Abstract
The Cdc6-1 and -2 proteins from the archaeon Methanothermobacter thermautotrophicus were previously shown to bind the minichromosome maintenance (MCM) helicase. It is shown here that Cdc6-2 protein dissociates the MCM complex. This observation supports the hypothesis that the Cdc6-2 protein functions as a helicase loader.
The minichromosome maintenance (MCM) helicase is thought to function as the replicative helicase in archaea. All archaeal species that have been sequenced contain at least one homologue of the MCM helicase. Biochemical studies of a number of archaeal MCM proteins demonstrated that the enzyme possesses an ATP-dependent 3′→5′ helicase activity and the ability to bind and translocate along single-stranded DNA (summarized in references 1 and 11). Studies with Methanothermobacter thermautotrophicus and Thermoplasma acidophilum MCM proteins showed that the enzymes could also bind and translocate along duplex DNA (8, 18). It was proposed that double-stranded DNA translocation plays an important role during the initiation (and perhaps elongation) process (13, 22). In addition, the M. thermautotrophicus MCM complex can displace proteins from DNA (18, 20) and unwind DNA-RNA hybrids while translocating along the DNA strand (19).
The mechanism of MCM helicase assembly at the archaeal origins of replication is currently unknown. It was suggested, however, that the Cdc6 proteins play a major role in the assembly process, functioning as a helicase loader. This is based on the similarity between the archaeal and eukaryal Cdc6 amino acid sequences (Cdc6 protein is the putative helicase loader in eukarya) (6). In addition, the Cdc6 proteins from several archaea were shown to interact with MCM and to regulate its helicase activity (5, 8, 10, 17). It was found that the interactions between a Cdc6 protein from T. acidophilum and its MCM protein stimulated helicase activity, while interaction between Cdc6 proteins from M. thermautotrophicus or Sulfolobus solfataricus and MCM protein inhibited the helicase (5, 8, 17). The inhibition of helicase activity is similar to the observation made with the Escherichia coli helicase loader DnaC, which inhibits DnaB helicase activity on an artificial substrate (23, 24).
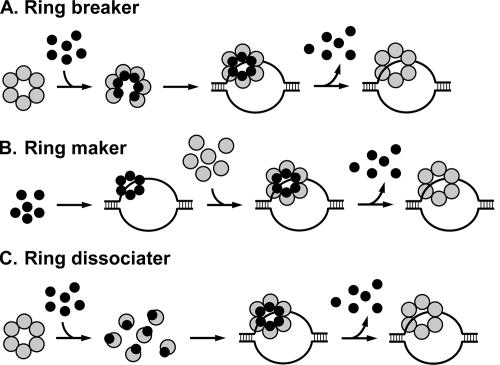
There are two main mechanisms used by the helicase loaders. They function either as a “ring breaker,” where the loader opens the hexameric ring of the helicase (e.g., E. coli) (Fig. 1A), or as a “ring maker,” where it builds the hexameric ring from monomers on the DNA (e.g., phage T4) (Fig. 1B) (summarized in reference 4).
FIG. 1.
Models for the mechanism of helicase assembly at the origin by various helicase loaders. Gray circles, helicase; black circles, helicase loader. See the text for details.
The archaeon M. thermautotrophicus contains two Cdc6 homologues, referred to as Cdc6-1 and Cdc6-2. Previous biochemical studies suggested that Cdc6-1 may function in origin recognition (2, 9, 15) while Cdc6-2 acts in helicase loading. If Cdc6-2 is involved in helicase assembly at the origin, it may have an effect on the oligomeric state of the MCM helicase and function as a ring breaker or ring maker.
In this study, the effect of the Cdc6-2 protein on MCM protein oligomeric structure was examined.
The Cdc6-2 protein can dissociate MCM multimers.
Although the Cdc6-1 protein was thought to function in origin recognition and Cdc6-2 in helicase loading, it is not clear whether Cdc6-1 is also involved in helicase loading. Thus, the effects of both proteins on MCM oligomerization were evaluated using size exclusion chromatography. Four hundred nanomolar MCM helicase (as a monomer) in 250 μl buffer (20 mM Tris-HCl [pH 7.5], 25 mM NaCl, and 10% glycerol) was incubated in the absence or presence of 8 μM Cdc6 (as a monomer) at 25°C for 1 h, followed by fractionation on a Superdex 200 gel filtration column. The column was preequilibrated with buffer containing 20 mM Tris-HCl (pH 7.5), 25 mM NaCl, 1 mM EDTA, and 10% glycerol. Five-hundred-microliter fractions were collected, and 100 μl of each was fractionated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed using Western blotting with rabbit anti-M. thermautotrophicus MCM helicase polyclonal antibodies.
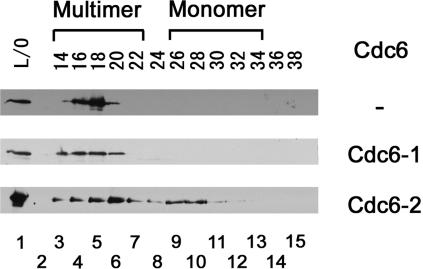
As shown in Fig. 2, and as previously reported (3, 12, 16), the MCM protein forms multimers in solution. When the Cdc6-2 protein is added to MCM helicase, the multimeric helicase is dissociated. Cdc6-1 protein, on the other hand, does not affect the oligomeric state of MCM helicase. It is important to point out that the concentrations of MCM protein in all experiments were the same. These data suggest that Cdc6-2 protein may function as a helicase dissociater and also support its role as a helicase loader. The data are compatible with the notion that Cdc6-1 protein may be involved in origin recognition but not in helicase loading.
FIG. 2.
Cdc6-2 protein dissociates the MCM helicase. MCM protein (400 nM as a monomer) was incubated in the absence or presence of Cdc6-1 or Cdc6-2 protein (8 μM as a monomer) for 1 h at 25°C, followed by fractionation on a Superdex 200 gel filtration column. Five-hundred-microliter fractions were collected, and 100 μl was separated on 10% SDS-PAGE gel, followed by Western analysis with MCM helicase polyclonal antibodies, and visualized by autoradiography. Lane 1 represents the gel filtration load-on sample, lane 2 represents no sample, and lanes 3 to 15 represent the different fractions.
ATP binding to Cdc6 is required for MCM helicase dissociation.
The Cdc6 proteins belong to the AAA+ family of ATPases. Therefore, ATP binding and/or hydrolysis may play a role in MCM helicase dissociation. Although the results described above (Fig. 2) were obtained in the absence of ATP, this does not exclude the possibility that nucleotide binding by Cdc6-2 is required for MCM helicase dissociation. It was previously shown that purified archaeal Cdc6 proteins are tightly bound to ADP and only denaturation in urea or guanidinium can remove the bound nucleotides (7, 14, 21). Thus, to determine whether ATP binding is required, the Cdc6-2 protein was denatured in buffer containing 6 M urea and 50 mM Tris-HCl (pH 7.5), followed by dialysis with buffer containing 20 mM Tris-HCl (pH 7.5), 25 mM NaCl, 1 mM EDTA, and 10% glycerol to refold the protein. Then, the effect of the Cdc6-2 protein without bound nucleotide (7) on MCM helicase oligomeric structure was determined. The experiment was performed under one of two conditions, in the absence or presence of 5 mM ATP. Both assays included 10 mM MgCl2 and were performed as described above.
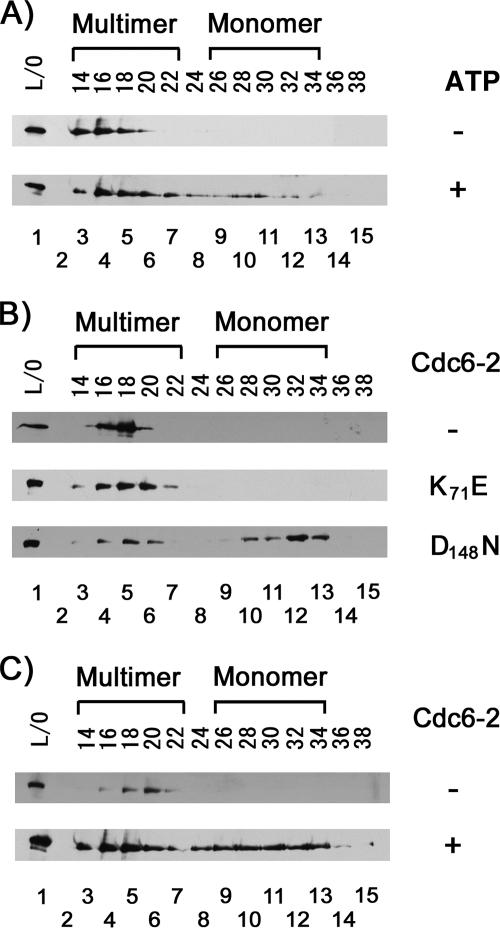
As shown in Fig. 3A, the refolded Cdc6-2 protein could not dissociate MCM helicase unless ATP was added to the reaction. No dissociation could be observed when ATP was omitted from the reaction. These results suggest that ATP (or ADP) binding by Cdc6-2 is required for MCM helicase dissociation.
FIG. 3.
ATP binding by Cdc6-2 protein, but not by MCM helicase, is required for MCM helicase dissociation. MCM protein (400 nM as a monomer) was incubated with Cdc6-2 protein (8 μM as a monomer) for 1 h at 25°C, followed by fractionation on a Superdex 200 gel filtration column. Five-hundred-microliter fractions were collected, and 100 μl was separated on 10% SDS-PAGE gel, followed by Western analysis with MCM helicase polyclonal antibodies, and visualized by autoradiography. Lane 1 represents the gel filtration load-on sample, lane 2 represents no sample, and lanes 3 to 15 represent the different fractions. (A) MCM protein was incubated with Cdc6-2 protein (8 μM as a monomer, which was denatured in 6 M urea, followed by refolding) in the absence or presence of 5 mM ATP. (B) MCM protein was incubated in the absence or presence of Cdc6-2(K71E) or Cdc6-2(D148N) mutant protein. (C) MCM mutant protein (K325A) was incubated in the absence or presence of Cdc6-2 protein.
An alternative approach to determining whether nucleotide binding by the Cdc6-2 protein is required for MCM helicase dissociation is to use Cdc6 proteins with mutations in the ATP binding or hydrolysis sites (7, 17). One mutant protein, Cdc6(K71E), carries a mutation in the Walker-A motif and cannot bind ATP (7). A second mutant protein, Cdc6(D148N), has a mutation in the Walker-B motif and can bind ATP but has very limited ATPase activity (7). Tests performed with the two mutant proteins determined that ATP binding by Cdc6-2 is required for MCM helicase dissociation, as Cdc6(K71E) cannot dissociate the helicase, while Cdc6(D148N) can (Fig. 3B).
The requirement for ATP binding by the putative loader in dissociating the helicase is reminiscent of the observation made with E. coli in which the DnaC protein has to be in the ATP-bound form in order to interact with and assemble the DnaB protein at the origin (25). The results are also consistent with the observation that ATP binding by the Cdc6-2 protein is required for the inhibition of MCM helicase activity (17).
MCM helicase does not require ATP binding for dissociation.
Like Cdc6, MCM helicase belongs to the AAA+ family of ATPases. The experiments whose results are shown in Fig. 2 and 3A and B were performed in the absence of ATP, suggesting that ATP binding by MCM helicase is not required for its dissociation. However, to address this question more directly, the experiments with Cdc6-2 were repeated as described above but with the substitution of a mutant MCM protein [MCM(K325A), previously shown to be devoid of ATP binding (3, 7, 16)] for the wild-type enzyme. As shown in Fig. 3C, Cdc6-2 protein dissociates the mutant MCM complex, demonstrating that ATP binding by the helicase is not required for its dissociation. These results are also similar to the observation made with the E. coli system in which ATP binding by the DnaB protein is not needed for its interaction and loading onto the origin by DnaC.
M. thermautotrophicus Cdc6-2 protein may function as a helicase loader.
The archaeal MCM helicase loader has not yet been identified. However, the data presented here, together with past observations, suggest that the M. thermautotrophicus Cdc6-2 protein may play an important role in helicase assembly at the origin. The Cdc6-2 protein possesses several biochemical properties similar to the E. coli helicase loader, DnaC. When Cdc6-2 protein binds to MCM helicase, it inhibits helicase activity, similar to the observation made with E. coli. Like DnaC, Cdc6-2 protein needs to bind ATP for efficient inhibition of MCM activity, but ATP hydrolysis is not required. These similarities in the biochemical properties of the DnaC and Cdc6-2 proteins lead to the proposal that Cdc6-2 may load the helicase similarly to DnaC.
However, the data shown here suggest some differences between E. coli and M. thermautotrophicus. While a stable complex between hexamers of DnaB and DnaC (forming a DnaB6-DnaC6 complex) can be readily detected (25), the data presented here show that the Cdc6-2 protein dissociates the MCM multimers. These observations may suggest a new mechanism for helicase loading that combines the properties of the ring breakers and makers. The Cdc6-2 protein may work as a ring dissociater (Fig. 1C).
It is not clear, however, how the MCM helicase assembles at the origin. One possibility is that another protein(s) assembles MCM helicase at the origin after its dissociation by Cdc6-2. Alternatively, Cdc6-2 protein may cooperate with the origin recognition protein Cdc6-1 in the assembly process. A similar situation, where the DnaC and DnaA proteins are both needed for helicase loading, occurs in E. coli.
It must be noted, however, that different archaea may possess different helicase loading mechanisms. As described above, in M. thermautotrophicus and S. solfataricus the Cdc6 protein inhibits helicase activity (5, 17) but in T. acidophilum it has a stimulatory effect (8). Thus, it is possible that in some cases, e.g., that for M. thermautotrophicus, the helicase loader works as a ring dissociater, while in other cases it functions as a ring maker or breaker. Only further studies with different archaea will shed light on the similarities and differences between the processes.
The mechanism of eukaryotic MCM helicase assembly is also not yet known. However, as many replication and initiation proteins, including Cdc6 and MCM helicase, are similar in archaea and eukarya, it is possible that one of the proteins involved in the loading process in eukarya (i.e., Cdc6) also dissociates the hexameric MCM helicase ring.
Acknowledgments
We thank Lori Kelman for her comments on the manuscript.
This work was supported by a research scholar grant from the American Cancer Society (RSG-04-050-01-GMC) to Z.K. and a Kyungpook National University Research Fund to J.-H.S. that allowed him to spend the summer of 2007 in the Kelman laboratory.
Footnotes
Published ahead of print on 4 April 2008.
REFERENCES
- 1.Barry, E. R., and S. D. Bell. 2006. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 70876-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capaldi, S. A., and J. M. Berger. 2004. Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic Acids Res. 324821-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong, J. P., M. K. Hayashi, M. N. Simon, R. M. Xu, and B. Stillman. 2000. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 971530-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davey, M. J., and M. O'Donnell. 2003. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13R594-R596. [DOI] [PubMed] [Google Scholar]
- 5.De Felice, M., L. Esposito, B. Pucci, F. Carpentieri, M. De Falco, M. Rossi, and F. M. Pisani. 2003. Biochemical characterization of a CDC6-like protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 27846424-46431. [DOI] [PubMed] [Google Scholar]
- 6.Giraldo, R. 2003. Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEBS Microbiol. Rev. 26533-554. [DOI] [PubMed] [Google Scholar]
- 7.Grabowski, B., and Z. Kelman. 2001. Autophosphorylation of the archaeal Cdc6 homologues is regulated by DNA. J. Bacteriol. 1835459-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haugland, G. T., J. H. Shin, N. K. Birkeland, and Z. Kelman. 2006. Stimulation of MCM helicase activity by a Cdc6 protein in the archaeon Thermoplasma acidophilum. Nucleic Acids Res. 346337-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasiviswanathan, R., J. H. Shin, and Z. Kelman. 2006. DNA binding by the Methanothermobacter thermautotrophicus Cdc6 protein is inhibited by the minichromosome maintenance helicase. J. Bacteriol. 1884577-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasiviswanathan, R., J. H. Shin, and Z. Kelman. 2005. Interactions between the archaeal Cdc6 and MCM proteins modulate their biochemical properties. Nucleic Acids Res. 334940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelman, L. M., and Z. Kelman. 2003. Archaea: an archetype for replication initiation studies? Mol. Microbiol. 48605-615. [DOI] [PubMed] [Google Scholar]
- 12.Kelman, Z., J. K. Lee, and J. Hurwitz. 1999. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum ΔH contains DNA helicase activity. Proc. Natl. Acad. Sci. USA 9614783-14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laskey, R. A., and M. A. Madine. 2003. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 426-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, J., C. L. Smith, D. DeRyckere, K. DeAngelis, G. S. Martin, and J. M. Berger. 2000. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell 6637-648. [DOI] [PubMed] [Google Scholar]
- 15.Majernik, A. I., and J. P. Chong. 2008. A conserved mechanism for replication origin recognition and binding in archaea. Biochem. J. 409511-518. [DOI] [PubMed] [Google Scholar]
- 16.Shechter, D. F., C. Y. Ying, and J. Gautier. 2000. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum ΔH minichromosome maintenance protein. J. Biol. Chem. 27515049-15059. [DOI] [PubMed] [Google Scholar]
- 17.Shin, J. H., B. Grabowski, R. Kasiviswanathan, S. D. Bell, and Z. Kelman. 2003. Regulation of minichromosome maintenance helicase activity by Cdc6. J. Biol. Chem. 27838059-38067. [DOI] [PubMed] [Google Scholar]
- 18.Shin, J. H., Y. Jiang, B. Grabowski, J. Hurwitz, and Z. Kelman. 2003. Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J. Biol. Chem. 27849053-49062. [DOI] [PubMed] [Google Scholar]
- 19.Shin, J. H., and Z. Kelman. 2006. The replicative helicases of bacteria, archaea and eukarya can unwind RNA-DNA hybrid substrates. J. Biol. Chem. 28126914-26921. [DOI] [PubMed] [Google Scholar]
- 20.Shin, J. H., T. J. Santangelo, Y. Xie, J. N. Reeve, and Z. Kelman. 2007. Archaeal minichromosome maintenance (MCM) helicase can unwind DNA bound by archaeal histones and transcription factors. J. Biol. Chem. 2824908-4915. [DOI] [PubMed] [Google Scholar]
- 21.Singleton, M. R., R. Morales, I. Grainge, N. Cook, M. N. Isupov, and D. B. Wigley. 2004. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol. 343547-557. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi, T. S., D. B. Wigley, and J. C. Walter. 2005. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem. Sci. 30437-444. [DOI] [PubMed] [Google Scholar]
- 23.Wahle, E., R. S. Lasken, and A. Kornberg. 1989. The dnaB-dnaC replication protein complex of Escherichia coli. I. Formation and properties. J. Biol. Chem. 2642463-2468. [PubMed] [Google Scholar]
- 24.Wahle, E., R. S. Lasken, and A. Kornberg. 1989. The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J. Biol. Chem. 2642469-2475. [PubMed] [Google Scholar]
- 25.Wickner, S., and J. Hurwitz. 1975. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc. Natl. Acad. Sci. USA 72921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]