Abstract
CreBC is a two-component system that controls the expression of a number of genes in Escherichia coli (called the cre regulon) that encode diverse functions, including intermediary metabolic enzymes. Using a reporter construct, we have shown that cre regulon gene expression is activated during growth in minimal media when glycolytic carbon sources are being fermented. It also is activated during aerobic growth when fermentation products are being used as carbon sources. CreB and CreC are essential for the activation of cre regulon gene expression, but CreA and CreD, encoded as part of the creABCD gene cluster, are not. CreB binds to a TTCACnnnnnnTTCAC direct repeat (the cre tag) in vitro, and this sequence, which is associated with cre regulon gene promoters, is required for the control of gene expression in vivo. These observations support the hypothesis that CreBC is a functional two-component system involved in the metabolic control of transcription in E. coli and confirm that CreB is a DNA binding transcriptional regulator.
Escherichia coli PhoBR is a paradigm two-component system (TCS). The sensor kinase (SK) PhoR responds to a decrease in the concentration of inorganic phosphate (Pi), and under conditions of Pi starvation the protein autophosphorylates and then trans-phosphorylates the response regulator (RR), PhoB (15, 25). In its phosphorylated form, PhoB binds more tightly to the pho box DNA sequence and interacts with RNA polymerase to stimulate the transcription of a regulon of genes that encode products that have a collective role in raising the concentration of cellular Pi (7-9). One of these genes, phoA, encodes bacterial alkaline phosphatase (Bap), and this enzyme activity generally is used as a reporter of pho regulon gene expression (21).
Mutants having the phoR68 allele, believed to encode a nonfunctional PhoR, express Bap production phenotype 1A, which is defined as moderate, constitutive Bap production at a level around one-third of that seen during Pi starvation of a phoR+ strain. The expression of the type 1A phenotype is dependent upon phoM; a phoM deletion, phoR68 derivative produces Bap constitutively at low levels (21). The cloning and sequencing of the phoM gene revealed that it is the third gene in a four-gene locus (2, 22). By sequence homology, phoM was predicted to encode an SK protein, and it was later shown that PhoM can autophosphorylate and then trans-phosphorylate PhoB in vitro. The second gene in the phoM locus (phoM-orf2) was predicted to encode an RR protein, and PhoM also was shown to trans-phosphorylate this putative RR in vitro (3). It is presumed, therefore, that PhoM responds to a signal other than Pi starvation, and in the absence of PhoR (i.e., in the phoR68 mutant background) it can regulate the production of Bap through trans-phosphorylating PhoB. In the presence of PhoR, however, PhoM is not able to affect the activity of PhoB (21); instead, it has long been thought to act through its cognate RR, which is encoded by phoM-orf2 (25).
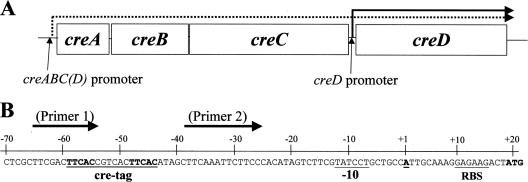
There are few clues about the activating signal for PhoM. It was shown early on that the PhoM-controlled production of Bap in a phoR68 mutant is responsive to the presence of glucose in the medium. Bap production was higher after glucose was added to nutrient agar plates (23). Accordingly, the phoM locus was renamed cre, for carbon source responsive; the four genes in the locus were renamed creA (phoM-orf1, a hypothetical open reading frame), creB (phoM-orf2), creC (phoM), and creD (phoM-orf4; also known as cet, encoding an inner membrane protein of uncertain function) (25) (Fig. 1A). It was originally supposed that the four genes form an operon (2), but Drury and Buxton showed that as well as being part of the operon, creD (cet) has its own promoter and is overexpressed in mutants expressing the Cet2 (colicin E2 tolerant) phenotype (11).
FIG. 1.
cre locus of E. coli and the creD upstream region. (A) The cre genes are represented to scale, as are their intergenic distances. It is proposed (2) that a single operonic promoter upstream of creA drives the transcription of creABCD, the product of which is marked with a dashed arrow. It is known (11) that creD also has its own promoter, and transcription from this point is marked with a solid arrow. (B) The sequence upstream from creD is highlighted. The creD transcriptional start site (TSS) was determined using the 5′ RACE method as set out in Materials and Methods. The TSS (denoted as +1, in boldface and underlined) is shown in context with the cre tag sequence, the putative −10 promoter element, the predicted ribosome binding site (RBS), and the translation initiation codon (in boldface). The binding positions of the PCR primers used to construct the pUB6070 (cre tag) and pUB6069 (no cre tag) reporter constructs used in this study are defined (cre tag, Primer 1) and (no-cre tag, Primer 2) by arrows above the sequence.
For many years following the discovery of CreBC, cre regulon genes, the expression levels of which are controlled by CreBC, proved elusive. The key to our discovery of the founder members of this regulon was the observation of significant sequence similarities between E. coli CreBC and a TCS from Aeromonas hydrophila named BlrAB, which regulates the expression of the blr regulon, which is comprised of three β-lactamase genes, cepH, imiH, and ampH, and at least one additional gene, blrD, which is a homologue of E. coli creD (4-6, 17). Following up on previous work by Rasmussen and colleagues (19), we found that the expression of cepH, imiH, and ampH comes under the control of CreBC when these genes are cloned into E. coli DH5α together with upstream sequences (4, 5). A so-called cre/blr tag sequence, TTCACnnnnnnTTCAC, located at around −60 relative to the transcriptional start site, is essential for the control of cepH expression by the RR, BlrA (6), so we postulated that CreB also controls cloned cepH expression through the cre/blr tag, and further, that the cre/blr tag is found upstream of CreB-regulated genes native to E. coli (5). A bioinformatic search revealed eight transcriptional units encoding diverse functions located downstream of cre/blr tags, and all eight were shown to be under CreBC-dependent control in DH5α (seven are activated by CreBC, and one is repressed), making them the first cre regulon genes to be identified (5). Interestingly, one of the most tightly controlled cre regulon genes is creD, and the creD-overexpressing (Cet2) mutant of Drury and Buxton (11) has a mutation in creC affecting an amino acid within the kinase domain, presumably activating it (5).
Given that there was some evidence from Bap assays using a phoR68 mutant that CreC is a carbon source-responsive SK (23, 24), we next investigated whether cre regulon gene expression is responsive to a carbon source. Using cultures grown in sealed universal bottles to an optical density at 600 nm (OD600) of 0.8, we found that cre regulon gene expression (monitored by reverse transcription-PCR [RT-PCR]) is least active during the growth of DH5α in complex medium, such as nutrient broth, and is significantly activated in minimal medium containing glucose as the carbon source (5). This gave us some clue to the possible CreC-activating ligand, but the primary aim of the work reported in this paper was to make a more detailed analysis of which carbon sources activate cre regulon gene expression and whether other factors are involved, e.g., respiratory or fermentative growth.
The presence of a cre tag upstream of all known cre regulon genes provides circumstantial evidence that the cre tag represents a CreB binding site and so a means by which CreBC can affect the transcription of cre regulon genes (5). However, the role of the cre tag in the CreBC-mediated control of cre regulon gene expression has not been examined experimentally, so it was the secondary aim of the work described in this paper to do so, by confirming that CreB binds to the cre tag sequence in vitro and that this sequence is essential for the CreBC-dependent control of gene expression in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli DH5α (F− endA1 hsdR17 supE44 thi recA1 gyrA96 relA1 spoT1 rbfD1 ΔlacU169 [Φ80ΔlacZM15]; Invitrogen Ltd., Paisley, United Kingdom) and its mutants (see below) were used throughout. Strains were routinely cultured in nutrient broth or on nutrient agar (Oxoid Ltd., Basingstoke, United Kingdom) supplemented with the appropriate antibiotic(s) when necessary. M9 minimal salts medium was prepared using a base of 6 g/liter Na2HPO4, 3 g/liter KH2PO4, 1 g/liter NH4Cl, and 0.5 g/liter NaCl in water. After being autoclaved, minimal medium was supplemented with thiamine (50 μg/ml), tetracycline (25 μg/ml), the appropriate carbon source (180 mM carbon atoms), and either sodium nitrate, sodium fumarate, or trimethylamine N-oxide (TMAO) when required. All chemicals were obtained from Sigma-Aldrich Ltd. (Poole, United Kingdom). Strains were routinely grown at 37°C with vigorous aeration (150 rpm) in 50-ml conical flasks with a foam bung, in which the culture occupied a volume of 10 ml. Anaerobic cultures (20 ml) were grown in tightly closed 20-ml glass universal bottles in a static incubator.
Construction of creD/lacZ reporter constructs and assay of β-galactosidase activity.
Two creD/lacZ fusion constructs were made using the reporter plasmid pRW50 (14). The regions of DNA starting at the immediate 5′ or 3′ end of the creD cre tag and the finishing 50 bp within the creD open reading frame (2) were amplified by PCR as described previously (4). For each PCR, one of two different forward primers (Fig. 1B), each inserting an EcoRI site at the 5′ end of the amplicon (underlined), was used (cre tag forward, 5′-CCTGAATTCGACTTCACC-3′; or Δcre tag forward, 5′-CATGAATTCAAATTCTTCCC-3′). In both cases, a single negative primer inserted a HindIII site (in boldface) at the 3′ end of each amplicon (cre tag reverse, 5′-TAAGCTTCACGTTCGACA-3′). The two PCR amplicons were ligated into pCR4.1 (Invitrogen Ltd., Paisley, United Kingdom) according to the manufacturer's instructions, and the inserts were sequenced. Correct inserts were excised from the vector using EcoRI/HindIII and were ligated into EcoRI/HindIII-linearized pRW50 (14). Recombinant plasmids pUB6070 (cre tag) and pUB6069 (Δcre tag) were used to transform E. coli DH5α cells to tetracycline resistance (25 μg/ml). β-Galactosidase assays in the presence of the reporter plasmid were carried out according to Miller's protocol (16) using 0.5 ml cells.
RT-PCR.
Total RNA was isolated using the Qiagen RNAeasy mini kit according to the manufacturer's instructions (Qiagen Ltd., Crawley, United Kingdom). Contaminating DNA was removed by treatment with RQ1 RNase-free DNase (Promega United Kingdom, Southampton); the absence of genomic DNA was confirmed by PCR. Total first-strand cDNA was synthesized from total RNA (200 ng) using Superscript III reverse transcriptase with random hexamer primers according to the manufacturer's instructions (Invitrogen Ltd., Paisley, United Kingdom). PCRs (25 μl) were performed using REDTAQ ReadyMix (Sigma-Aldrich Ltd., Poole, United Kingdom) from the cDNA template (2 μl). PCRs for the 16S rRNA and creD cDNAs were performed using a 1/1,000-fold and 1/5-fold dilution of total cDNA in UltraPure water, respectively. The primers for 16S were 16S_RT_F (5′-GGTGCAAGCGTTAATCGGAA-3′) and 16S_RT_R (5′-CTTCCGTGGATGTCAAGACC-3′); creC and creD primers were as reported previously (5). Following an initial denaturation at 95°C for 5 min, PCR (comprising the following sequential segments: 95°C for 30 s, 54°C for 30 s, and 72°C for 1 min) was performed for 30 cycles. Changes in the expression of creC and creD were inferred from the intensity of the resultant PCR products under UV transillumination following agarose gel electrophoresis; the expression of the control 16S rRNA housekeeping gene was used as a cDNA loading control.
Deletion of cre genes.
The creA, creB, creC, and creD genes were disrupted and deleted individually using the PCR-based method of Datsenko and Wanner (10). Deletion primers were designed to replace the full coding sequence of each gene with the kanamycin resistance gene amplified from pKD4 (10). The sequences of primers used to disrupt the genes (_KO primers) are shown in Table 1. The insertion of the kanamycin resistance gene at each locus was confirmed by PCR using an upstream, gene-specific forward primer with the reverse primer k1 (10). To check the other junction, the primer kan_F_seq (5′-GAAGAGCTTGGCGGCGAATG-3′) was used with a downstream, gene-specific, reverse primer. Gene-specific checking primers (_chk) are shown in Table 1. Unmarked deletions were generated using the pCP20-encoded FLP recombinase to excise the kanamycin resistance gene as described elsewhere (10). The amplimer generated from the excised sequence using the gene-specific forward and reverse primers was sequenced to confirm the genetic structure (ABC Sequencing, London, United Kingdom).
TABLE 1.
Sequences of oligonucleotides used for the disruption of the cre genes
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| creA_F_KO | CTAATTGCTCGTGTAATAGATAAAAATGGTAACAATGTGTAGGCTGGAGCTGCTTC |
| creA_R_KO | TAACCAGACCGTTTCCCGTTGCATAAATCGCCTCTGCATATGAATATCCTCCTTAG |
| creB_F_KO | CCTGTCATGCCGTGGCGGCAATAACAGAGGCGATTTGTGTAGGCTGGAGCTGCTTC |
| creB_R_KO | AAAAATAGCCCAGCAACAACCGCATGCCGATACGCACATATGAATATCCTCCTTAG |
| creC_F_KO | TCATCGCGGCATGGGATATAGCCTGAGGGGCCTGTAGTGTAGGCTGGAGCTGCTTC |
| creC_R_KO | AGCAGGATACGAAGACTATGTGGGAAGAATTTGAAGCATATGAATATCCTCCTTAG |
| creD_F_KO | TCTTCGTATCCTGCTGCCATTGCAAAGGAGAAGACTGTGTAGGCTGGAGCTGCTTC |
| creD_R_KO | AACGGCGCTTTTTAGCGCCGTTTTTATTTTTCAACCCATATGAATATCCTCCTTAG |
| creA_F_chk | CTTTCAACAACGAGCACCTG |
| creA_R_chk | TATCCGGCAGACCAACATCG |
| creB_F_chk | ACGGCAAAGCTCAGGGCGAG |
| creB_R_chk | GCAACGTTGCGGTGTCGATC |
| creC_F_chk | TTGAAGACGTTACTCAAGTC |
| creC_R_chk | AATCGCATCTTCCACATCGC |
| creD_F_chk | GTTTGTTTAACGGCGAAGTC |
| creD_R_chk | CCGTATTCGTAAACATTTCG |
Transcriptional start site analysis for creD.
Total RNA was isolated from DH5α (5 ml of a 50-ml culture) grown aerobically to mid-logarithmic growth phase (A600 = 0.5 to 0.6) in pyruvate-M9 minimal medium supplemented with thiamine by using the Qiagen RNAeasy mini kit according to the manufacturer's instructions (Qiagen Ltd., Crawley, United Kingdom). Contaminating DNA was removed by DNase treatment (Promega United Kingdom, Southampton). The determination of the 5′ end of creD mRNA was done using version 2.0 of the 5′ rapid amplification of cDNA ends (5′ RACE) system according to the manufacturer's instructions (Invitrogen Ltd., Paisley, United Kingdom). First-strand cDNA synthesis was primed from the RNA preparation by using the antisense creD gene-specific primer 1 (5′-CTAAGACGCGAAAC-3′). The resulting cDNA was purified using a SNAP column, and a 5′ cytidine homopolymeric tail was added using terminal deoxynucleotidyl transferase. The 5′ RACE abridged anchor primer (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) was used with creD gene-specific primer 2 (5′-CGTATAAAGCTCGGTCAC-3′) to amplify the tailed cDNA using an annealing temperature of 54°C. To enrich the product, it was reamplified using the 5′ RACE universal amplification primer (5′-CUACUACUACUAGGCCACGCGTCGACTAGTAC-3′) and creD gene-specific primer 2. The largest specific product was purified by gel extraction (QIAquick gel extraction kit; Qiagen, Crawley, United Kingdom), and the product(s) was ligated into the pCR2.1 TOPO cloning vector according to the manufacturer's instructions (Invitrogen Ltd., Paisley, United Kingdom). The insert was amplified using M13 primers (Invitrogen Ltd., Paisley, United Kingdom) and sequencing in both orientations (ABC Sequencing, London, United Kingdom). Ten clones were sequenced in order to confirm that the correct transcriptional start site had been identified.
Cloning and transient overproduction of CreB and CreB-His using pBAD.
The creB gene was amplified from E. coli DH5α by PCR using creB forward primer (5′-ATGCAACGGGAAACGGTC-3′) with either the creB reverse primer (5′-TTACAGGCCCCTCAGGC-3′) or the creB no-stop codon primer (5′-CAGGCCCCTCAGGCTAT-3′). These amplicons were individually ligated into the pBAD vector (Invitrogen Ltd.), rescued in E. coli TOP10 chemically competent cells, and selected with 50 μg/ml ampicillin. The orientation of the inserts was checked by PCR using M13-forward and the appropriate creB reverse primer (as described above). The plasmids were named pUB6073 (creB amplicon) and pUB6074 (creB no-stop codon amplicon); they were purified from 1 ml of an overnight culture using the QIAprep spin miniprep kit (Qiagen Ltd.) and electroporated into electrocompetent E. coli DH5α cells, with transformants being selected using 50 μg/ml ampicillin. The overproduction of CreB (from pUB6073) or CreB-His (from pUB6074) was induced by adding 0.2% (wt/vol) arabinose to glucose-M9 minimal medium cultures at a starting OD600 of 0.3. Induction was continued for 2 h prior to cell extraction and the assay of the appropriate reporter.
Purification of recombinant CreB-His.
CreB-His was produced by growing DH5α(pUB6074) in nutrient broth at 37°C with aeration to an OD600 of 1.2 to 1.3 in 1 liter of nutrient broth supplemented with 50 μg/ml thiamine, 30 mM glucose, 0.2% (vol/vol) Casamino Acids, 1% (wt/vol) arabinose, and 50 μg/ml ampicillin. The cell suspension was centrifuged at 4,500 rpm for 30 min and the supernatant removed, and the pellet was resuspended in 50 ml binding buffer (20 mM sodium phosphate, 500 mM NaCl, 5 mM imidazole, pH 7.4; filtered through a 0.22-μm nitrocellulose filter) and chilled on ice before being sonicated in a Sonics Vibra-cell sonicator (Sonics and Materials Inc., Newtown, CT) eight times for 30 s, delivered as 60 cycles of 0.5-s pulses with 0.5-s rest periods at 60% amplitude. The sample was centrifuged at 18,000 rpm for 30 min (Sorvall RC5C with an SS34 rotor; Kendro Laboratory Products, Hertfordshire, United Kingdom). Meanwhile, a 5-ml HiTrap chelating HP column (Amersham Biosciences, Little Chalfornt, Bucks, United Kingdom) was prepared according to the manufacturer's instructions. The supernatant was loaded onto the column at a rate of 4 ml/min. The column then was washed with 70 ml binding buffer, and bound proteins were eluted over 330 ml with a gradually increasing gradient of elution buffer (20 mM sodium phosphate, 500 mM NaCl, 5 mM imidazole, pH 7.4; filtered through a 0.22-μm nitrocellulose filter), thereby producing 82.5 fractions of 4 ml (1 min) each. Fractions were stored at 4°C. Fractions containing essentially pure CreB-His, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting using an anti-His antibody according to the manufacturer's instructions (Qiagen Ltd.), were concentrated using a Vivaspin 5000 column according to the manufacturer's instructions (Vivascience, Lincoln, United Kingdom) and dialyzed into a storage buffer (50 mM Tris-HCl, 250 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 2 mM dithiothreitol [DTT], 50% [vol/vol] glycerol). The concentration of CreB-His was determined using the Bio-Rad protein assay dye reagent concentrate and Bio-Rad quick start bovine serum albumen standards (Bio-Rad Laboratories Ltd., Hemel Hempstead, United Kingdom).
Radioactive labeling of oligonucleotide probes.
Two pairs of 28-bp oligonucleotides (Table 2) were designed based on the sequence spanning the creD upstream cre tag sequence. Each labeling reaction mixture contained 5 μl of 3.5 μM forward oligonucleotide, 4 μl Redivue adenosine 5′-[γ-32P]triphosphate, triethylammonium salt (GE Healthcare United Kingdom Ltd., Buckinghamshire), 2 μl T4 polynucleotide kinase 10× reaction buffer, 1 μl T4 polynucleotide kinase (Roche Diagnostics GmbH, Mannheim, Germany), and 8 μl water. The reaction mixture was incubated at 37°C for 30 min before 30 μl of the reverse annealing oligonucleotide (100 μM) was added. The mixture was incubated at 90°C for 10 min in a heat block, which then was switched off; the reaction mixture was left to cool in the block overnight. The resulting double-stranded, labeled oligonucleotides were purified using Micro-bio spin columns (Bio-Rad Laboratories Ltd., Hertfordshire, United Kingdom) according to the manufacturer's instructions. For cold competition, double-stranded oligonucleotides were made and purified exactly as set out above, but without the 30-min labeling reaction.
TABLE 2.
Sequences of oligonucleotides used as substrates for CreB-His binding in EMSA experiments
| EMSA oligonucleotide | Sequence (5′-3′) |
|---|---|
| creD wild-type cre tag forward | TTCGACTTCACCGTCACTTCACATAGCT |
| creD wild-type cre tag reverse | AGCTATGTGAAGTGACGGTGAAGTCGAA |
| creD no cre tag forward | TTCGACCCTGTCGTCACCCTGTATAGCT |
| creD no cre tag reverse | AGCTATACAGGGTGACGACAGGGTCGAA |
Electrophoretic mobility shift assay (EMSA).
Reactions mixtures (10 μl) were prepared with 1 nM radiolabeled double-stranded 28-bp oligonucleotide (described above), 50 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 100 μM EDTA, 1 mM DTT, and 4% (vol/vol) glycerol, with various amounts of purified CreB-His protein. For cold competition experiments, the appropriate cold oligonucleotide (described above) also was added to a concentration of 1 or 10 nM. In all cases, reaction mixtures were incubated at 37°C for 30 min before being loaded into 1.5-mm nondenaturing gels made with 10% acrylamide/bisacrylamide (37.5:1), 90 mM Tris base, 90 mM sodium borate, 2 mM EDTA, 0.05% (wt/vol) ammonium persulfate, 0.1% (vol/vol) N,N,N,N-tetramethylethylenediamine, which had been prerun in 90 mM Tris-base, 90 mM sodium borate, 2 mM EDTA in water for at least 30 min. Once loaded, gels were run at 100 V for 65 min before exposure to a phosphor storage screen (GE Healthcare) overnight. Phosphor storage screens were visualized using a Typhoon scanner (GE Healthcare).
RESULTS
cre regulon gene expression during growth on different carbon sources.
Our previously published work using RT-PCR showed that creD is part of the cre regulon, and its expression is strongly induced upon the activation of CreBC (5). In order to extend that work and to carefully monitor the effects of various well-defined growth conditions on cre regulon gene expression, we decided to make a β-galactosidase reporter of creD expression by splicing the sequence upstream of creD to a promoterless lacZ in the reporter plasmid pRW50. The result was plasmid pUB6070.
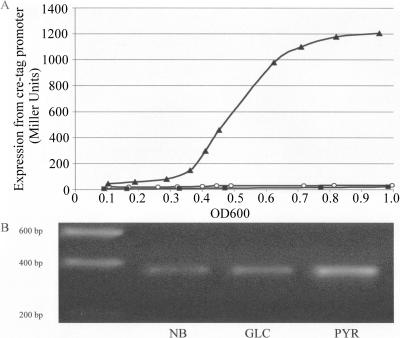
β-Galactosidase activity in E. coli DH5α(pUB6070) grown in nutrient broth or in minimal medium with glucose or pyruvate as the sole carbon source was measured at intervals during a period of time from the initial inoculation of the culture to early stationary phase (OD600 = 0.9 to 1.0) (Fig. 2A). These growth curves revealed that representative data could be collected at mid-log phase (OD600 = 0.5 to 0.6), so this growth point was used for future studies. Growth in either glucose or pyruvate minimal medium previously has been shown to cause a significant increase in cre regulon gene expression relative to that of growth in nutrient broth (5). Consistently with our previous RT-PCR studies, considerable activation of β-galactosidase reporter enzyme activity occurred during growth in pyruvate minimal medium relative to that observed during growth in nutrient broth (Fig. 2A). However, inconsistently with our previous creD RT-PCR work, a similarly low level of β-galactosidase activity was observed during growth on glucose minimal medium, as seen for nutrient broth. To validate these results, RNA was purified from these same cultures at an OD600 of 0.5 to 0.6, and RT-PCR was used to monitor creD transcription from the chromosome. The RT-PCR data mirrored the LacZ reporter data for cultures grown under the same conditions (Fig. 2B).
FIG. 2.
creD expression during aerobic growth in different media. Expression was measured using β-galactosidase reporter plasmid pUB6070 (A) and RT-PCR (B). DH5α(pUB6070) was grown aerobically for an entire growth curve (A) or to mid-log phase (OD600 = 0.5 to 0.6) (B) in nutrient broth (NB; black squares), in M9 minimal medium containing glucose (GLC; open circles), or with pyruvate (PYR; black triangles) as the sole carbon source. (B) Bands represent the RT-PCR product separated using agarose gel electrophoresis and visualized under UV transillumination. The data are averages of (A) or representative of (B) three individual biological replicates.
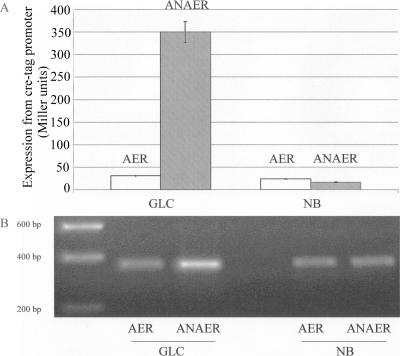
Our previously published data reporting the activation of cre regulon gene expression during growth on glucose minimal medium came from cultures of logarithmic-phase DH5α cells grown in sealed universal bottles (5). The data shown in Fig. 2 come from cultures grown in flasks with foam bungs. Accordingly, we hypothesized that the reason why we were not able to replicate the results of our earlier study was that the oxygen tension of cultures in the current experiment was far higher. If correct, this would mean that during growth on glucose minimal medium, cre regulon gene expression is responsive to a switch from aerobic to anaerobic conditions, and so it is possibly activated when cells are fermenting glucose. To test this, we grew cultures anaerobically as set out in Materials and Methods. There was a 10-fold increase in cre regulon reporter gene expression during anaerobic growth on glucose-M9 minimal medium (measured at an OD600 of 0.5 to 0.6) compared with that seen during aerobic growth (Fig. 3A). However, there was no significant induction of cre regulon reporter gene expression during anaerobic growth on the complex medium, nutrient broth (Fig. 3A), confirming our earlier finding that during growth in complex media, cre regulon gene expression is not activated (5). Again, the reporter gene assays correlated well with RT-PCR measurements of creD transcript levels from the chromosome (Fig. 3B), so we can be confident that pUB6070 accurately reports creD expression levels.
FIG. 3.
Effect of anaerobic growth on creD expression. Expression was measured using β-galactosidase reporter plasmid pUB6070 (A) and RT-PCR (B). DH5α(pUB6070) was grown aerobically (AER) or anaerobically (ANAER) to mid-log phase (OD600 = 0.5 to 0.6) in nutrient broth (NB) or in M9 minimal medium containing glucose (GLC) as the sole carbon source. (A) Error bars indicate standard deviations from at least three independent experiments. (B) Bands represent the RT-PCR product separated using agarose gel electrophoresis and visualized under UV transillumination. RT-PCR data are representative of three biological replicates.
Adding increasing concentrations of nitrate, fumarate, or TMAO to anaerobic glucose-M9 minimal medium cultures as alternative terminal electron acceptors resulted in a dosage-dependent reduction of cre regulon reporter gene expression (Fig. 4), which further supports our hypothesis that the activation of cre regulon gene expression occurs when cells are fermenting glucose.
FIG. 4.
Effect of alternative electron acceptors on anaerobic activation of creD expression. Expression was measured using β-galactosidase reporter plasmid pUB6070. DH5α(pUB6070) was grown anaerobically to mid-log phase (OD600 = 0.5 to 0.6) in M9 minimal medium containing glucose as the sole carbon source in the presence of different concentrations of three alternate electron acceptors: sodium nitrate (black triangles), sodium fumarate (open circles), and TMAO (black squares). Error bars indicate standard deviations from at least three independent experiments.
During aerobic growth, high levels of cre regulon reporter gene expression were seen during growth in M9 minimal medium containing all single or short-chain carbon sources tested (formate, acetate, lactate, pyruvate, and malate) but not with any of the longer five- and six-carbon compounds tested (arabinose, fucose, gluconate, and glucose) (Fig. 5).
FIG. 5.
creD expression during aerobic growth in M9 minimal medium supplemented with different carbon sources. Expression was measured using β-galactosidase reporter plasmid pUB6070. DH5α(pUB6070) was grown anaerobically to mid-log phase (OD600 = 0.5 to 0.6) in M9 minimal medium containing formate (FOR), acetate (ACE), lactate (LAC), pyruvate (PYR), malate (MAL), arabinose (ARA), fucose (FUC), gluconate (GLU), or glucose (GLC) as the sole carbon source (180 mM carbon atoms in each case) or in nutrient broth (NB). Error bars indicate standard deviations from at least three independent experiments.
Role of cre locus genes in the carbon source-mediated activation of cre regulon expression.
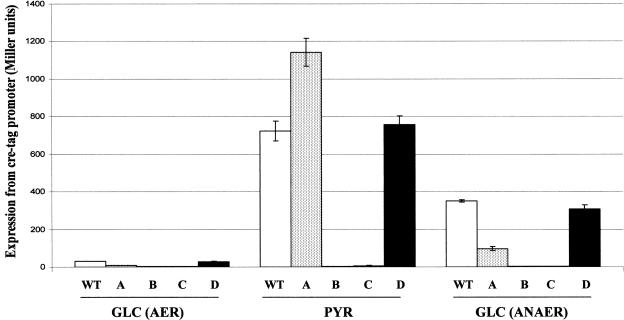
Genes encoding the CreBC TCS, which is believed to control cre regulon gene expression, are encoded as part of the creABCD genetic locus. The systematic deletion of creA, creB, creC, and creD was undertaken to assess the roles of their protein products on the activation of cre regulon gene expression (measured using the cre regulon reporter plasmid, pUB6070) during aerobic growth on glucose or pyruvate minimal medium (Fig. 6). In all cases, the resistance gene cassette used to disrupt the genes then was excised, creating deletions without polar effects; for example, RT-PCR showed that downstream creC expression is not significantly reduced in the creB disruption mutant (data not shown). The deletion of creB or creC completely abolished any activation of cre regulon reporter gene expression during growth on pyruvate. These data confirm that CreB and CreC both are essential for controlling the expression of cre regulon genes. In contrast, the disruption of creD did not alter the activation of cre regulon reporter gene expression upon a switch from glucose to pyruvate minimal medium, showing that CreD is not itself essential for the activation of CreBC. Interestingly, the creA deletion mutant showed a slightly enhanced expression of the reporter gene under these conditions (Fig. 6).
FIG. 6.
Effect of disruption of genes of the cre locus on creD expression. Expression was measured using β-galactosidase reporter plasmid pUB6070. DH5α (wild type [WT]), DH5αΔcreA (A), DH5αΔcreB (B), DH5αΔcreC (C), and DH5αΔcreD (D), each carrying pUB6070, were grown aerobically (AER) or anaerobically (ANAER) to mid-log phase (OD600 = 0.5 to 0.6) in M9 minimal medium containing glucose (GLC) or pyruvate (PYR) as the sole carbon source. Error bars indicate standard deviations from at least three independent experiments.
During anaerobic growth on glucose minimal medium, intact creB and creC also were found to be essential for the activation of cre regulon gene expression, but again, neither creA nor creD is required for this process (Fig. 6). Interestingly, although the deletion and excision of creA caused a slight increase in cre regulon gene expression during aerobic growth on pyruvate minimal medium, its deletion was found to cause a threefold decrease in the activation of cre regulon gene expression anaerobically on glucose (Fig. 6). Given our current understanding of how CreC becomes activated, we cannot explain this observation, and indeed, it may be simply a minor effect of the genetic lesion created, which could conceivably alter the production of CreBC through introducing or removing some cis-acting element within the 5′ end of the creABCD operon. We can state clearly, however, that CreD and CreA are not required for the activation of cre regulon gene expression under any of the conditions tested in this study.
Confirmation of a role for the cre tag sequence in transcriptional activation of creD in vivo.
We previously hypothesized that the cre tag sequence, TTCACnnnnnnTTCAC, located upstream of all known cre regulon genes, is important for controlling their expression by CreBC (5). To test this hypothesis, we made a new creD/lacZ reporter construct that was identical to pUB6070, except that the 16-bp cre tag sequence upstream of creD was absent. The resultant plasmid was called pUB6069. β-Galactosidase activity in DH5α(pUB6069) cells did not increase significantly during aerobic growth in pyruvate minimal medium or during anaerobic growth on glucose minimal medium compared to that of aerobic growth on glucose minimal medium (data not shown), confirming that the cre tag is essential for the control of creD expression and adding further evidence for its role in the CreBC-mediated control of cre regulon gene expression per se.
These studies were complemented by determining the creD transcriptional start site in E. coli DH5α during aerobic growth in pyruvate minimal medium using 5′ RACE as set out in Materials and Methods. The transcriptional start site was located 18 nucleotides upstream of the ATG initiation codon for creD (Fig. 1B). An appropriately positioned 4 of 6 matching −10 sequence (TATCCT) is located upstream of this transcriptional start site, but there is no appropriately positioned −35 element having the requisite 17- to 18-nucleotide separation from this −10 element. This experiment revealed that the cre tag is centered at −51.5 with respect to the creD transcriptional start site (Fig. 1B).
Interaction of CreB with the cre tag sequence in vitro and in vivo.
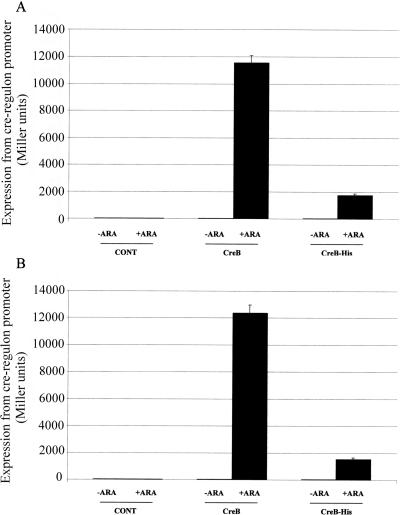
It has been established that pho box DNA binding by E. coli PhoB is not dependent upon PhoB phosphorylation, but that phosphorylation increases the affinity of PhoB for the pho box. Hence, the reduced affinity of unphosphorylated PhoB for the pho box can be mitigated by the overproduction of PhoB, which activates pho regulon gene expression even in the absence of an active PhoR signal sensor (12). Given that PhoB and CreB are highly similar (25), we next tested whether CreB overproduction in E. coli DH5α would stimulate cre regulon gene expression even during aerobic growth on glucose, when the CreBC TCS is not activated. To this end, creB was overexpressed using the arabinose-inducible pBAD system; the recombinant plasmid was designated pUB6073. This plasmid was electroporated into DH5α(pUB6070), and β-galactosidase assays were used to measure cre regulon reporter gene expression during aerobic growth on glucose minimal medium with or without 0.2% (wt/vol) arabinose (2 h) to stimulate the expression of CreB from pUB6073. These results (Fig. 7A) clearly demonstrate that the overproduction of CreB activates cre regulon gene expression. In parallel, pUB6073 was introduced into a creC deletion derivative of DH5α(pUB6070). The activation of LacZ reporter production was similar to that for creC+ cells (Fig. 7B), confirming that CreC is not required for this process, as expected given that, under these growth conditions, CreC is minimally active.
FIG. 7.
Effect of CreB and CreB-His overproduction on creD expression. Expression was measured using β-galactosidase reporter plasmid pUB6070. DH5α(pUB6070) (A) or DH5α(ΔcreC)(pUB6070) (B) containing either empty pBAD vector (CONT), pUB6073 (CreB), or pUB6074 (CreB-His) were grown to early log phase (OD600 = 0.3) in M9 minimal medium containing glucose, and then 0.2% (wt/vol) arabinose was added (+ARA) to induce CreB or CreB-His production, and the cells were grown for a further 2 h. Alternatively, no arabinose was added (−ARA). Error bars indicate standard deviations from at least three independent experiments.
In order to purify recombinant CreB for in vitro EMSAs, a new creB overexpression vector was constructed using pBAD in such a way that a C-terminal hexahistidine tag was introduced into the protein. When overproduced, CreB-His is able to strongly stimulate cre regulon reporter gene expression in vivo (approximately 60-fold), though not as strongly as the untagged protein under the same induction conditions (Fig. 7). This may be for a number of reasons, including the reduced expression and/or reduced stability of the His-tagged version of CreB. However, since CreB-His clearly is biologically active and more easily purified than native CreB, it was used to test for the ability of CreB to bind to the cre tag sequence in vitro.
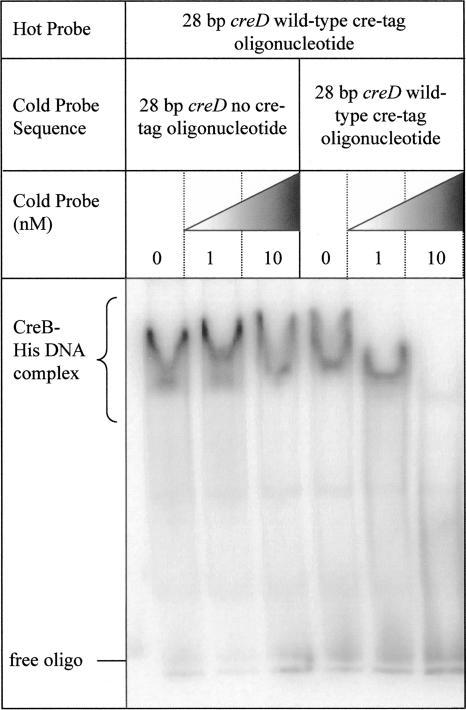
CreB-His was purified using nickel affinity chromatography and used in EMSAs together with 28-bp radiolabeled oligonucleotide probes either containing or specifically lacking the cre tag sequence upstream of creD. There was a clear band shift associated with CreB-His binding to the cre tag-containing probe (Fig. 8A). At higher concentrations of protein, a supershift is observed. When a probe lacking the cre tag (i.e., the probe was identical to that described above, except that the two TTCAC sequences were converted to CCTGT) was used, no band shift was observed (Fig. 8B). The addition of the cold cre tag probe in competition with the same concentration as the radioactive cre tag probe (1 nM) caused a significant change in the association of CreB-His with the radioactive probe, with the supershift no longer occurring. At a 10-fold molar excess, cold cre tag-containing probe blocked the binding of CreB-His to the radioactive cre tag probe to below detectable levels (Fig. 9). Competition using a cold probe lacking the cre tag, however, did not affect the binding of CreB-His to the radioactive cre tag probe even at a 10-fold molar excess (Fig. 9). These data confirm that CreB binds specifically to the cre tag sequence in vitro and, taken together with the data presented in Fig. 7 and the determined transcriptional start site for creD (Fig. 1B), provide strong evidence that CreB is a transcriptional activator of creD and other cases of cre regulon gene expression through interaction with the cre tag.
FIG. 8.
Binding of CreB-His to the cre tag sequence in vitro. EMSAs were performed as set out in Materials and Methods using increasing amounts of purified CreB-His with 1 nM radiolabeled double-stranded oligonucleotide (oligo) substrates with either no cre tag (A) or a wild-type cre tag (B). The free oligonucleotides and shifted forms are labeled. This figure is representative of three experiments.
FIG. 9.
Specificity of CreB-His binding to the cre tag sequence. EMSAs were performed using 1 nM radiolabeled double-stranded cre tag oligonucleotide (oligo) substrate and 1 μg of CreB-His. Cold competition was with double-stranded no cre tag oligonucleotide at 0, 1, or 10 nM (lanes 1 to 3) or cre tag oligonucleotide at 0, 1, or 10 nM (lanes 4 to 6). The free oligonucleotides and shifted forms are labeled. This figure is representative of three experiments.
DISCUSSION
In our earlier study (5), we reported that CreBC regulates the expression of cre regulon genes in response to a switch from complex to minimal medium. We hypothesized that there is some component common to complex media that switches off CreBC and that in minimal media this component is absent, so CreBC becomes activated. Following work reported in this paper, we must refine our earlier hypothesis. We have confirmed that, during growth in complex media, cre regulon gene expression is not stimulated. However, simply growing cells in minimal medium does not activate CreBC. The data reported in this study show that CreBC is inactive when cells are generating energy by respiration (even in the absence of oxygen) during growth on glycolytic carbon sources. In fact, the activation of CreBC occurs when cells are fermenting glycolytic carbon sources. The fermentation of glucose primarily causes a buildup of acetate and lactate, which is made from pyruvate (1). It is interesting, therefore, that CreBC also is activated during aerobic growth on low-molecular-weight, gluconeogenic carbon sources such as pyruvate, acetate, and lactate. Accordingly, it may be that the ligand causing the activation of CreC is a metabolic intermediate found at high concentrations during fermentation and during growth on gluconeogenic carbon sources. The reason why growth on nutrient broth does not activate CreBC, even during fermentation, presumably either is because the carbon source used for growth (catabolyzable amino acids [20]) cannot lead to a buildup of the appropriate activating ligand or because there is a separate repressive input into cre regulon gene expression that relies on the presence of complex media.
Fifteen years ago, Wanner and Wilmes-Riesenberg presented data that suggested that CreC responds to changes in carbon source (24). To measure CreC activation, they exploited the ability of active CreC to trans-phosphorylate PhoB (and so stimulate Bap production) in a phoR68 mutant background. They showed that Bap production was increased during growth on acetate or pyruvate minimal media compared to that on glucose minimal medium, suggesting, as we do now, that CreC is activated in these growth conditions. However, while the acetate growth effect on Bap production was entirely CreC dependent, the effect of pyruvate was only partially reduced upon the deletion of creC. This observation led the authors to conclude that there is a further mechanism by which Bap production can be regulated that is independent of both PhoR and CreC. They proposed, and later proved, that this regulator is acetyl phosphate, which is a phosphoryl donor for PhoB. They showed that during growth on pyruvate minimal medium, acetyl phosphate levels build up, causing PhoB to become phosphorylated and inducing Bap production (24, 25).
The fact that Wanner and Wilmes-Reisenberg (24) showed that the pyruvate-induced production of Bap is not entirely CreC dependent in no way detracts from our conclusion above that CreC is activated during growth on pyruvate, since they clearly showed that acetyl phosphate takes over to activate Bap production in the absence of CreC and PhoR during pyruvate growth. We must conclude, however, that acetyl phosphate accumulation does not lead to the phosphorylation and activation of the CreB RR under these conditions, because even if cellular acetyl phosphate levels are high during our experiments, as might be expected (13), the induction of cre regulon gene expression during pyruvate growth is entirely CreC dependent.
We have confirmed here that the activation of cre regulon gene expression is absolutely dependent upon creB and creC being intact and the presence of a 16-bp TTCACnnnnnnTTCAC cre tag sequence. CreC and CreB are known to form an active TCS in vitro (3), and we have shown here that CreB binds to the cre tag in vitro and specifically stimulates cre regulon gene expression in vivo when overproduced in E. coli. Therefore, we are confident in proposing the following model: CreC responds to changes in the growth medium and/or metabolite pool levels and autophosphorylates; this phosphate is passed on to CreB, which binds to cre tag sequences; at some promoters (e.g., creD or talA), CreB works as a transcriptional activator by recruiting RNA polymerase in a manner typical of other winged-helix RR proteins, such as PhoB (7); at other promoters (e.g., malE), the binding of CreB to the cre tag represses transcription through promoter occlusion. The reason for this response is presumably to tailor the needs of the cell to fermentative and/or gluconeogenic growth, in which many intermediary metabolic pathways must be rerouted. We do not know yet the size of the cre regulon, though even now there are hints that it does have a role in modifying intermediary metabolism. As well as including genes that encode proteins with no characterized function, such as creD, yieI, and yidS, the gene encoding TalA (transaldolase) is part of the cre regulon (5). TalA is the committed enzyme in the nonoxidative branch of the pentose phosphate pathway. This pathway is essential for the production of precursors for vitamin, nucleotide, and aromatic amino acid biosynthesis. Moreover, it is used predominantly during the fermentation of glycolytic carbon sources and during growth on low-molecular-weight carbon sources. These are the conditions found to activate cre regulon gene expression in this study.
In 2002, Oshima et al. (18) performed microarray transcriptomic experiments following the individual deletion of all known TCS genes. The deletion of creBC had no discernible effect. However, their experiments were performed using cells grown aerobically on complex medium. Under these conditions, CreBC is totally inactive. Accordingly, the full extent of the cre regulon is not known. Our future work will specifically address this issue and will try to determine what specific ligand(s) activates CreC.
Acknowledgments
This work was funded by a project grant to M.B.A. from the Biotechnology and Biological Sciences Research Council. A.E.T. is in receipt of a University of Bristol postgraduate scholarship.
We are grateful to Patrick Walter, Steven Pullan, and Rachel Horton for technical assistance and to Abigail Smith and Nigel Savery (Department of Biochemistry, University of Bristol, United Kingdom), who provided advice and reagents for the EMSA experiments.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Aguayo, J. B., M. P. Gamcsik, and J. D. Dick. 1988. High resolution deuterium NMR studies of bacterial metabolism. J. Biol. Chem. 26319552-19557. [PubMed] [Google Scholar]
- 2.Amemura, M., K. Makino, H. Shinagawa, and A. Nakata. 1986. Nucleotide sequence of the phoM region of Escherichia coli: four open reading frames may constitute an operon. J. Bacteriol. 168294-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amemura, M., K. Makino, H. Shinagawa, and A. Nakata. 1990. Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM-open reading frame 2. J. Bacteriol. 1726300-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avison, M. B., P. Niumsup, T. R. Walsh, and P. M. Bennett. 2000. Aeromonas hydrophila AmpH and CepH beta-lactamases: derepressed expression in mutants of Escherichia coli lacking creB. J. Antimicrob. Chemother. 46695-702. [DOI] [PubMed] [Google Scholar]
- 5.Avison, M. B., R. E. Horton, T. R. Walsh, and P. M. Bennett. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J. Biol. Chem. 27626955-26961. [DOI] [PubMed] [Google Scholar]
- 6.Avison, M. B., P. Niumsup, K. Nurmahomed, T. R. Walsh, and P. M. Bennett. 2004. Role of the 'cre/blr-tag’ DNA sequence in regulation of gene expression by the Aeromonas hydrophila β-lactamase regulator, BlrA. J. Antimicrob. Chemother. 53197-202. [DOI] [PubMed] [Google Scholar]
- 7.Bachhawat, P., G. V. T. Swapna, G. T. Montelione, and A. M. Stock. 2005. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 131353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baek, J. H., and S. Y. Lee. 2006. Novel gene members in the Pho regulon of Escherichia coli. FEMS Microbiol. Lett. 264104-109. [DOI] [PubMed] [Google Scholar]
- 9.Blanco, A. G., M. Sola, F. X. Gomis-Ruth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10701-713. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drury, L. S., and R. S. Buxton. 1988. Identification and sequencing of the Escherichia coli cet gene which codes for an inner membrane protein, mutation of which causes tolerance to colicin E2. Mol. Microbiol. 2109-119. [DOI] [PubMed] [Google Scholar]
- 12.Ellison, D. W., and W. R. McCleary. 2000. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J. Bacteriol. 1826592-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein, A. H., A. Shulla, S. A. Reimann, D. H. Keating, and A. J. Wolfe. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 1895574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74271-276. [DOI] [PubMed] [Google Scholar]
- 15.Makino, K., H. Shinagawa, M. Amemura, T. Kawamoto, M. Yamada, and A. Nakata. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210551-559. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Niumsup, P., A. M. Simm, K. Nurmahomed, T. R. Walsh, P. M. Bennett, and M. B. Avison. 2003. Genetic linkage of the penicillinase gene, amp, and blrAB, encoding the regulator of β-lactamase expression in Aeromonas spp. J. Antimicrob. Chemother. 511351-1358. [DOI] [PubMed] [Google Scholar]
- 18.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Micro. 46281-291. [DOI] [PubMed] [Google Scholar]
- 19.Rasmussen, B. A., D. Keeney, Y. Yang, and K. Bush. 1994. Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob. Agents Chemother. 382078-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sezonov, G., D. Joseleau-Petit, and R. D'Ari. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 1898745-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanner, B. L., and P. Latterell. 1980. Mutants Affected in alkaline phosphatase expression: evidence for multiple positive regulators of the phosphate regulon in Escherichia coli genetics. 96353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanner, B. L., M. R. Wilmes, and E. Hunter. 1988. Molecular cloning of the wild-type phoM operon in Escherichia coli K-12. J. Bacteriol. 170279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wanner, B. L., M. R. Wilmes, and D. C. Young. 1988. Control of bacterial alkaline phosphatase synthesis and variation in an Escherichia coli K-12 phoR mutant by adenyl cyclase, the cyclic AMP receptor protein, and the phoM operon. J. Bacteriol. 1701092-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wanner, B. L., and M. R. Wilmes-Riesenberg. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 1742124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 5147-54. [DOI] [PubMed] [Google Scholar]