Abstract
In Escherichia coli and Salmonella enterica, RyeA and RyeB RNAs are encoded on opposite DNA strands at the same locus. We present evidence indicating that the last 23 bp of the ryeB gene, corresponding to an internal portion of the ryeA gene, served repeatedly as the integration site for exogenous DNA during Salmonella evolution and still act as an attachment site for present-day bacteriophages. Interestingly, ryeA sequence and expression are modified upon lysogenization.
Lateral gene transfer is a prolific source of evolutionary changes in microorganisms and is thought to have had a major impact in the emergence of bacterial pathogens. In particular, the acquisition of so-called pathogenicity islands is regarded as being a key event in the conversion of ancestral extracellular bacteria into intracellular pathogens (2, 9, 11, 15). A common pathway to DNA acquisition involves the integrative recombination of circular DNA molecules into the host genome. The step is catalyzed by integrases, a class of site-specific recombinases encoded by temperate bacteriophages and plasmids. Integrases introduce staggered cuts at specific sequences on both donor and host DNAs and promote strand exchange and ligation (8, 14). As a result, the sequences recognized by the integrase are duplicated at each end of the inserted DNA. When conserved, such directed repeats allow a precise definition of the site of the original integration event.
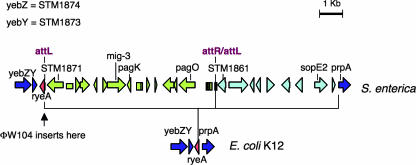
Structure of the Salmonella CS 40 island.
Salmonella enterica serovars harbor a panoply of genomic islands and islets. Some such elements are associated with prophage remnants, suggesting that they were incorporated as a result of lysogenization events. This is the case for a 16-kb insert lying at about centisome (CS) 40 on the chromosome map. The CS 40 island contains various loci linked to pathogenicity, such as mig-3 (18) pagK-pagO (10), and sopE2 (1, 16), interspersed with sequences reminiscent of phage genes. Among the latter is a presumptive integrase gene (STM1871) (12) located near one end of the island and oriented outwards. As in lambdoid prophage maps (int gene near attL), the STM1871-proximal end is hereafter designated the “left” end of the island (Fig. 1). Alignment with the Escherichia coli K-12 genome sequence reveals that the Salmonella CS 40 island is inserted into the intergenic region between the pprA gene (also named pphA) and the ortholog of the yebY locus (STM1873) (Fig. 1). Recently, the prpA-yebY intergenic region of E. coli was shown to contain two small-RNA (sRNA) genes with opposite polarities, ryeA and ryeB, transcribed from opposite DNA strands, with the ryeB sequence entirely contained within the larger ryeA gene (19, 20). The core region of this locus, a 146-bp segment comprising the entirety of ryeB, is highly conserved in Salmonella and is located immediately to the left of the CS 40 island near the end of the putative integrase gene.
FIG. 1.
Organization of the CS 40 island in Salmonella enterica serovar Typhimurium and of the corresponding region in the E. coli chromosome. Horizontal arrows represent open reading frames. The arrow clusters depicted in green and light blue correspond to segments proposed to originate from separate insertion events (see text). Phage ΦW104 inserts at the left boundary of the island. The diagram is based on data from references 10, 12, 16, 19, and 20).
Since we wondered if the interval between ryeB and STM1871 contained the integrase recognition site, we examined whether a portion of the sequence was repeated on the opposite end of the island. No such repeat could be identified at the right end of the element; however, a sequence identical to the last 23 bp of the ryeB gene, and in the same orientation, was located inside the island, approximately 10 kb from the left end. Interestingly, this sequence lies adjacent to an open reading frame (STM1861) whose putative product shares 77% identity (89% similarity) with the C-terminal half of the STM1871-encoded integrase. This strongly suggests that the CS 40 island is in fact made of two separate islets lying side by side, one carrying mig-3 and pagKO (left) and the other containing the sopE2 gene (right) (Fig. 1). The lack of a recognizable attachment site at the right end of the insert, as well as the apparent defective nature of STM1861, suggests that the sopE2 islet was acquired earlier and has since suffered extensive decay. Consistent with this idea, Salmonella bongori, a lineage sharing a common ancestor with Salmonella enterica, carries sopE2 (13) but lacks the mig-3-pagKO islet (data not shown).
The above-described findings tentatively define the last 23 bp of the ryeB gene as the core region of the integration site. Since this sequence is reconstituted upon integration, the acquisition of the CS 40 islets is not expected to have affected ryeB gene structure or expression. In contrast, if the ryeA gene is positioned in Salmonella as in E. coli, the incorporation of the islets should have separated the structural portion of the gene from its original promoter. Thus, it seemed relevant to determine the ryeA transcriptional status in Salmonella. For this purpose, RNA extracted from S. enterica serovar Typhimurium strain LT2 was subjected to Northern hybridization analysis using oligonucleotides complementary to the predicted sequences of RyeA and RyeB RNAs as probes. As shown in Fig. 2, both probes gave positive signals. In the case of RyeA, the most intense band corresponded to an RNA of 250 to 300 nucleotides, while the RyeB analysis detected an RNA of approximately 100 nucleotides. These sizes are closely comparable to those of RyeA and RyeB RNAs in E. coli (19, 20). As observed in E. coli (20), additional fainter signals were detected with both probes. These minor bands are likely to represent processing products.
FIG. 2.

Northern blot analysis of RyeA and RyeB RNAs in Salmonella enterica serovar Typhimurium LT2. Cultures grown overnight in LB were diluted 1:200 in fresh LB and grown to an optical density at 600 nm of 0.35. RNA was extracted as previously described (4), fractionated on a 6% polyacrylamide-8 M urea gel (lane 1) or on an 8% polyacrylamide-8 M urea gel (lane 2), transferred onto a Hybond-N+ membrane, and hybridized to 32P-labeled oligonucleotides complementary to RyeA (pp925 [5′-GGAAAACCTGGCGTCGTCATCTATTCTTAAAGGGCAAGGCGA-3′]) and RyeB (pB13 [5′-GATTCCTGTATTCGGTCCAGGGAAATGGCTCTTGGGAGAGAG-3′]). Sizes were estimated from migration distances of tmRNA and 5S RNA (not shown).
A survey of the Salmonella genome sequences revealed the occurrence of yet another insertion event at the ryeA/ryeB locus. Some strains carry a prophage-related insert between the end of ryeB and the left boundary of the mig-3-pagKO islet (Fig. 1). The length and structure of the Salmonella ryeA/ryeB locus insert vary considerably, from a full-length prophage in some isolates (e.g., S. enterica serovar Typhi strain CT18) (17) to a shortened and scrambled version in others (e.g., S. enterica serovar Enteritidis strain LK5) (7). In all instances, the terminal 23 to 26 bp of ryeB are found duplicated at the two ends of the element, indicating that this portion of the gene serves as the attachment site. Thus, in strains carrying the ryeB-linked prophage, ryeA transcription is expected to originate from within phage DNA.
While tRNA and tmRNA genes constitute a favored target for temperate phages and other integrative elements (21), to our knowledge, only one example of the phage insert in an sRNA gene has been reported. Wassarman and colleagues found the previously mapped attB site for bacteriophage P2 in E. coli to correspond with the 3′ end of the ryeE gene, which encodes an Hfq-binding sRNA of unknown function (20). In both the ryeB and ryeE genes, the attachment site lies within the sequence encoding the stem-loop of the transcription terminator, suggesting that the region of dyad symmetry participates in integrase recognition (14, 21).
Effect of phage integration on ryeA/ryeB expression.
As part of a separate study, we examined the occupancy of the ryeA/ryeB att site in 84 Salmonella enterica serovar Typhimurium isolates of human or animal origin using a three-primer-based PCR assay. This analysis revealed the presence of a DNA insert in a fraction of the strains. Interestingly, all positive strains belonged to the DT104 phage type, suggesting that the acquisition of the insert occurred recently, possibly coinciding with the emergence of the virulent epidemic clone (3; N. Figueroa-Bossi, F. X. Weill, P. A. Grimont, and L. Bossi, unpublished data). Sequence data from the Sanger Institute website (http://www.sanger.ac.uk/Projects/Salmonella/) show the ryeA/ryeB-associated element to be a full-size lambdoid prophage. To assess its functional state, we deleted the prophage from the genome of a DT104 strain in our collection, MA6711 (5), and used the resulting strain (MA7860) as a host for monitoring the release of plaque-forming particles in cultures of the MA6711 parent. Tiny plaques from which active virus could be isolated and characterized were detected. The phage, hereafter named ΦW104, proved capable of infecting and lysogenizing a variety of serovar Typhimurium strains including LT2, ATCC 14028, and SL1344. The isolation of such lysogenic derivatives provided a system for studying how ΦW104 integration affected the expression of the ryeA/ryeB locus. To identify the ryeA promoter, RNA preparations from exponential cultures of strain LT2 and its ΦW104-lysogenized derivative MA7833 were subjected to primer extension analysis (Fig. 3A). These experiments located the 5′ end of RyeA RNA at identical positions in the two strains, approximately 80 bp to the right of the ryeB gene (Fig. 4). The presence of sequences resembling the −10 and −35 consensus motifs of σ70-dependent promoters immediately upstream from the 5′-end position is consistent with this being the ryeA transcription start site. Overall, the sequences around this region in naive and lysogenic strains are highly conserved (Fig. 4), with the identity extending to the ΦW104 putative int gene that strongly resembles STM1871 (see above). Nonetheless, the difference in the intensities of the primer extension bands shown in Fig. 3A suggested that RyeA RNA might be more abundant in strain LT2 than in its lysogenic derivative. The difference was confirmed by Northern blot hybridization analysis (Fig. 3B). Interestingly, RyeB followed an opposite trend, being synthesized at a lower level in LT2 than in MA7833 (Fig. 3B). Since the ryeB sequence is unaffected by the lysogenization event, the observed difference might reflect the change in ryeA transcription associated with such an event. Conceivably, RNA polymerases transcribing the ryeA gene could dislodge polymerases bound to the ryeB promoter, causing the activity of the latter to negatively correlate with that of the ryeA promoter. Some variability in the sequence upstream from the ryeA promoter, particularly a 9-bp deletion/insertion at position −49 (Fig. 4), might account for the difference in ryeA transcription rates.
FIG. 3.
Effect of phage ΦW104 integration on ryeA/ryeB expression. Bulk RNA was extracted from strains LT2 and MA7833 (ΦW104 lysogen) as described in the legend to Fig. 2. (A) Primer extension analysis of RyeA RNA from the lysogenic strain (lane 1) and from wild-type LT2 (lane 2). Reverse transcriptase reactions were carried out using primer ppB12 (5′-CCCTGGACCGAATACAGGA-3′) as previously described (6). Sequencing reactions were performed with the fmol DNA cycle sequencing system from Promega according to the manufacturer's protocol. The template was a DNA fragment obtained by PCR amplification of chromosomal DNA from strain MA7833 with oligonucleotides pp490 (5′-TGGCGTCGTCATCTATTC-3′) and pp491 (5′-CAGGGACGCTATCACACA-3′) as primers. (B) Northern blot quantification of RyeA and RyeB levels in strains MA7833 (lane 1) and wild-type LT2 (lane 2). Bulk RNA was fractionated on an 8% polyacrylamide-8 M urea gel. Membranes were probed with 32P-labeled oligonucleotides pp925 and ppB13 (see the legend to Fig. 2). 5S RNA and tmRNA probed with ppB10 (5′-ACACTACCATCGGCGCTACG-3′) and pp813 (5′-GCGGAGGCTAGGGAGAGAGG-3′), respectively, were used as internal controls. Due to the higher gel concentration, the two RyeA bands are less separated than in Fig. 2.
FIG. 4.
Sequence alignment of ryeA/ryeB chromosomal regions in LT2 and ΦW104 lysogen. Underlined triplets indicate the translation termination codon of yebY and the complements of termination codons of STM1871 (LT2) (12) and the ΦW104 int gene (MA7833). The sequence of the ryeB gene is in italics and shaded. Gene boundaries are inferred from the 96% identical E. coli sequence (19, 20). Facing arrows indicate the transcription terminator stem sequence. Green underlining indicates the sequence found duplicated at the two ends of the ΦW104 prophage. The left-pointing arrow defines the ryeA transcription start site. Purple boxes indicate ryeA promoter elements.
Biological significance of prophage insertion at the ryeA/ryeB locus.
The above-described data suggest that the integration of the ΦW104 prophage “resets” the levels of RyeA and RyeB RNAs in the cell. In addition, the 5′ portion of RyeA RNA is changed upon lysogenization. It is temping to speculate that these modifications might have physiological consequences. Unfortunately, the lack of information on the physiological roles of RyeA and RyeB does not offer much grounds for such speculation. RyeB RNA was shown to strongly bind the Hfq protein (20), suggesting its involvement in some Hfq-mediated regulatory mechanism. In contrast, RyeA bound Hfq with low affinity (20). Possibly, the role of this RNA is limited to regulating RyeB levels through transcriptional interference, as suggested above, or by a direct RNA-RNA interaction (19).
Acknowledgments
This work was supported by French National Research Agency (ANR) grant BLAN07-1_187785 and by grant BIO2004-3455-CO2-02 from the Spanish Ministry of Education and Science and the European Regional Fund.
Footnotes
Published ahead of print on 4 April 2008.
REFERENCES
- 1.Bakshi, C. S., V. P. Singh, M. W. Wood, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2000. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 1822341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumler, A. J. 1997. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 5318-322. [DOI] [PubMed] [Google Scholar]
- 3.Bossi, L., and N. Figueroa-Bossi. 2005. Prophage arsenal of Salmonella enterica serovar Typhimurium, p. 165-186. In M. Waldor, D. Friedman, and S. Adhya (ed.), Phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC.
- 4.Bossi, L., and N. Figueroa-Bossi. 2007. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 65799-810. [DOI] [PubMed] [Google Scholar]
- 5.Bossi, L., J. A. Fuentes, G. Mora, and N. Figueroa-Bossi. 2003. Prophage contribution to bacterial population dynamics. J. Bacteriol. 1856467-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueroa, N., N. Wills, and L. Bossi. 1991. Common sequence determinants of the response of a prokaryotic promoter to DNA bending and supercoiling. EMBO J. 10941-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueroa-Bossi, N., S. Ammendola, and L. Bossi. 2006. Differences in gene expression levels and in enzymatic qualities account for the uneven contribution of superoxide dismutases SodCI and SodCII to pathogenicity in Salmonella enterica. Microbes Infect. 81569-1578. [DOI] [PubMed] [Google Scholar]
- 8.Grainge, I., and M. Jayaram. 1999. The integrase family of recombinase: organization and function of the active site. Mol. Microbiol. 33449-456. [DOI] [PubMed] [Google Scholar]
- 9.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87791-794. [DOI] [PubMed] [Google Scholar]
- 10.Gunn, J. S., W. J. Belden, and S. I. Miller. 1998. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb. Pathog. 2577-90. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54641-679. [DOI] [PubMed] [Google Scholar]
- 12.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 13.Mirold, S., K. Ehrbar, A. Weissmuller, R. Prager, H. Tschape, H. Russmann, and W. D. Hardt. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J. Bacteriol. 1832348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nash, H. A. 1996. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments, p. 2363-2376. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 15.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 1714-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. D. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 361206-1221. [DOI] [PubMed] [Google Scholar]
- 17.Thomson, N., S. Baker, D. Pickard, M. Fookes, M. Anjum, N. Hamlin, J. Wain, D. House, Z. Bhutta, K. Chan, S. Falkow, J. Parkhill, M. Woodward, A. Ivens, and G. Dougan. 2004. The role of prophage-like elements in the diversity of Salmonella enterica serovars. J. Mol. Biol. 339279-300. [DOI] [PubMed] [Google Scholar]
- 18.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 2772007-2011. [DOI] [PubMed] [Google Scholar]
- 19.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 316435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 151637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams, K. P. 2002. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 30866-875. [DOI] [PMC free article] [PubMed] [Google Scholar]