Abstract
Epidemiological evidence links high-salt diets and Helicobacter pylori infection with increased risk of developing gastric maladies. The mechanism by which elevated sodium chloride content causes these manifestations is unclear. Here we characterize the response of H. pylori to temporal changes in sodium chloride concentration and show that growth, cell morphology, survival, and virulence factor expression are all altered by increased salt concentration.
Helicobacter pylori is a microaerophilic, gram-negative bacterium whose presence in the gastric environment is correlated with diseases such as gastritis, peptic ulcer disease, mucosa-associated lymphoid tissue lymphoma, and gastric adenocarcinoma (7). Several epidemiological studies have shown that secondary risk factors affect the occurrence of certain gastric maladies associated with H. pylori infection (for a review, see reference 30). In these studies, dietary sodium chloride intake emerged as a key player in enhancing the likelihood of severe disease outcomes (25, 30). In support of this, experimental animal infections have shown that there is a synergistic effect between H. pylori and salt in terms of disease progression (2, 8, 21, 23). Thus, the bacterium likely senses fluctuations in salt concentration due to dietary intake and alters its growth and gene expression accordingly.
The ability to adapt to changing environments is critical for bacterial survival, and thus sodium chloride and osmotic stress have been shown to affect bacterial gene expression in a number of bacterial species (for reviews, see references 12 and 32). In several of these bacteria, salt resistance involves changes in expression of outer membrane proteins, such as the transporters ProP, ProU, BetT, and BetU in Escherichia coli (32). For pathogenic bacteria, osmotic stress can also serve as a signal that controls expression of virulence factors. Specifically, it has been suggested that sodium chloride concentration serves as a cue for the regulation of virulence in Listeria monocytogenes (10, 20) and Leptospira interrogans (17). In keeping with this, a recent study showed that elevated salt concentrations result in alterations in expression of the virulence factor CagA in H. pylori strain 26695 (16). Loh et al. showed that an increased salt concentration slows H. pylori growth and affects expression of a large number of genes (16). Concomitant studies by our group that explored this effect in further detail are described here.
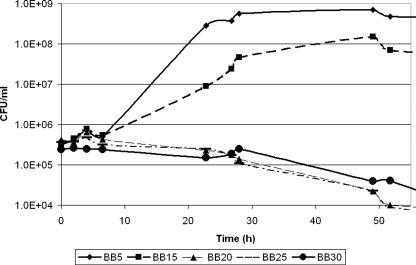
Given the fact that H. pylori is adept at survival in the tumultuous environment of the stomach and the fact that there is a strong epidemiological link between salt intake and H. pylori-induced disease, we first sought to determine the effect of increasing the salt concentration on bacterial growth and survival of H. pylori. Strain G27 (Table 1) liquid cultures were grown at 37°C with shaking at 100 rpm in brucella broth (BB) containing various NaCl concentrations. The salt concentrations ranged from 5 g/liter (BB5) (which is the normal salt concentration found in BB) to 25 g/liter (BB25). All liquid growth media were supplemented with fetal bovine serum (FBS) (10%) and 10 μg/ml vancomycin, and cultures were grown in gas evacuation jars under microaerophilic conditions (5% O2, 10% CO2, 85% N2) generated with an Anoxomat gas evacuation and replacement system (Spiral Biotech) as previously described (3). Since in our hands we often find that optical density readings do not provide an adequate assessment of bacterial numbers due to cell clumping and transition to the coccoid form at later time points, we measured growth and survival by plating on blood agar plates as previously described (9). We found that as the concentration of NaCl in the liquid growth media increased, H. pylori cells stopped multiplying and eventually died (Fig. 1). This effect was also evident when the bacteria were exposed to increasing levels of salt both in F-12 defined media (27) and in F-12 media supplemented with 1% FBS (data not shown).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Descriptiona | Reference |
|---|---|---|
| Plasmids | ||
| pTM117 | Modified pHP666 including E. coli origin of replication, aphA-3 cassette (Kanr), multiple cloning site, and promoterless gfpmut3 | 3 |
| pDSM196 | pTM117 cagA promoter::gfpmut3 fusion | This study |
| pDSM463 | pTM117 ureA promoter::gfpmut3 fusion | This study |
| pDSM223 | pTM117 vacA promoter::gfpmut3 fusion | This study |
| pDSM134 | pGEM-T Easy::internal fragment of ureA for riboprobe generation | This study |
| pDSM393 | pGEM-T Easy::internal fragment of cagA for riboprobe generation | This study |
| pDSM394 | pGEM-T Easy::internal fragment of vacA for riboprobe generation | This study |
| H. pylori strains | ||
| G27 | Wild-type H. pylori | 5 |
| 26695 | Wild-type H. pylori | 29 |
| J99 | Wild-type H. pylori | 1 |
| HPAG1 | Wild-type H. pylori | 22 |
| SS1 | Wild-type H. pylori | 14 |
| J166 | Monkey-colonizing strain, isolate 316-3.4 | J. Solnick lab collection |
| K6 | Clinical isolate from a 45-year-old female gastric cancer patient | J. Cha and I. Chung lab collection |
| K112 | Clinical isolate from a 57-year-old male gastritis patient | J. Cha and I. Chung lab collection |
| DSM195 | G27(pDSM196), Kanr (25 μg/ml) | This study |
| DSM215 | G27(pTM117), Kanr (25 μg/ml) | 3 |
| DSM241 | G27(pDSM223), Kanr (25 μg/ml) | This study |
| DSM464 | G27(pDSM463), Kanr (25 μg/ml) | This study |
Relevant antibiotic concentrations are indicated in parentheses.
FIG. 1.
Growth and survival of H. pylori G27 in liquid cultures containing different concentrations of NaCl. A broth-grown, overnight culture of H. pylori G27 was diluted into fresh growth media containing different concentrations of NaCl (BB5, BB15, BB20, BB25, and BB30). Growth and survival were monitored by plating.
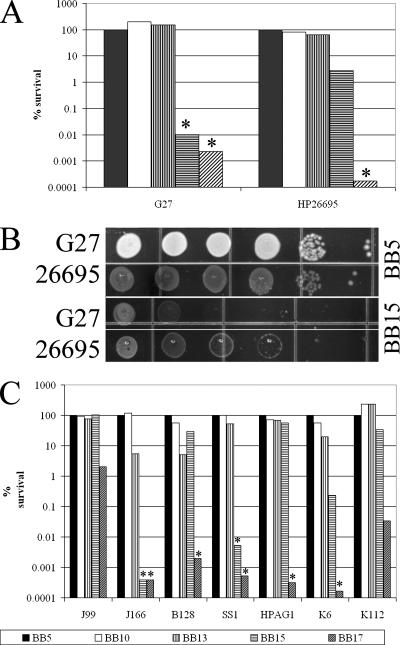
Our data differ from the data presented by Loh et al., who found that similar salt concentrations slowed the growth of strain 26695 but did not have a bactericidal effect on the H. pylori cells (16). While the difference could be attributed to the use of slightly different medium formulations and growth conditions, we wondered if strain-specific differences in salt resistance between G27 and 26695 were responsible for this phenotype. To assess strain-to-strain variability in the ability to grow in the presence of elevated concentrations of NaCl, we decided to directly compare G27 and 26695 (Table 1). The strains were first grown in liquid BB5 overnight and then serially diluted and plated side by side on clear agar-based medium mimicking the composition of the liquid medium (BB supplemented with 1.2% Agar-agar, 10% FBS, and a standard H. pylori antibiotic mixture [9]). Plates contained the normal concentration of NaCl (5 g/liter) or 10, 13, 15, or 17 g/liter of NaCl. Salt sensitivity was assessed by enumerating single colonies after growth for 7 to 10 days in the conditions described above. The data are expressed as percentages of survival relative to the growth on medium containing the normal salt concentration. As shown in Fig. 2A and B, strain 26695 was able to grow in the presence of 15 g/liter NaCl, while strain G27 could not. This shows that 26695 is more resistant to an increased salt concentration than G27. This fact may partially explain why no bactericidal effect of salt was observed in the study of Loh et al. (16). We next wondered whether there was variation in the level of salt resistance in other H. pylori strains. To study this, we examined strains J99, HPAG1, SS1, J166, and B128, as well as K6 and K112, which are two recent Korean clinical isolates (Table 1). We found a wide range of sensitivity to increased salt concentrations (Fig. 2C). Some strains (HPAG1, J99, B128, and K112) exhibited virtually no decrease in viable counts at salt concentrations that completely inhibited the growth of or killed other H. pylori strains (J166, SS1, and K6). This variation may imply that different H. pylori strains have evolved to withstand different NaCl pressures that could arise from specific host dietary habits. The strain difference in sensitivity to salt is particularly interesting when it is considered in the context of the epidemiological findings concerning salt and H. pylori infection; the salt resistance phenotype of the infecting strain may be important for determining the disease outcome.
FIG. 2.
Relative NaCl sensitivities of various H. pylori strains. (A) H. pylori cultures of strains G27 and 26695 were grown in liquid BB5. The numbers of starting cells were normalized by using optical density at 600 nm. The relative salt sensitivity was assessed by serial dilution and plating on BB-based agar plates containing increasing concentrations of salt. CFU were enumerated, and the percentages of survival relative to the number of CFU on the BB5 plates are presented. (B) Relative growth of H. pylori strains G27 and 26695 on BB5 and BB15 plates. Serial dilutions of cultures normalized using optical density at 600 nm were plated side by side on BB5 and BB15. (C) H. pylori strains J99, J166, B128, SS1, HPAG1, K6, and K112 were examined as described above. An asterisk indicates that no single colonies were detected, and the bars below the asterisks indicate the limits of detection at the concentrations used. The results are representative of multiple biological repeats.
Since H. pylori has been shown to modulate virulence factor expression in response to low pH, iron limitation, and other stressful environments (18, 19), we wondered if an increased salt concentration would affect expression of the virulence factors cagA (HP0527), vacA (HP0887), and ureA (HP0073) (4). To determine if there was any effect on expression of any of these three genes, transcriptional fusions of each gene to the promoterless gfpmut3 carried on pTM117 (3) were made. The transcriptional fusions were constructed by amplification of the predicted promoters of ureA and vacA using primers ureA_promoter_F and ureA_promoter_R and primers VAF1 and VAR1, respectively (Table 2). To facilitate directional ligation into pTM117, primers ureA_promoter_F and VAF1 have KpnI restriction sites at the 5′ end, and primers ureA_promoter_R and VAR1 have XbaI restriction sites at the 3′ end. In a similar manner, the cagA promoter was amplified using primers cagA_promoter_F and cagA_promoter_R, which have SacII and BamHI restriction sites. The reporter plasmids constructed were moved into wild-type strain G27 by natural transformation, and transformants were selected on plates containing 25 μg/ml kanamycin.
TABLE 2.
Primers used in this study
| Primer (restriction endonuclease) | Sequence (5′-3′)a | Reference |
|---|---|---|
| Promoter::gfp fusion primers | ||
| cagA-promoter_F (SacII) | CCGCGGCAAATTGAAATCAATCG | This study |
| cagA_Promoter_R (BamHI) | GGATCCTACTATACCTAGTTTCATAC | This study |
| ureA_Promoter_F (KpnI) | GGTACCCAAAAACAAAACAAAATTAAGGCATA | This study |
| ureA_Promoter_R (XbaI) | TCTAGATGGGGTGAGTTTCATCTCATT | This study |
| VAF1 (KpnI) | GGTACCGCGCCTATTTCCAATTAGGCG | This study |
| VAR1 (XbaI) | TCTAGAGGGTGCGACTTTAGACTAG | This study |
| RPA primers | ||
| cagA-RPA-F | AATGTTGTCGCTTCATTTGATCCT | This study |
| cagA-RPA-R | GATAGGGGGTTGTATGATATTTTC | This study |
| vacA-RPA-F | CACACCGCAAAATCAATCGCC | This study |
| vacA-RPA-R | CCATTGCCCGCCTTTGACCC | This study |
| ureA-RPA-F | CGGATGATGTGATGGATGG | This study |
| ureA-RPA-R | TTCTACGGATTTTTCTTCG | This study |
| Sequencing primer | ||
| apha3-2 | CGGTGATATTCTCATTTTAGCC | 3 |
Restriction endonuclease sites are indicated by bold type.
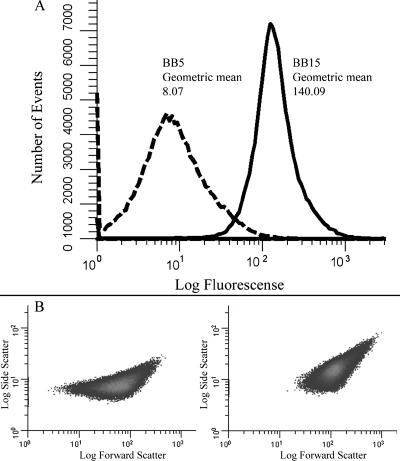
Strains DSM195, DSM464, and DSM241 (Table 1) were grown as described above in BB5 and BB15 for 16 to 20 h. At this point, cells were harvested by centrifugation and resuspended in phosphate-buffered saline (pH 7.0), and 100,000 events were analyzed using a Beckman Coulter Epics XL-MCL flow cytometer with a laser setting of 750 V. The resulting data were analyzed using WinList 3D, version 6.0 (Verity Software House). We found that the level of green fluorescent protein (GFP) expression was significantly elevated for both the ureA and vacA reporter strains but only moderately increased for the cagA reporter strain (Fig. 3A and data not shown). Interestingly, we observed a marked change in the forward scatter of each of the H. pylori strains that were grown in BB15 (Fig. 3B and data not shown). Since forward scatter is a relative indication of cell size and shape (6), we concluded that salt might have affected the morphology of the H. pylori cells. Thus, observed changes in the level of GFP expression may be influenced by changes in cell size or shape and not solely attributed to true changes in transcription. We therefore decided to monitor expression of the genes directly using RNase protection assays (RPAs). We also investigated alterations in cell morphology arising from exposure to NaCl.
FIG. 3.
Flow cytometry analysis of the vacA GFP transcriptional fusion. Strain DSM241 bearing a vacA promoter fusion was grown in either BB5 or BB15 for 20 h and analyzed by flow cytometry. (A) Fluorescence histograms for cells grown in BB5 (dashed line) and for cells grown in BB15 (solid line). Fluorescence is expressed in relative units, and the geometric mean is indicated. The data are representative of multiple independent flow analyses. (B) Density plots of side scatter versus forward scatter for DSM241. The density plot for cells grown in BB5 is on the left, and the density plot for cells grown in BB15 is on the right.
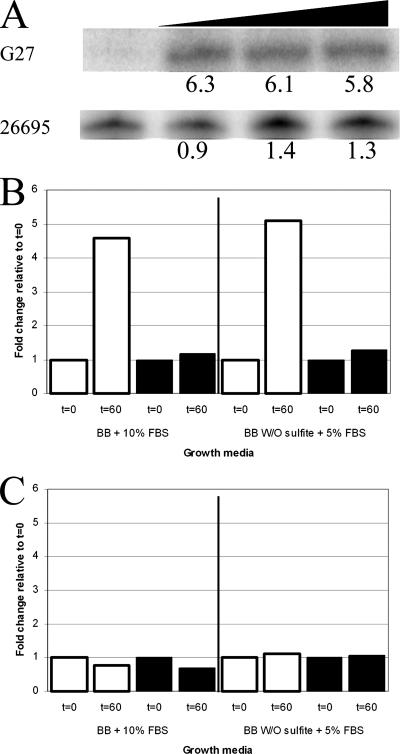
RPAs were conducted with RNA from overnight H. pylori G27 cultures grown in BB5, which had been harvested by centrifugation and then exposed to a salt shock in BB25, whose salt concentration completely halts H. pylori growth but does not result in immediate cell death (Fig. 1). Cells were harvested for RNA isolation at 0, 15, 30, and 60 min after resuspension. The RNA was extracted as previously described (28), and 1 or 2 μg of RNA was used in each RPA reaction using riboprobes for cagA, ureA, and vacA. For each gene, the riboprobe template was generated by PCR using the primer pairs listed in Table 2. Radiolabeled riboprobes were produced, and the RPAs were performed as previously described (9). ureA and cagA showed no change in expression in response to salt shock (Fig. 4C and data not shown). However, as shown in Fig. 4A and B, expression of the vacA transcript was significantly increased upon exposure to salt. These results differ from the results of Loh et al., who found that the salt concentration affected expression of cagA but not expression of vacA in H. pylori strain 26695 (16). To determine whether salt shock resulted in strain-specific differences in gene expression in G27 and 26695, we repeated our experiment using our media as well as the sulfite-free media used by Loh et al. As shown in Fig. 4A and B, we observed strain-specific differences in response to salt shock; expression of the vacA transcript increased only in G27, and expression of the cagA transcript changed in neither strain. While the reason for the difference between our results and the results of Loh et al. is not completely clear, the findings likely reflect the fact that we used short-term salt shock to assess changes in gene expression, whereas Loh et al. assessed extended growth in the presence of salt (16). Moreover, it should be noted that Loh et al. reported that they found strain-specific differences in the effect of salt on cagA expression (16); this fact, along with the strain-specific differences in survival in response to salt stress that we observed (Fig. 2C), may suggest that the specific response to salt stress of the infecting strain may be important for determining the H. pylori-induced disease outcome.
FIG. 4.
Transcription of vacA and cagA following NaCl shock. (A) Logarithmically growing liquid cultures of G27 and 26695 were salt shocked by replacing BB5 with BB25. Samples were taken at 0, 15, 30, and 60 min after the change of medium. RNA was harvested, and RPAs were conducted as described in the text. Protected bands for the vacA transcript for G27 and 26695 and the changes (fold) relative to the zero-time results are shown. Increasing length of the salt shock is indicated by the triangle. (B) vacA transcript in G27 (open bars) and 26695 (filled bars). The changes in expression at 60 min are relative to the data for zero time. Cells were grown in BB5 and shocked in BB25 or in the comparable sulfite-free BB-based media containing 5% FBS used by Loh et al. (16). (C) cagA transcript in G27 (open bars) and 26695 (filled bars). The changes in expression at 60 min are relative to the data for zero time. Cells were grown in BB5 and shocked in BB25 or in the comparable sulfite-free BB-based media containing 5% FBS used by Loh et al. (16).
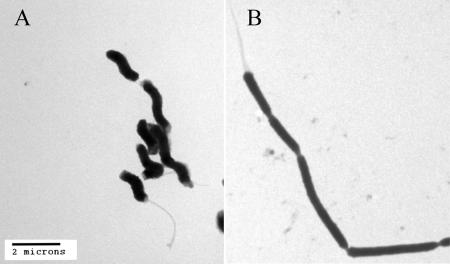
Finally, given the pronounced effect on growth of G27 that we observed (Fig. 1) and the fact that we observed large differences in the flow cytometry profiles of cells exposed to salt (Fig. 3B), we sought to determine the specific effect that salt had on the bacterial cells. To this end, electron microscopy was performed with samples of cells grown overnight in BB5 or BB15. The cells were harvested by centrifugation and then resuspended in 2% electron microscopy grade glutaraldehyde in phosphate-buffered saline for ∼1 h. Subsequently, samples were washed three times with double-distilled H2O, dried, and examined with a Philips CM100 electron microscope. As show in Fig. 5, H. pylori grown in BB5 exhibited a characteristic spiral shape and possessed multiple unipolar flagella. Conversely, the bacterial cells grown in BB15 showed marked morphological changes; they lost the spiral shape, became elongated, and formed chains. This suggests that salt stress results in a delay in septum formation. Interestingly, despite this significant effect on cell morphology, the cells still had unipolar flagella (Fig. 5). However, perhaps not surprisingly, there was a clear reduction in cell motility when the cells were observed by bright-field microscopy (data not shown). Similar salt-induced changes in cell morphology were also observed when we examined bacteria grown in F-12 and F-12 containing 1% FBS (data not shown). Additionally, we observed the same morphological changes for H. pylori strains HPAG1, J99, SS1, and J166, and a similar effect has previously been shown for strain HPK5 (26). This suggests that despite strain-specific differences in gene expression and survival of H. pylori, high salt concentrations have a physiological effect that results in changes in cell morphology. Similar results have also been obtained for Salmonella enterica serovar Enteritidis and L. monocytogenes (11), which suggests that salt concentration may significantly impact a diverse number of bacteria. Interestingly, in recent studies to assess the ability of H. pylori to survive in seawater, Konishi et al. observed multiple elongated cells among the spiral and coccoid H. pylori forms (13). These authors suggested that these cells were bacterial contamination; however, the elongated forms closely resemble the salt stress phenotype that we observed here.
FIG. 5.
Transmission electron microscopy of H. pylori cells grown under normal conditions or with an increased NaCl concentration. H. pylori cells from overnight liquid cultures in either BB5 (left panel) or BB15 (right panel) were examined using transmission electron microscopy. Bar = 2 μm.
H. pylori resides in an ever-changing environment, where pH, osmotic pressure, ionic composition, and other host-dependent factors are constantly changing. Given the fact that H. pylori is able to persistently colonize its human host, it must be particularly adept at responding to environmental stressors, such as osmotic or salt fluctuations. Thus, an understanding of stress adaptation is particularly important for the study of this bacterium. Mechanistically, other bacteria have been shown to utilize typical response regulatory proteins and a stress-related sigma factor (rpoS or σ38) in adaptation to osmotic or salt stress (15, 24). However, relatively few response regulators and two-component systems have been described for H. pylori (31), and no rpoS homologue appears to be encoded in the H. pylori genome. Based on these limitations, it is possible that alternative effects on regulation may involve DNA supercoiling (33) or forms of posttranscriptional regulation. This mechanism has also been suggested by Wang et al. to be important for regulation of antioxidant proteins (31). Regardless, adaptation to the tumultuous environment of the stomach requires the ability to sense and respond to the myriad of environmental cues in a timely manner. Diet composition has been shown to be one factor that contributes to the outcome of H. pylori infection. Here we show that H. pylori responds to changes in the concentration of NaCl in its environment in such a way that growth, cell morphology, survival, and virulence factor expression are all altered by increased salt concentration. Moreover, the strain-specific differences that we observed provide increasing evidence suggesting that diet and the strain of H. pylori with which a person is infected may greatly affect the disease outcome.
Acknowledgments
We thank Dennis McDaniel for preparation and acquisition of the electron microscopy images and members of the Merrell lab for useful discussions. We also thank Jeong-Heon Cha and In-Sik Chung for providing the Korean H. pylori clinical isolates, J. Solnick for providing J166, Tim McDaniel for construction of pDSM196, and Beth Carpenter for critical reading of the manuscript.
This publication was made possible by grant AI065529 from the NIAID. Research in the laboratory of D. Scott Merrell is also supported by grant R073LA from USUHS.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397176-180. [DOI] [PubMed] [Google Scholar]
- 2.Bergin, I. L., B. J. Sheppard, and J. G. Fox. 2003. Helicobacter pylori infection and high dietary salt independently induce atrophic gastritis and intestinal metaplasia in commercially available outbred Mongolian gerbils. Dig. Dis. Sci. 48475-485. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter, B. M., T. K. McDaniel, J. M. Whitmire, H. Gancz, S. Guidotti, S. Censini, and D. S. Merrell. 2007. Expanding the Helicobacter pylori genetic tool box: a modified, endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl. Environ. Microbiol. [DOI] [PMC free article] [PubMed]
- 4.Clyne, M., B. Dolan, and E. P. Reeves. 2007. Bacterial factors that mediate colonization of the stomach and virulence of Helicobacter pylori. FEMS Microbiol. Lett. 268135-143. [DOI] [PubMed] [Google Scholar]
- 5.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, and N. Figura. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 905791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davey, H. M., and D. B. Kell. 1996. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol. Rev. 60641-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn, B. E., N. B. Vakil, B. G. Schneider, M. M. Miller, J. B. Zitzer, T. Peutz, and S. H. Phadnis. 1997. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect. Immun. 651181-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, J. G., C. A. Dangler, N. S. Taylor, A. King, T. J. Koh, and T. C. Wang. 1999. High-salt diet induces gastric epithelial hyperplasia and parietal cell loss, and enhances Helicobacter pylori colonization in C57BL/6 mice. Cancer Res. 594823-4828. [PubMed] [Google Scholar]
- 9.Gancz, H., S. Censini, and D. S. Merrell. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garner, M. R., K. E. James, M. C. Callahan, M. Wiedmann, and K. J. Boor. 2006. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl. Environ. Microbiol. 725384-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazeleger, W. C., M. Dalvoorde, and R. R. Beumer. 2006. Fluorescence microscopy of NaCl-stressed, elongated Salmonella and Listeria cells reveals the presence of septa in filaments. Int. J. Food Microbiol. 112288-290. [DOI] [PubMed] [Google Scholar]
- 12.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170319-330. [DOI] [PubMed] [Google Scholar]
- 13.Konishi, K., N. Saito, E. Shoji, H. Takeda, M. Kato, M. Asaka, and H. K. Ooi. 2007. Helicobacter pylori: longer survival in deep ground water and sea water than in a nutrient-rich environment. APMIS 1151285-1291. [DOI] [PubMed] [Google Scholar]
- 14.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 1121386-1397. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. J., and J. D. Gralla. 2004. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cells 14153-162. [DOI] [PubMed] [Google Scholar]
- 16.Loh, J. T., V. J. Torres, and T. L. Cover. 2007. Regulation of Helicobacter pylori cagA expression in response to salt. Cancer Res. 674709-4715. [DOI] [PubMed] [Google Scholar]
- 17.Matsunaga, J., M. Lo, D. M. Bulach, R. L. Zuerner, B. Adler, and D. A. Haake. 2007. Response of Leptospira interrogans to physiologic osmolarity: relevance in signaling the environment-to-host transition. Infect. Immun. 752864-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 713529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrell, D. S., L. J. Thompson, C. C. Kim, H. Mitchell, L. S. Tompkins, A. Lee, and S. Falkow. 2003. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect. Immun. 716510-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers, E. R., A. W. Dallmier, and S. E. Martin. 1993. Sodium chloride, potassium chloride, and virulence in Listeria monocytogenes. Appl. Environ. Microbiol. 592082-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nozaki, K., N. Shimizu, K. Inada, T. Tsukamoto, M. Inoue, T. Kumagai, A. Sugiyama, T. Mizoshita, M. Kaminishi, and M. Tatematsu. 2002. Synergistic promoting effects of Helicobacter pylori infection and high-salt diet on gastric carcinogenesis in Mongolian gerbils. Jpn. J. Cancer Res. 931083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh, J. D., H. Kling-Backhed, M. Giannakis, J. Xu, R. S. Fulton, L. A. Fulton, H. S. Cordum, C. Wang, G. Elliott, J. Edwards, E. R. Mardis, L. G. Engstrand, and J. I. Gordon. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA 1039999-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers, A. B., N. S. Taylor, M. T. Whary, E. D. Stefanich, T. C. Wang, and J. G. Fox. 2005. Helicobacter pylori but not high salt induces gastric intraepithelial neoplasia in B6129 mice. Cancer Res. 6510709-10715. [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal, A. Z., M. Hu, and J. D. Gralla. 2006. Osmolyte-induced transcription: −35 region elements and recognition by σ38 (rpoS). Mol. Microbiol. 591052-1061. [DOI] [PubMed] [Google Scholar]
- 25.Shikata, K., Y. Kiyohara, M. Kubo, K. Yonemoto, T. Ninomiya, T. Shirota, Y. Tanizaki, Y. Doi, K. Tanaka, Y. Oishi, T. Matsumoto, and M. Iida. 2006. A prospective study of dietary salt intake and gastric cancer incidence in a defined Japanese population: the Hisayama study. Int. J. Cancer. 119196-201. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi, H., M. Shirai, J. K. Akada, M. Tsuda, and T. Nakazawa. 1998. Nucleotide sequence and characterization of cdrA, a cell division-related gene of Helicobacter pylori. J. Bacteriol. 1805263-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Testerman, T. L., D. J. McGee, and H. L. Mobley. 2001. Helicobacter pylori growth and urease detection in the chemically defined medium Ham's F-12 nutrient mixture. J. Clin. Microbiol. 393842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 712643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388539-547. [DOI] [PubMed] [Google Scholar]
- 30.Tsugane, S., and S. Sasazuki. 2007. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 1075-83. [DOI] [PubMed] [Google Scholar]
- 31.Wang, G., P. Alamuri, and R. J. Maier. 2006. The diverse antioxidant systems of Helicobacter pylori. Mol. Microbiol. 61847-860. [DOI] [PubMed] [Google Scholar]
- 32.Wood, J. M. 2007. Bacterial osmosensing transporters. Methods Enzymol. 42877-107. [DOI] [PubMed] [Google Scholar]
- 33.Ye, F., T. Brauer, E. Niehus, K. Drlica, C. Josenhans, and S. Suerbaum. 2007. Flagellar and global gene regulation in Helicobacter pylori modulated by changes in DNA supercoiling. Int. J. Med. Microbiol. 29765-81. [DOI] [PubMed] [Google Scholar]