachenberg, D. E. Johnson, and H. L. Mobley. 2002. Visualization of Proteus mirabilis within the matrix of urease-induced bladder stones during experimental urinary tract infection. Infect. Immun. 70389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liaw, S. J., H. C. Lai, S. W. Ho, K. T. Luh, and W. B. Wang. 2000. Inhibition of virulence factor expression and swarming differentiation in Proteus mirabilis by p-nitrophenylglycerol. J. Med. Microbiol. 49725-731. [DOI] [PubMed] [Google Scholar]
- 42.Liaw, S. J., H. C. Lai, and W. B. Wang. 2004. Modulation of swarming and virulence by fatty acids through the RsbA protein in Proteus mirabilis. Infect. Immun. 726836-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 961071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloyd, A. L., D. A. Rasko, and H. L. Mobley. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 1893532-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maroncle, N., C. Rich, and C. Forestier. 2006. The role of Klebsiella pneumoniae urease in intestinal colonization and resistance to gastrointestinal stress. Res. Microbiol. 157184-193. [DOI] [PubMed] [Google Scholar]
- 46.Massad, G., F. K. Bahrani, and H. L. Mobley. 1994. Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infect. Immun. 621989-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massad, G., J. F. Fulkerson, Jr., D. C. Watson, and H. L. Mobley. 1996. Proteus mirabilis ambient-temperature fimbriae: cloning and nucleotide sequence of the atf gene cluster. Infect. Immun. 644390-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massad, G., C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 1994. Proteus mirabilis fimbriae: construction of an isogenic pmfA mutant and analysis of virulence in a CBA mouse model of ascending urinary tract infection. Infect. Immun. 62536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 994620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, V. L., J. B. Bliska, and S. Falkow. 1990. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J. Bacteriol. 1721062-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller, W. L., K. Pabbaraju, and K. E. Sanderson. 2000. Fragmentation of 23S rRNA in strains of Proteus and Providencia results from intervening sequences in the rrn (rRNA) genes. J. Bacteriol. 1821109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mobley, H. L., and R. Belas. 1995. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 3280-284. [DOI] [PubMed] [Google Scholar]
- 53.Mobley, H. L., R. Belas, V. Lockatell, G. Chippendale, A. L. Trifillis, D. E. Johnson, and J. W. Warren. 1996. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 645332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mobley, H. L., and G. R. Chippendale. 1990. Hemagglutinin, urease, and hemolysin production by Proteus mirabilis from clinical sources. J. Infect. Dis. 161525-530. [DOI] [PubMed] [Google Scholar]
- 55.Mobley, H. L., G. R. Chippendale, K. G. Swihart, and R. A. Welch. 1991. Cytotoxicity of the HpmA hemolysin and urease of Proteus mirabilis and Proteus vulgaris against cultured human renal proximal tubular epithelial cells. Infect. Immun. 592036-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mobley, H. L., and R. P. Hausinger. 1989. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 5385-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mobley, H. L., and J. W. Warren. 1987. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J. Clin. Microbiol. 252216-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ness, J., A. Ahmed, and W. S. Aronow. 2004. Demographics and payment characteristics of nursing home residents in the United States: a 23-year trend. J. Gerontol. A Biol. Sci. Med. Sci. 591213-1217. [DOI] [PubMed] [Google Scholar]
- 59.O'Hara, C. M., F. W. Brenner, and J. M. Miller. 2000. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin. Microbiol. Rev. 13534-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413848-852. [DOI] [PubMed] [Google Scholar]
- 61.Pearson, M. M., and H. L. Mobley. 2007. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J. Med. Microbiol. 561277-1283. [DOI] [PubMed] [Google Scholar]
- 62.Rashid, T., and A. Ebringer. 2007. Rheumatoid arthritis is linked to Proteus—the evidence. Clin. Rheumatol. 261036-1043. [DOI] [PubMed] [Google Scholar]
- 63.Rather, P. N. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 71065-1073. [DOI] [PubMed] [Google Scholar]
- 64.Römling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57629-639. [DOI] [PubMed] [Google Scholar]
- 65.Rubin, R. H., N. E. Tolkoff-Rubin, and R. S. Cotran. 1986. Urinary tract infection, pyelonephritis, and reflux nephropathy, p. 1085-1141. In B. M. Brenner and F. C. Rector (ed.), The kidney. W. B. Saunders Co., Philadelphia, PA.
- 66.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16944-945. [DOI] [PubMed] [Google Scholar]
- 67.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Sandt, C. H., J. E. Hopper, and C. W. Hill. 2002. Activation of prophage eib genes for immunoglobulin-binding proteins by genes from the IbrAB genetic island of Escherichia coli ECOR-9. J. Bacteriol. 1843640-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38779-786. [DOI] [PubMed] [Google Scholar]
- 70.Sturgill, G., and P. N. Rather. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51437-446. [DOI] [PubMed] [Google Scholar]
- 71.Swihart, K. G., and R. A. Welch. 1990. Cytotoxic activity of the Proteus hemolysin HpmA. Infect. Immun. 581861-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40440-450. [DOI] [PubMed] [Google Scholar]
- 73.Trollfors, B., M. Jertborn, J. Martinell, G. Norkrans, and G. Lidin-Janson. 1982. Mecillinam versus cephaloridine for the treatment of acute pyelonephritis. Infection 1015-17. [DOI] [PubMed] [Google Scholar]
- 74.Vernikos, G. S., and J. Parkhill. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 222196-2203. [DOI] [PubMed] [Google Scholar]
- 75.Vigneux, F., R. Zumbihl, G. Jubelin, C. Ribeiro, J. Poncet, S. Baghdiguian, A. Givaudan, and M. Brehelin. 2007. The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J. Biol. Chem. 2829571-9580. [DOI] [PubMed] [Google Scholar]
- 76.Walker, K. E., S. Moghaddame-Jafari, C. V. Lockatell, D. Johnson, and R. Belas. 1999. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 32825-836. [DOI] [PubMed] [Google Scholar]
- 77.Warren, J. W., D. Damron, J. H. Tenney, J. M. Hoopes, B. Deforge, and H. L. Muncie, Jr. 1987. Fever, bacteremia, and death as complications of bacteriuria in women with long-term urethral catheters. J. Infect. Dis. 1551151-1158. [DOI] [PubMed] [Google Scholar]
- 78.Warren, J. W., J. H. Tenney, J. M. Hoopes, H. L. Muncie, and W. C. Anthony. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146719-723. [DOI] [PubMed] [Google Scholar]
- 79.Wray, S. K., S. I. Hull, R. G. Cook, J. Barrish, and R. A. Hull. 1986. Identification and characterization of a uroepithelial cell adhesin from a uropathogenic isolate of Proteus mirabilis. Infect. Immun. 5443-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zabłotni, A., K. Zych, A. N. Kondakova, M. Siwińska, Y. A. Knirel, and Z. Sidorczyk. 2007. Serological and structural characterization of the O-antigens of the unclassified Proteus mirabilis strains TG 83, TG 319, and CCUG 10700 (OA). Arch. Immunol. Ther. Exp. 55347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao, H., X. Li, D. E. Johnson, I. Blomfield, and H. L. Mobley. 1997. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol. Microbiol. 231009-1019. [DOI] [PubMed] [Google Scholar]
- 82.Zhao, H., X. Li, D. E. Johnson, and H. L. Mobley. 1999. Identification of protease and rpoN-associated genes of uropathogenic Proteus mirabilis by negative selection in a mouse model of ascending urinary tract infection. Microbiology 145185-195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.