Abstract
We determined the complete genome sequence of Streptomyces griseus IFO 13350, a soil bacterium producing an antituberculosis agent, streptomycin, which is the first aminoglycoside antibiotic, discovered more than 60 years ago. The linear chromosome consists of 8,545,929 base pairs (bp), with an average G+C content of 72.2%, predicting 7,138 open reading frames, six rRNA operons (16S-23S-5S), and 66 tRNA genes. It contains extremely long terminal inverted repeats (TIRs) of 132,910 bp each. The telomere's nucleotide sequence and secondary structure, consisting of several palindromes with a loop sequence of 5′-GGA-3′, are different from those of typical telomeres conserved among other Streptomyces species. In accordance with the difference, the chromosome has pseudogenes for a conserved terminal protein (Tpg) and a telomere-associated protein (Tap), and a novel pair of Tpg and Tap proteins is instead encoded by the TIRs. Comparisons with the genomes of two related species, Streptomyces coelicolor A3(2) and Streptomyces avermitilis, clarified not only the characteristics of the S. griseus genome but also the existence of 24 Streptomyces-specific proteins. The S. griseus genome contains 34 gene clusters or genes for the biosynthesis of known or unknown secondary metabolites. Transcriptome analysis using a DNA microarray showed that at least four of these clusters, in addition to the streptomycin biosynthesis gene cluster, were activated directly or indirectly by AdpA, which is a central transcriptional activator for secondary metabolism and morphogenesis in the A-factor (a γ-butyrolactone signaling molecule) regulatory cascade in S. griseus.
The gram-positive, soil-inhabiting, filamentous bacterial genus Streptomyces is characterized by its ability to produce a wide variety of secondary metabolites, such as antibiotics, parasiticides, herbicides, and pharmacologically active substances, including antitumor agents and immunosuppressants. Another characteristic feature of the genus is its complex multicellular development. Spores germinate to form a branched, multinucleoid substrate mycelium, which then produces an aerial mycelium. After septa have been formed at regular intervals along the aerial hyphae, long chains of uninucleoid spores are formed. Because of its complex morphogenesis and industrial and medical importance, Streptomyces has become a model prokaryote for the study of multicellular differentiation and secondary metabolism. The complete genomic sequences of two Streptomyces species, a model strain, Streptomyces coelicolor A3(2) (3), and an industrial strain, Streptomyces avermitilis (20, 36), have been published.
Unlike most other eubacterial chromosomes, the chromosome of Streptomyces is linear and contains a centrally located origin of replication (oriC) and unique terminal inverted repeats (TIRs) with terminal proteins (Tpgs) covalently bound to the 5′ ends. Replication proceeds bidirectionally from oriC, and a terminal single-stranded gap on the discontinuous lagging strand is filled in by DNA synthesis primed by Tpg (1). A telomere-associated protein (Tap) which binds specifically to the terminal single-stranded DNA is also involved in the replication of telomeres (2). Tpg and Tap are conserved among several Streptomyces species; accordingly, the terminal sequences of linear chromosomes and some linear plasmids of Streptomyces show extensive homology (19).
Streptomycin, the first aminoglycoside antibiotic, was discovered in S. A. Waksman's laboratory more than 60 years ago (41). This antibiotic, which has saved many people from tuberculosis, is produced industrially by Streptomyces griseus. S. griseus also provided arguably the first well-studied bacterial example of extracellular signaling by a diffusible low-molecular-weight compound. In Streptomyces, chemical-signaling molecules that have a γ-butyrolactone are commonly used as the switch for secondary metabolite production and/or morphological development. A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) of S. griseus, originally discovered by Khokhlov et al. (25) 40 years ago, is one such γ-butyrolactone (16, 17). When A-factor produced in a growth-dependent manner reaches a critical concentration as low as 10−9 M, it binds an A-factor-specific receptor (ArpA) that has bound to the promoter of adpA and dissociates ArpA from the promoter (34). The transcriptional activator AdpA then activates the transcription of a number of genes that are required for secondary metabolism and morphological differentiation, forming an AdpA regulon (33). strR, which encodes the pathway-specific transcriptional activator for streptomycin biosynthesis, is one of the targets of AdpA, which explains how A-factor induces streptomycin production (34).
Here, we present the complete nucleotide sequence of the S. griseus IFO 13350 genome and compare it with the two other sequenced Streptomyces genomes. The S. griseus genome sequence has revealed a novel pair of telomeric proteins, which makes us speculate that S. griseus acquired the unique telomere from a linear plasmid during evolution. In addition, comparison of the genome sequences of three Streptomyces species has revealed a core genome sequence of Streptomyces, as well as genes specific to S. griseus, including gene clusters for secondary metabolite biosynthesis. We also describe the results of the S. griseus transcriptome analysis using a DNA microarray to show genome-wide transcriptional regulation by A-factor.
MATERIALS AND METHODS
Strain.
The S. griseus strain IFO 13350 was originally transferred from the IMRU (Institute of Microbiology, Rutgers, The State University of New Jersey) as Streptomyces bikiniensis IMRU 3514 to the IFO (Institute for Fermentation, Culture Collection of Microorganisms, Osaka, Japan), via the National Institute of Health (Tokyo, Japan); however, it was later shown not to be S. bikiniensis IMRU 3514 but rather S. griseus (Krainsky) Waksman and Henrici 1948 (38). To avoid confusion, the genome-sequenced strain has been deposited as S. griseus strain NBRC 102592 in the Biological Resource Center of the National Institute of Technology and Evaluation (NITE; Chiba, Japan).
Genome sequencing and assembly.
The nucleotide sequence of the S. griseus IFO 13350 genome was determined using a whole-genome shotgun strategy. We constructed small-insert (2-kb) and large-insert (10-kb) genomic libraries and generated approximately 110,000 sequences, giving 9.5-fold coverage, from both ends of each of the genomic clones. The sequence data were assembled using the Phred/Phrap/Consed package and in-house scripts. Sequence gaps were closed by transcriptional sequencing (Nippon GeneTech) or by primer walking. The left and right TIR sequences were determined to be a single sequence.
The telomere sequence was determined as follows. Adenine-homopolymer [poly(A)] tails were attached to the naked 3′ ends of the linear chromosome by incubating the purified chromosomal DNA (1 μg) with 15 units of terminal deoxynucleotidyl transferase (Invitrogen) at 37°C for 15 min, according to the manufacturer's instructions. After being heated at 60°C for 10 min, with the reaction mixture as a template, the terminal sequence was amplified by PCR using primers 5′-ACGAGCACAACGCTGATC-3′ (430R; positions 430 to 413) and 5′-GTAAAACGACGGCCAGTATAGGGTTTTTTTTTTTT-3′, followed by nested PCR using primers 5′-GTAAAACGACGGCCAG-3′ (M13) and 5′-AAATGATGGATTCTCATATCCG-3′ (182R; positions 182 to 161). The amplified fragment was cloned into pCR4Blunt-TOPO (Invitrogen) and sequenced. Similarly, thymine-homopolymer [poly(T)] was attached to the 3′ ends of the chromosomal DNA using dTTP instead of dATP. The terminal sequence was amplified by PCR using primers 430R and 5′-GTAAAACGACGGCCAGTATAGGGAAAAAAAAAAAA-3′, followed by nested PCR using primers M13 and 182R, and sequenced.
According to the method described by Ômura et al. (36), we also cloned every DraI linking clone and constructed a DraI physical map of the chromosome to evaluate the validity of the final sequence assembly (Fig. 1, line i). Finally, the entire sequence was estimated to have an error rate of <1 per 10,000 bases (Phrap score, ≥40).
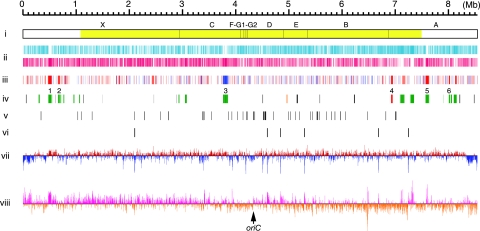
FIG. 1.
Schematic representation of the S. griseus chromosome. (i) DraI physical map. Fragments observed after pulsed-field gel electrophoresis are indicated above the bar. The largest fragment, X, was not detected by Southern blotting using linkage clones as probes. The yellow color indicates the “core region.” (ii) Distribution of ORFs according to direction of transcription (positive strand, upper line; negative strand, lower line). (iii) Distribution of genes that were affected by adpA disruption shown by DNA microarray analysis (P < 0.05, n = 4). Red, >2-fold upregulated in the wt strain in comparison with the ΔadpA mutant; blue, >2-fold downregulated in comparison with the ΔadpA mutant. (iv) Distribution of secondary metabolite gene clusters. Red, streptomycin (str, sts); orange, grixazone (gri); green, PKS and/or NRPS; black, other. Six secondary metabolite gene clusters, the transcription of which is affected by adpA disruption, are indicated as numerals above the line. 1, SGR443 to SGR455 (NRPS); 2, SGR574 to SGR593 (NRPS); 3, SGR3239 to SGR3288 (type II PKS and NRPS); 4, SGR5914 to SGR5940 (streptomycin); 5, SGR6360 to SGR6387 (type I PKS); 6, SGR6709 to SGR6717 (NRPS). (v) Distribution of tRNA genes. (vi) Distribution of rRNA operons. (vii) G+C content percentage variation for nonoverlapping 5-kb window. Red and blue are above and below the mean, respectively. (viii) GC-skew for 3-kb window and 1-kb step. Deep pink and dark orange are above and below zero, respectively. The putative oriC gene is indicated by an arrow.
Genome annotation and analysis.
Putative protein-coding sequences were predicted using the GLIMMER program trained with all annotated open reading frames (ORFs) of S. coelicolor A3(2) and S. avermitilis and then manually examined using the BLASTP program and the FRAME PLOT program optimized to handle genomic-size sequences. The tRNA and transfer-messenger RNA genes were predicted using the tRNAscan-SE and ARAGORN programs, respectively. Proteins were clustered using the BLASTCLUST program.
Microarray preparation.
An array of 13,042 specific oligonucleotides (45-mer or 35-mer; melting temperature, 72 ± 5°C) was designed based on the S. griseus genome sequence. Sequences from the 7,138 genes that encoded putative ORFs were used in the design. The oligonucleotides were designed and synthesized by Sigma-Genosys (Sigma-Aldrich, Japan) and printed onto a glass slide produced by KakenGeneqs (Chiba, Japan) according to the manufacturer's protocol. As a positive control, all of the oligonucleotides were mixed and printed at the corners of each subgrid.
DNA microarray analysis.
S. griseus wild-type (wt) and adpA deletion (ΔadpA) (34) strains were precultured at 30°C for 2 days in 100 ml of YMPD medium containing 0.5% glycine (in a 500-ml shaking [Sakaguchi] flask) with reciprocal shaking (120 rpm). YMPD is a nutrient-rich medium (pH 7.2) consisting of 0.2% yeast extract, 0.22% meat extract, 0.4% Bacto peptone, 0.5% NaCl, 0.2% MgSO4·7H2O, and 1% glucose. Two milliliters of each culture was inoculated into 100 ml of YMPD medium (in a 500-ml shaking [Sakaguchi] flask). The culture was incubated at 30°C with reciprocal shaking (120 rpm) until the cells entered the early stationary phase (wt, 30 h; ΔadpA mutant, 19 h). Note that the difference in the cultivation times resulted from the following two reasons. (i) The growth lag time of the wt strain was approximately 3 h longer than that of the ΔadpA mutant, probably due to the difference in the physiological conditions of the inoculated cells; some of the wt cells produced submerged spores in the preculture, but the ΔadpA cells did not. (ii) The maximum cell mass of the ΔadpA mutant in the culture was reproducibly smaller than that of the wt strain, resulting in an apparently earlier entry into the stationary phase. The difference in maximum cell mass between the wt and an A-factor-deficient mutant (strain HH1) has long been observed (our unpublished observation), although the reason is not yet known. The cells were harvested by centrifugation. To preserve RNA, RNAlater (Ambion) was added to the cells, and the mixture was incubated at 4°C for 12 h according to the manufacturer's instructions. The cells were collected by centrifugation and stored at −80°C. Total RNA was isolated from the cells using the RNAqueous mini kit (Ambion) according to the manufacturer's instructions. The RNA was quantified by measuring the absorbance at 260 nm. The cDNA probes were indirectly labeled using reverse transcription in the presence of amino allyl dUTP (GE Healthcare). Six micrograms of total RNA was mixed with 0.75 μg of random hexamers (Invitrogen) and 0.75 μg of high-GC-content (70%) random hexamers and made up to 13.3 μl with diethyl pyrocarbonate-treated water. The RNA was denatured at 70°C for 10 min and cooled on ice for 5 min. The following were then added to the RNA sample: 3 mM (final concentration) (each) dATP, dCTP, and dGTP; 1.2 mM dTTP; 1.8 mM amino allyl dUTP (Ambion); 0.01 mM dithiothreitol; 40 U cloned RNase inhibitor (Takara); 6 μl of 5× reverse transcriptase (RT) buffer; and 380 U Superscript II RT (Invitrogen). After the mixture had been incubated at 42°C for 2 h for cDNA synthesis, the RNA template was hydrolyzed by adding 10 μl of 1.0 M NaOH and 10 μl of 0.5 M EDTA and incubated at 65°C for 30 min to degrade the RNA. The sample was then neutralized with 25 μl of 1 M HEPES (pH 7.5), purified using a Microcon YM30 column (Amicon), and dried in a vacuum. Either Cy3 or Cy5 dye was coupled to the amino allyl dUTP in the cDNA in the presence of 4.5 μl of 0.1 M sodium bicarbonate (pH 9.0) for 1 h. The labeled probe was purified using the QIAquick PCR purification system (Qiagen) and concentrated using a Microcon YM30 column.
Microarray hybridization was carried out at 42°C for 18 h by mixing using 120 μl per slide of SlideHyb no. 1 hybridization solution (Ambion) in an automated hybridization machine, GeneTac HybStation (Genomic Solutions). The posthybridization washing consisted of three cycles of 20-s incubations with each of the following solutions: 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% sodium dodecyl sulfate (medium stringency) at 42°C, 0.1× SSC plus 0.05% sodium dodecyl sulfate (high stringency) at 25°C, and 0.1× SSC (low stringency) at 25°C. The slides were washed, dried by centrifugation at 350 × g for 5 min at room temperature, and scanned using a model 428 array scanner (Affymetrix). The spot intensities were quantified using Imagene 6.1 (BioDiscovery). RNA samples isolated from four independent cultures of each strain were used for cDNA synthesis. Two cDNA samples of each strain were labeled with Cy5, and the other two were labeled with Cy3. We performed duplicate competitive hybridization experiments using Cy3-labeled wt cDNA plus Cy5-labeled ΔadpA mutant cDNA or Cy5-labeled wt cDNA plus Cy3-labeled ΔadpA mutant cDNA. Equal amounts of Cy3- and Cy5-labeled cDNA were used in all hybridizations. The expression ratios (wt strain to ΔadpA mutant) were normalized by the LIMMA (linear model for microarray analysis) loess (subgrid) method using ArrayPipe 2.0. Normalized expression ratios were calculated for each gene and tested for significance (expression ratio of either >2.0 or <0.5 and a P of <0.05). We designed two or more specific oligonucleotides for each of the 5,643 ORFs and one oligonucleotide for each of the 1,495 ORFs of small size. When an ORF had two or more oligonucleotides, distinctive change values were obtained for the ORF due to the difference in signal intensity caused by the distinct hybridization efficiencies of the oligonucleotides. Among them, the change value with the lowest P value in a statistical analysis (t test) was employed as the most reliable one.
Quantitative RT-PCR.
For validation of the microarray data, transcripts from 17 genes that encoded known or putative regulatory proteins were quantified using RT-PCR. Primers for each gene (Table 1) were designed for use with the Sybr green method. As an internal standard, we used hrdB(SGR1701), which encodes a principal sigma factor of RNA polymerase, because this gene is presumably transcribed similarly throughout growth in the wt and ΔadpA strains. The cDNA was synthesized using the ThermoScript RT-PCR system with random hexamers according to the manufacturer's instructions (Invitrogen). All reactions were performed in the Takara Sybr Premix Ex Taq II (Takara) reaction mixture using a SmartCycler II real-time PCR system (Cepheid) under the following conditions: 10 s at 95°C, followed by 50 cycles of 5 s at 95°C for denaturing and 30 s at 60°C for annealing and extension. All reactions were performed in triplicate, and the data were normalized using the average for the internal standard.
TABLE 1.
Primers used for quantitative RT-PCR
| Primer | Positionsa | Sequence (5′ to 3′) |
|---|---|---|
| SGR1701 hrdB F | 1,182 to 1,203 | GAAGGTCATCGAGGTCCAGAAG |
| SGR1701 hrdB R | 1,506 to 1,488 | GTGGCGGAGCTTCGACATC |
| SGR2064 F | 195 to 213 | CGGCATGATCCACCAACTG |
| SGR2064 R | 669 to 649 | TTCGGAGTACGAGGTCTTCTG |
| SGR2271 sigH F | 1 to 17 | GTGAGCGACGGGAACGG |
| SGR2271 sigH R | 303 to 285 | CATCCGCACCAGTCTGTTG |
| SGR2274 F | 83 to 103 | TGCAGATCGAGGAAGCCAAGG |
| SGR2274 R | 257 to 241 | CAGGCGCTTGCGTTGCG |
| SGR2378 F | 256 to 276 | CGCATCACGACCAATCTCTTC |
| SGR2378 R | 575 to 558 | CGGTGCTTGAGCGCCTTG |
| SGR2760 F | 4 to 23 | GCAGATTTCTCCCGCCTTCC |
| SGR2760 R | 275 to 259 | CTGGCCCGGCCCATCAG |
| SGR3271 F | 50 to 69 | GTATGGATCAGGCGGCGAAG |
| SGR3271 R | 203 to 186 | GCGGACGGCAGGATCATG |
| SGR3340 F | 21 to 39 | GAGCCTCGATTCGGGAGAG |
| SGR3340 R | 194 to 178 | GCATCGGCCAGGCACTC |
| SGR3555 F | 106 to 122 | TCCGCCGCCGTACTGAC |
| SGR3555 R | 470 to 454 | CTGCACTCGCCGCTGTG |
| SGR3905 atrA F | 259 to 275 | CTGGTGCGCCGGATAGC |
| SGR3905 atrA R | 625 to 607 | CCACGACCTCCAACAGCTC |
| SGR4240 griR F | 215 to 233 | GCCTCCGAAGACTCCTCTC |
| SGR4240 griR R | 599 to 583 | AGGCACAGCGACTCCTG |
| SGR4276 hrdD F | 675 to 693 | CCCGGTCAGCCTGAACATG |
| SGR4276 hrdD R | 982 to 964 | CCGTGTCGTGAGCCATTCG |
| SGR5049 F | 685 to 702 | CAGCCGTCCTTCTTCTGC |
| SGR5049 R | 993 to 976 | GGTCAGTGTGTGCAGGAC |
| SGR5931 strR F | 86 to 106 | AATTATCCGCCGTGACAATGG |
| SGR5931 strR R | 422 to 404 | GGATGGGTCTCCAGGACAC |
| SGR6369 F | 473 to 489 | GCGTGGAGGCGGAGTTC |
| SGR6369 R | 942 to 924 | CACCACCCAGCGGATCTTG |
| SGR6380 F | 170 to 188 | TCGCCTGCCTCGACAAGTC |
| SGR6380 R | 469 to 453 | GTGCCGAGCGCCTCCTG |
| SGR6385 F | 345 to 365 | CCAATGGGAGCTGAACGAAGG |
| SGR6385 R | 680 to 659 | ATGCCGTAGAGGTTCTGGTAGG |
Nucleotide positions are relative to the first nucleotide of the start codon, which is 1.
Accession numbers.
The nucleotide sequence has been deposited in the DDBJ DNA database under accession no. AP009493. The fully annotated genomic sequence is also available at http://park.itc.u-tokyo.ac.jp/hakko/genome. Details of the microarray design, transcriptome experimental design, and transcriptome data have been deposited in the NCBI Gene Expression Omnibus under accession no. GSE9882.
RESULTS AND DISCUSSION
General features of the S. griseus genome.
The complete genome sequence indicated a single linear chromosome composed of 8,545,929 bp with no plasmids. The main features of the S. griseus IFO 13350 chromosome are summarized in Table 2 and Fig. 1. Table 2 also includes those of the chromosomes of S. coelicolor A3(2) and S. avermitilis for comparison. The S. griseus chromosome contains at least 7,138 ORFs (Fig. 1, line ii). A total of 4,464 ORFs (62.6%) were assigned to known or putative functions, and 2,674 ORFs (37.4%) were annotated as genes for hypothetical proteins. The chromosome also contains six rRNA operons (16S-23S-5S) (Fig. 1, line vi) and 66 tRNA genes (42 species) (Fig. 1, line v). The average G+C content of the chromosome is 72.2%, but the approximately 300-kb regions (including the 133-kb TIR sequence) at both ends show a remarkably lower G+C content (Fig. 1, line vii). The replication origin, oriC (positions 4,324,631 to 4,325,203), which contains 19 DnaA box-like sequences (22), was predicted to be located in the middle of the chromosome (52 kb away from the center toward the right end). A GC-skewed inversion observed between the left and right arms of the chromosome supports this prediction (Fig. 1, line viii).
TABLE 2.
General features of the chromosomes of three Streptomyces speciesc
| Species | Length (bp) | TIR (bp) | Avg G+C content (%) | No. of protein-coding genes | Avg CDS length (bp) | Coding density (%) | No. of rRNA (16S-23S-5S) operons | No. of tRNA genes | No. of tmRNAs | No. of 4.5S RNAs |
|---|---|---|---|---|---|---|---|---|---|---|
| S. griseus | 8,545,929 | 132,910 | 72.2 | 7,138 | 1,055 | 88.1 | 6 | 66 | 1 | 1 |
| S. coelicolor A3(2)a | 8,667,507 | 21,653 | 72.1 | 7,825 | 991 | 88.9 | 6 | 63 | 1 | 1 |
| S. avermitilisb | 9,025,608 | 49 | 70.7 | 7,583 | 1,027 | 86.3 | 6 | 68 | 1 | 1 |
Data reported by Bentley et al. (3).
Up-to-date annotation data are shown.
tmRNAs, transfer-messenger RNAs; CDS, protein coding sequence. The topology of the chromosome each species is linear.
Telomere structure of the S. griseus chromosome.
Since we could not clone the terminal fragments carrying covalently bound Tpgs by the glass bead binding method (19), we determined the telomere sequence of the S. griseus chromosome by a newly devised method. Briefly, either poly(A) or poly(T) was attached to the naked 3′ end of the chromosome using terminal deoxynucleotidyl transferase, and the terminal DNA fragment was amplified by PCR and then sequenced. Only one guanine was found between the poly(A) [or poly(T)] sequence and the tentative S. griseus chromosomal end that had been determined using whole-genome shotgun sequencing. Thus, we concluded that the additional guanine was the true 3′ end. We also confirmed that specific amplification of the terminal sequence of the S. coelicolor A3(2) chromosome was achieved by this method (data not shown). Our new method for the determination of the telomere sequence of a given linear chromosome is very convenient once the sequence near the chromosomal end is available.
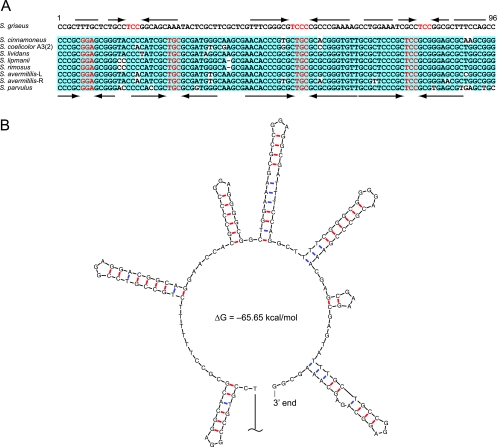
The telomere sequence of the S. griseus IFO 13350 chromosome shows no similarity to the typical telomere sequences conserved among many linear chromosomes and some linear plasmids of Streptomyces (Fig. 2A). The telomere has several palindromes whose loop sequences are 5′-GGA-3′ (Fig. 2B), whereas typical Streptomyces telomeres carry 5′-GCA-3′ loops in their palindromes (19). The positions of the palindromes also do not correspond to those of palindromes in other Streptomyces telomeres. S. griseus seems not to produce any conserved Tpg or Tap because the chromosome has pseudogenes for them (SGR6986 and SGR6987, respectively). These pseudogenes have apparently undergone rearrangements and mutations. SGR6986, which once encoded a Tpg protein conserved in typical Streptomyces telomeres, is disrupted by a 73-bp deletion. SGR6987, with its upstream region, was assumed to be a ruin of the conserved tac gene; several mutations in both the 5′ and the 3′ portions of SGR6987, including deletions and frameshift mutations, are found. Recently, a novel pair of Tpg and Tap proteins for the S. coelicolor A3(2) linear plasmid SCP1, the telomere of which is also unique, was identified to be encoded by the plasmid (18). The diversity of telomere palindromic sequences among Streptomyces linear plasmids was also shown (46). Therefore, we assumed that S. griseus acquired the unique telomere together with a novel pair of Tpg and Tap proteins from a certain linear plasmid during its evolution and that the original tpg and tap genes became unnecessary and decayed.
FIG. 2.
Telomere of S. griseus. (A) Comparisons of the S. griseus telomere sequence with the typical telomere sequences of seven other Streptomyces species. Conserved nucleotides among eight telomeres of the seven Streptomyces species are highlighted in light blue. Palindromes are indicated by converging arrows. Nucleotides in the possible loop of the palindromes are colored red. L and R, left and right TIRs, respectively. (B) Secondary structure of the 3′-terminal 164 nucleotides of the S. griseus chromosome predicted by the Mfold program.
We expected that a novel pair of Tpg and Tap proteins must be encoded as an operon near the chromosomal ends. In fact, we found two copies of candidates for such an operon in both TIRs (SGR98t to SGR97t and SGR7041t to SGR7042t). We found that SGR97t (SGR7042t), a 185-amino-acid (aa) protein showing very low similarity (18% identity) to conserved Tpgs, was attached to the chromosome ends and that SGR98t (SGR7041t), an 837-aa protein with a C-terminal DnaB-like-helicase domain, was able to bind specifically to the single-stranded DNA of the chromosomal end (H. Suzuki, K. Marushima, Y. Ohnishi, and S. Horinouchi, submitted for publication). We therefore propose that SGR97t (SGR7042t) and SGR98t (SGR7041t) code for functional Tpg and Tap, respectively, in S. griseus.
The telomere of S. griseus IFO 13350 was also shown to be different from the unique telomere of S. griseus 2247 (14). The length of the TIRs of S. griseus 2247 (24 kb) is much shorter than that of the TIRs of S. griseus IFO 13350 (133 kb). We also found that the DraI physical map of S. griseus IFO 13350 (Fig. 1, line i) was somewhat different from that of S. griseus 2247 (27), implying the heterogeneity of species S. griseus.
Comparative analysis of the S. griseus genome against the genomes of S. coelicolor A3(2) and S. avermitilis.
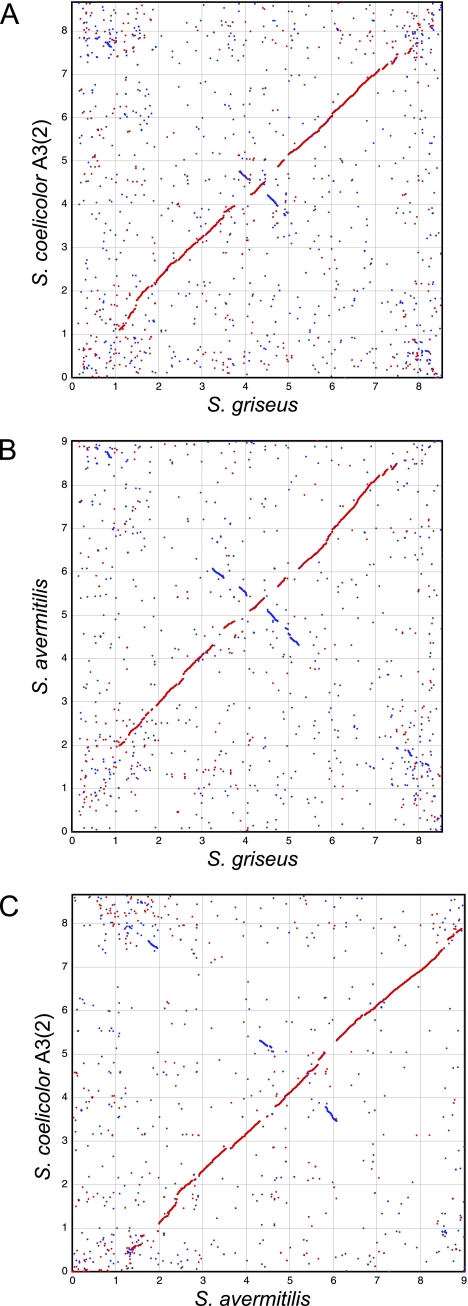
We compared all annotated proteins on the chromosomes of three Streptomyces species, namely, S. griseus, S. coelicolor A3(2), and S. avermitilis (SGR, SCO, and SAV, respectively, are used in their locus tags). Ortholog (reciprocal best-hit pair) plots using pairwise BLASTP searches clearly demonstrated the existence of a highly conserved internal core region of each chromosome, and several large inversions centered at oriC were found (Fig. 3). Comparison of the results from the three chromosomes predicted the core regions to be SGR923 to SGR6311 (6.39 Mb) (Fig. 1, line i), SCO6804 to SCO1209 (6.28 Mb), and SAV1625 to SAV7128 (6.50 Mb). The subtelomeric regions were less conserved among these three chromosomes with regard to sequence and ortholog distribution. Variability in the subtelomeric regions was also found in the Streptomyces ambofaciens chromosome (7). Thus, a conserved internal core region of approximately 6.4 Mb, with a couple of variable subtelomeric regions of 1 to 2 Mb, seems to be a common feature of Streptomyces linear chromosomes.
FIG. 3.
Ortholog plots for S. griseus versus S. coelicolor A3(2) (A), S. griseus versus S. avermitilis (B), and S. avermitilis versus S. coelicolor A3(2) (C). Dots represent reciprocal best matches (by BLAST comparison) between orthologs. Matches on the same strand are in red, and those on the opposite strand are in blue.
We clustered S. griseus, S. coelicolor A3(2), and S. avermitilis proteins using the BLASTCLUST program with a threshold of 60% identity plus 70% length coverage (Table 3). Despite these strict conditions, 3,204 S. griseus proteins (45% of the total proteins), 3,280 S. coelicolor A3(2) proteins (42%), and 3,240 S. avermitilis proteins (43%) were classified into 3,039 clusters that are commonly present among these three species. Because these three species are taxonomically distant from one another within the genus Streptomyces (see Fig. S1 in the supplemental material), it is probable that most of these proteins are also common in any other Streptomyces species. Thus, 45% of the proteins encoded on the S. griseus chromosome are likely to be “common Streptomyces proteins” that have essential or important functions in the biology of Streptomyces. We then searched for Streptomyces-specific proteins among the 3,204 S. griseus proteins. These proteins were compared with the proteins of nine other actinobacteria, i.e., Nocardia farcinica IFM 10152 (21), Mycobacterium tuberculosis H37Rv (8), Corynebacterium glutamicum ATCC 13032 (24), Rhodococcus jostii RHA1 (30), Propionibacterium acnes KPA 171202 (5), Frankia alni ACN14a (32), Thermobifida fusca YX (28), Saccharopolyspora erythraea NRRL23338 (35), and Salinispora tropica CNB-440 (39), using the BLASTCLUST program with a threshold of 30% identity plus 50% length coverage. In total, 240 S. griseus proteins were not clustered with any of the proteins in these nine actinobacteria. We carried out BLAST analyses of each of these 240 S. griseus proteins and selected 24 S. griseus proteins (≥60 aa) that were highly conserved among only the three Streptomyces species (≥75% identity); their significant homologs were never found in any other organisms (Table 4). The functions of all of these 24 S. griseus proteins and their S. coelicolor A3(2) and S. avermitilis homologs are not yet known. However, their high-level conservation and their unique distribution among the Streptomyces species suggest that these proteins have some important functions in the nature of the genus Streptomyces.
TABLE 3.
Comparative analysis of ORFs encoded on the chromosomes of three Streptomyces species
| Distribution classa | No. of proteins classified byb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BLASTP with threshold of indicated E value
|
BLASTCLUSTc
|
|||||||||
| <0.1
|
<10−20
|
|||||||||
| SGR | SCO | SAV | SGR | SCO | SAV | Clustersd | SGR | SCO | SAV | |
| SGR(+) SCO(+) SAV(+) | 6,586 | 7,203 | 7,004 | 4,944 | 5,191 | 5,111 | 3,039 | 3,204 | 3,280 | 3,240 |
| SGR(+) SCO(+) SAV(−) | 180 | 190 | 455 | 454 | 469 | 487 | 493 | |||
| SGR(+) SCO(−) SAV(+) | 153 | 173 | 319 | 304 | 302 | 310 | 307 | |||
| SGR(−) SCO(+) SAV(+) | 196 | 253 | 672 | 685 | 941 | 976 | 967 | |||
| SGR(+) SCO(−) SAV(−) | 219 | 1,420 | 2,998 | 3,137 | ||||||
| SGR(−) SCO(+) SAV(−) | 234 | 1,506 | 2,973 | 3,074 | ||||||
| SGR(−) SCO(−) SAV(+) | 153 | 1,483 | 2,992 | 3,069 | ||||||
+ and - indicate the presence and the absence of ORFs, respectively.
A total of 7,138 S. griseus, 7,823 S. coelicolor A3(2), and 7,583 S. avermitilis proteins are classified.
Proteins were clustered by the BLASTCLUST program with a threshold of 60% identity plus 70% length coverage.
The number of clusters, not proteins, is shown.
TABLE 4.
Twenty-four S. griseus proteins selected based on BLAST analyses
| S. griseus protein (aa) |
S. coelicolor A3(2)
|
S. avermitilis
|
Other hit (non-Streptomyces protein)
|
|||
|---|---|---|---|---|---|---|
| Protein (aa) | E value | Protein (aa) | E value | Source (aa) | E value | |
| SGR1707 (212) | SCO5814 (204) | 1.1e−83 | SAV2451 (211) | 2.4e−86 | Roseiflexus castenholzii (111) | 8.9e−01 |
| SGR2042 (244) | SCO5475 (265) | 1.3e−111 | SAV2770 (268) | 4.7e−114 | Alcanivorax borkumensis (982) | 2.2e−02 |
| SGR3064 (148) | SCO4402 (166) | 1.1e−62 | SAV3847 (166) | 1.0e−68 | Aspergillus terreus (419) | 4.0e−01 |
| SGR3164 (69) | SCO4335 (62) | 2.5e−22 | SAV3896 (60) | 1.6e−21 | Pyrobaculum arsenaticum (157) | 2.0e+00 |
| SGR3380 (216) | SCO3624 (221) | 5.4e−102 | SAV4548 (216) | 2.6e−104 | Gloeobacter violaceus (223) | 7.5e−03 |
| SGR3428 (106) | SCO3662 (111) | 8.0e−45 | SAV4512 (111) | 1.5e−46 | Roseovarius nubinhibens (350) | 6.2e−05 |
| SGR3657 (70) | SCO3924 (70) | 1.3e−26 | SAV4269 (70) | 1.1e−27 | Saccharopolyspora erythraea (492) | 1.8e−01 |
| SGR3786 (155) | SCO3799 (156) | 1.1e−72 | SAV4391 (154) | 9.7e−72 | Chlamydomonas reinhardtii (1216) | 9.0e−01 |
| SGR3920 (102) | SCO4131 (102) | 6.0e−45 | SAV4098 (102) | 5.8e−48 | Pseudomonas aeruginosa (667) | 9.1e−01 |
| SGR4040 (165) | SCO4269 (166) | 2.5e−79 | SAV3953 (166) | 3.4e−81 | Aspergillus clavatus (664) | 3.9e+00 |
| SGR4191 (145) | SCO4467 (145) | 4.1e−78 | SAV4793 (145) | 4.6e−77 | Lactobacillus plantarum (603) | 2.0e+00 |
| SGR4388 (325) | SCO3116 (335) | 6.6e−158 | SAV3557 (328) | 1.4e−163 | Clostridium thermocellum (566) | 9.7e−08 |
| SGR4514 (215) | SCO3022 (204) | 1.1e−94 | SAV5054 (204) | 1.0e−100 | Nocardia farcinica (699) | 2.4e−01 |
| SGR4720 (113) | SCO2820 (115) | 2.8e−42 | SAV5240 (121) | 7.3e−43 | Stigmatella aurantiaca (431) | 9.0e−01 |
| SGR4722 (539) | SCO2809 (547) | 0.0e+00 | SAV5242 (531) | 0.0e+00 | Rhodobacter sphaeroides (651) | 3.3e−02 |
| SGR4723 (335) | SCO2808 (342) | 1.2e−130 | SAV5243 (346) | 5.0e−132 | Psychromonas ingrahamii (920) | 1.2e−05 |
| SGR4972 (77) | SCO2574 (77) | 1.7e−39 | SAV5484 (77) | 1.7e−39 | Aedes aegypti (887) | 2.7e+00 |
| SGR5248 (60) | SCO2261 (85) | 1.4e−14 | SAV5936 (60) | 5.2e−17 | Pseudomonas stutzeri (97) | 7.6e+00 |
| SGR5318 (66) | SCO2195 (71) | 7.0e−17 | SAV6008 (65) | 1.8e−20 | Salinispora tropica (77) | 1.6e+00 |
| SGR5784 (87) | SCO1721 (89) | 4.7e−29 | SAV6584 (89) | 1.7e−31 | Mus musculus (3291) | 1.6e+00 |
| SGR6060 (187) | SCO1474 (183) | 2.9e−73 | SAV6876 (180) | 7.8e−71 | Rhodococcus jostii RHA1 (193) | 4.0e−06 |
| SGR6119 (472) | SCO1413 (472) | 0.0e+00 | SAV6933 (472) | 0.0e+00 | Anaeromyxobacter dehalogenans (690) | 3.2e−03 |
| SGR6277 (75) | SCO1249 (75) | 4.3e−22 | SAV7086 (79) | 5.2e−28 | Prochlorococcus marinus (227) | 2.1e+00 |
| SGR6836 (166) | SCO7249 (165) | 2.0e−81 | SAV1239 (165) | 4.2e−79 | Aeromicrobium erythreum (3574) | 2.7e−01 |
We also found 941 clusters that are present in both S. coelicolor A3(2) and S. avermitilis but absent in S. griseus by the BLASTCLUST program with a threshold of 60% identity plus 70% length coverage (Table 3). S. griseus lacks the whiE gene cluster (SCO5320 to SCO5214, SAV2836 to SAV2841) that is involved in the biosynthesis of an aromatic-polyketide spore pigment (10). Consistent with this, a type III polyketide synthetase (PKS), RppA (13), and a cytochrome P-450 monooxygenase, P-450 mel (12), are involved in the biosynthesis of a different spore pigment (hexahydroxyperylenequinone [HPQ] melanin) in S. griseus. Concerning developmental genes, the bldK genes (SCO5112 to SCO5116) (31), which encode a peptide transporter involved in morphological development in S. coelicolor A3(2), are also present in S. avermitilis (two copies, SAV3176 to SAV3172 and SAV3150 to SAV3154) but not in S. griseus. S. griseus contains at least six putative peptide transporters. The lack of the BldK transporter might be compensated for by one of these transporters in S. griseus. Alternatively, the BldK system involved in morphological development by means of a putative extracellular signaling peptide may not be used by S. griseus. Putative gene clusters for gas vesicle synthesis are duplicated in S. coelicolor A3(2) (SCO0649 to SCO0658 and SCO6499 to SCO6508) and triplicated in S. avermitilis (SAV599 to SAV591, SAV1890 to SAV1881, and SAV2356 to SAV2347) but are never found in S. griseus. Although the role of gas vesicle proteins in the complex biology of Streptomyces has been discussed (40, 42), these proteins do not seem to be essential in Streptomyces.
We also compared S. griseus, S. coelicolor A3(2), and S. avermitilis proteins using the BLASTP program with two different thresholds (E values, <0.1 and <10−20) (Table 3). With the relatively strict conditions under which the E value was <10−20, 1,420 S. griseus, 1,506 S. coelicolor A3(2), and 1,483 S. avermitilis proteins did not show sufficient similarity to any proteins of the other two strains. The number of strain-specific proteins greatly decreased under the relaxed condition where the E value was <0.1; 219 S. griseus, 234 S. coelicolor A3(2), and 153 S. avermitilis proteins are classified as strain-specific proteins. In S. griseus, these include biosynthesis proteins for some specific secondary metabolites, such as streptomycin (SGR5918, SGR5933, SGR5935, and SGR5936), a putative enediyne compound (SGR604 and SGR606), and a putative lantibiotic compound (SGR4418), in addition to many hypothetical proteins. A putative restriction enzyme (a SacI isoschizomer, SGR3359) was also found in these strain-specific proteins. Just upstream of the SacI isoschizomer gene, a putative cytosine-specific DNA methylase (SGR3358) is encoded. It is likely that the SGR3358 protein has SacI-methylase activity because DNA extracted from S. griseus IFO 13350 is resistant to SacI digestion. Interestingly, a transmissible-plasmid-like segment is integrated just upstream of SGR3358. S. griseus may acquire this restriction/modification system by virtue of the transmissible integrating plasmid.
Gene clusters for secondary metabolites.
The biosynthetic gene cluster for streptomycin has been analyzed previously (11). We found that the cluster (str and sts, SGR5914 to SGR5940) is located 560 kb interior to the right end of the conserved core region. We previously analyzed three other gene clusters for secondary metabolites, specifically, grixazone (gri, SGR4238 to SGR4250) (15), a carotenoid (crt, SGR6824 to SGR6830) (26), and the HPQ melanin (P-450mel-rppA, SGR6619 to SGR6620) (12, 13). Genomic sequencing revealed a further 30 gene clusters or genes for putative secondary metabolites (Table 5). Of the total 34 clusters, 18 occur outside the conserved core region of the chromosome (Fig. 1, line iv), supporting the idea that the variable subtelomeric regions of linear chromosomes contribute to the metabolic diversity of Streptomyces (3, 20).
TABLE 5.
Secondary metabolite-biosynthetic gene clusters (or genes) of S. griseus IFO 13350
| Gene clustera | Product and/or type |
|---|---|
| Known | |
| SGR5914-SGR5940 | Streptomycin (aminoglycoside) |
| SGR4238-SGR4250 | Grixazone (phenoxazinone) |
| SGR6824-SGR6830 | Isorenieratene (carotenoid) |
| SGR6619-SGR6620 | HPQ melanin (by type III PKS) |
| SGR470-SGR472 | Alkylresorcinol (by type III PKS) |
| SGR1268-SGR1269 | 2-Methylisoborneol (by terpene cyclase) |
| Predicted | |
| Terpene | |
| SGR54t-SGR60t | Carotenoid |
| SGR7085t-SGR7079t | Carotenoid |
| SGR962-SGR966 | Hopanoid |
| SGR2079 | Terpene |
| SGR6065 | Terpene |
| SGR6839 | Germacradienol/geosmin |
| PKS and NRPS | |
| SGR443-SGR455 | NRPS for siderophore |
| SGR574-SGR593 | NRPS |
| SGR653-SGR656 | NRPS |
| SGR895-SGR901 | NRPS |
| SGR2586-SGR2598 | NRPS |
| SGR6709-SGR6717 | NRPS for siderophore |
| SGR6730-SGR6742 | NRPS for siderophore |
| SGR6071-SGR6083 | Type I PKS |
| SGR6177-SGR6183 | Type I PKS |
| SGR6360-SGR6387 | Type I PKS |
| SGR604-SGR611 | Enediyne PKS |
| SGR278-SGR283 | PKS-NRPS hybrid |
| SGR810-SGR815 | PKS-NRPS hybrid |
| SGR2482-SGR2489 | Type I PKS, NRPS |
| SGR6776-SGR6786 | Type I PKS, PKS-NRPS hybrid |
| SGR3239-SGR3288 | Type II PKS, NRPS |
| Others | |
| SGR4747-SGR4751 | Nocardamine (siderophore) |
| SGR527-SGR528 | Melanin |
| SGR2446-SGR2447 | Melanin |
| SGR3845-SGR3849 | Lantibiotic |
| SGR4408-SGR4421 | Lantibiotic |
| SGR5285-SGR5295 | Unknown |
The range of clusters for unknown compounds was predicted on the basis of possible gene function, putative transcriptional units, synteny, and/or our transcriptome data.
In addition to the crt cluster described above, another crt cluster (SGR54t to SGR60t or SGR7079t to SGR7085t) is found in the TIR sequence. Thus, S. griseus has three copies of crt clusters on the chromosome. However, S. griseus is unable to produce carotenoids under any conditions thus far examined (26; H. Ikeda, unpublished data), representing an example of gene clusters for secondary metabolism that are cryptic. SGR962, encoding a putative squalene-hopene cyclase, seems to be a member of the hopanoid biosynthesis gene cluster (hopABCDE, SGR962 to SGR966), which is also found in S. coelicolor A3(2) (37) and S. avermitilis (36). Four putative terpene cyclases containing a metal binding motif (SGR1269, SGR2079, SGR6065, and SGR6839) are found in the S. griseus genome. According to the homology with the germacradienol/geosmin synthases SCO6073 (23) and SAV2163 (6), SGR6839 seems to be involved in the biosynthesis of germacradienol/geosmin; germacradienol/geosmin production by S. griseus has already been confirmed (H. Ikeda, unpublished data). Recently, SGR1269 has been identified as a 2-methylisoborneol synthase (25a). SGR2079 and SGR6065 seem to be sesquiterpene cyclases (25a), but their products remain to be elucidated. S. griseus has no mevalonate pathway, which sometimes occurs in a flanking region of a biosynthesis gene cluster for terpenoids such as furaquinocin A and terpentecin (9). S. griseus has a type III PKS (SGR472) other than RppA. SGR472 has been demonstrated to be an alkylresorcinol synthase (13a). Like S. coelicolor A3(2) and S. avermitilis, S. griseus has putative melanin-biosynthetic gene clusters (mel, SGR527 to SGR528 and SGR2446 to SGR2447) and a gene cluster (SGR4747 to SGR4751) for nocardamine (desferrioxamine E). Nocardamine production by S. griseus has been confirmed (44).
Similar to other actinomycete strains, S. griseus has many gene clusters that contain a putative PKS, nonribosomal peptide synthetase (NRPS), and PKS-NRPS hybrid genes. An NRPS gene cluster (SGR6709 to SGR6717) is homologous with SCO499 to SCO491 of S. coelicolor A3(2). The gene organization of an enediyne gene cluster (SGR604 to SGR611) is similar to that of many enediyne gene clusters found in several actinomycete strains (45). The other 14 PKS and/or NRPS gene clusters seem to be specific to S. griseus. Therefore, these clusters probably direct the synthesis of some novel compounds. Three NRPS gene clusters (SGR443 to SGR455, SGR6709 to SGR6717, and SGR6730 to SGR6742) appear to be involved in the biosynthesis of siderophores because they contain some genes for a siderophore-dependent iron transport system. The presence of IdeR (iron-dependent transcription regulator) box-like sequences (29) in the upstream regions of many genes within these three gene clusters (data not shown) also supports this idea.
Two putative gene clusters (SGR3845 to SGR3849 and SGR4408 to SGR4421) for lantibiotics are found in S. griseus. SGR3848 and SGR4418 seem to be lantibiotic precursors because these small proteins contain many Cys, Thr, and Ser residues in their C-terminal regions, most of which could be modified to produce lanthionine and methyllanthionine bridges (43). We annotated SGR5285 to SGR5295, containing a putative 5-aminolevulinic acid synthase gene (SGR5295), as a gene cluster for an unknown secondary metabolite. A similar gene cluster (CV0801 to CV0808) is found in Chromobacterium violaceum (4).
Genes possibly under the control of A-factor.
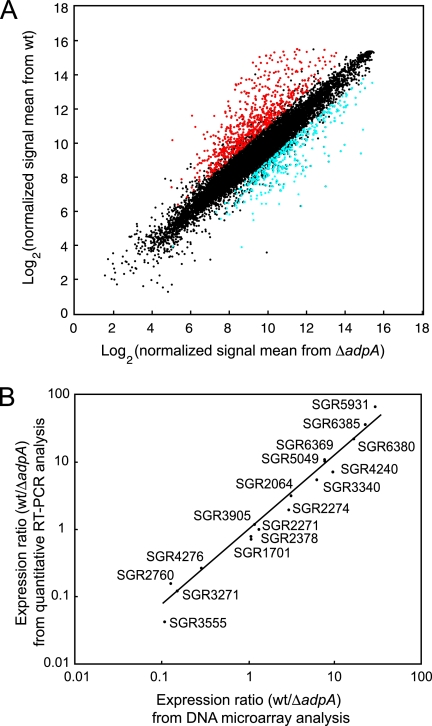
In S. griseus, A-factor induces morphological development and secondary metabolite formation (16, 17). We have revealed the A-factor regulatory cascade in which AdpA functions as a central transcriptional regulator that has multiple target genes. Although we have already identified 14 AdpA target genes, many other genes are expected to be targets of AdpA. Furthermore, the transcription of a much greater number of genes seems to be activated or repressed indirectly by AdpA. For the first step in analyzing global gene regulation by A-factor in S. griseus, we compared the transcriptomes of the wt and ΔadpA mutant strains. The total RNA of both strains was extracted from the cells in the early stationary phase in liquid culture because AdpA begins to trigger the transcription of target genes in the middle of the exponential growth phase. To evaluate the accuracy of our DNA microarray analysis, we randomly selected 17 regulator genes and analyzed their transcription using quantitative RT-PCR. There was a good correlation between the ratios measured using the DNA microarray and quantitative RT-PCR analyses (Fig. 4B). Therefore, we regarded genes with an increase in expression of more than twofold and a P value of <0.05 when analyzed with a t test (n = 4) as genes with significant variation in transcriptional level. When this threshold was used, 639 and 373 genes were transcriptionally upregulated and downregulated, respectively, in the wt strain, in comparison with the ΔadpA mutant (Fig. 4A). These genes are dispersed over the whole chromosome (Fig. 1, line iii). This result indicates that the disruption of adpA had a great influence on the transcription of many genes in S. griseus, as expected. Of 14 known AdpA target genes, six genes (strR [change, 29.3-fold increase], sgiA [14.2-fold], orf1-AdBS2 [SGR6559] [4.19-fold], sprD [3.77-fold], adsA [3.52-fold], and orfA-AdBS3 [SGR4617] [2.57-fold]) were detected to be upregulated. A similar DNA microarray analysis using RNA from cells grown on a solid medium indicated that all of the 14 known AdpA target genes were upregulated in the wt strain (details will be published elsewhere).
FIG. 4.
DNA microarray analysis of S. griseus gene expression. (A) Scatter plot for DNA microarray analysis of S. griseus gene expression affected by the deletion of the adpA gene. The scatter plot shows the averages of normalized signal means from the wt strain and those from the ΔadpA mutant (n = 4). The red dots indicate genes that were >2-fold upregulated (P < 0.05), and the blue dots indicate genes that were >2-fold downregulated (P < 0.05) in the wt strain. (B) Comparison of results obtained by DNA microarray and quantitative RT-PCR analyses. The ratios of gene expression (wt strain to ΔadpA mutant) are plotted. The results obtained by the two methods show a high level of correlation (r2 = 0.93).
In conventional nutrient-rich liquid medium, at least four secondary metabolite gene clusters (SGR443 to SGR455 [NRPS], SGR574 to SGR593 [NRPS], SGR6360 to SGR6387 [type I PKS], and SGR6709 to SGR6717 [NRPS]), as well as the streptomycin-biosynthetic gene cluster, were upregulated in the wt strain (Fig. 1, lines iii and iv), indicating that these four gene clusters were activated directly or indirectly by AdpA. A-factor probably induces the production of some peptide and polyketide compounds that are synthesized by these gene clusters. In contrast, one large gene cluster (SGR3239 to SGR3288 [type II PKS and NRPS]) was downregulated in the wt strain (Fig. 1, lines iii and iv). To confirm the apparent repression of this gene cluster by AdpA, further experiments are required. We are now analyzing the changes in the transcriptome triggered by the exogenous addition of A-factor to an A-factor-deficient mutant using the same DNA microarray. The overall picture of genome-wide gene regulation by A-factor will be determined through a combination of several DNA microarray analyses.
Concluding remarks.
A striking feature of the S. griseus genome is the unique telomere sequence. The complete genome sequence revealed the absence of conserved Tpg and Tap in their functional forms and allowed us to find a novel pair of Tpg and Tap proteins encoded in the TIRs. Further analysis of the novel pair of Tpg and Tap proteins will provide insights into the evolution of the linear chromosome of Streptomyces. Comparisons of the S. griseus genome with the genomes of S. coelicolor A3(2) and S. avermitilis clearly showed a common feature of the Streptomyces linear chromosome—a conserved internal core region of approximately 6.4 Mb with a couple of variable subtelomeric regions of 1 to 2 Mb. It also allowed us to find 24 Streptomyces-specific proteins. S. griseus has at least 34 gene clusters (or genes) for the production of known or unknown secondary metabolites, although a considerable number of these gene clusters (or genes) are presumably transcriptionally inactive under the conventional culture conditions. Novel secondary metabolic enzymes or pathways can be found through studies of these gene clusters (or genes); the SGR470-to-SGR472 and SGR1268-to-SGR1269 proteins have already been demonstrated to be responsible for the biosynthesis of alkylresorcinol and 2-methylisoborneol, respectively. We constructed a DNA microarray that allowed us to analyze the transcriptome of S. griseus. For the first step of the analysis of global gene regulation by A-factor, we compared the transcriptomes of the wt strain and the ΔadpA mutant. The transcription of approximately 1,000 genes was affected by adpA disruption, demonstrating a large effect of A-factor in the biology of S. griseus. Importantly, we found that four secondary metabolite gene clusters for unknown peptide and polyketide compounds were activated by A-factor. Thus, the genome sequence will greatly facilitate future studies of the molecular genetics of S. griseus.
Supplementary Material
Acknowledgments
This research was supported by a Grant-in-Aid for Scientific Research on Priority Area “Applied Genomics” (S.H.) and a Grant-in-Aid for Scientific Research on Priority Area “Comprehensive Genomics” (M.H.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank K. Furuya, C. Yoshino, H. Inaba, K. Motomura, and Y. Hattori (University of Tokyo) and A. Tamura (Kitasato University) for technical assistance in the genome sequencing.
Footnotes
Published ahead of print on 28 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 151518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, K., and S. N. Cohen. 2003. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 17774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 4.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. USA 10011660-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brüggemann, H., A. Henne, F. Hoster, H. Liesegang, A. Wiezer, A. Strittmatter, S. Hujer, P. Dürre, and G. Gottschalk. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305671-673. [DOI] [PubMed] [Google Scholar]
- 6.Cane, D. E., X. He, S. Kobayashi, S. Ômura, and H. Ikeda. 2006. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J. Antibiot. 59471-479. [DOI] [PubMed] [Google Scholar]
- 7.Choulet, F., B. Aigle, A. Gallois, S. Mangenot, C. Gerbaud, C. Truong, F.-X. Francou, C. Fourrier, M. Guérineau, B. Decaris, V. Barbe, J.-L. Pernodet, and P. Leblond. 2006. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol. Biol. Evol. 232361-2369. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 9.Dairi, T. 2005. Studies on biosynthetic genes and enzymes of isoprenoids produced by actinomycetes. J. Antibiot. 58227-243. [DOI] [PubMed] [Google Scholar]
- 10.Davis, N. K., and K. F. Chater. 1990. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol. Microbiol. 41679-1691. [DOI] [PubMed] [Google Scholar]
- 11.Distler, J., K. Mansouri, G. Mayer, M. Stockmann, and W. Piepersberg. 1992. Streptomycin biosynthesis and its regulation in streptomycetes. Gene 115105-111. [DOI] [PubMed] [Google Scholar]
- 12.Funa, N., M. Funabashi, Y. Ohnishi, and S. Horinouchi. 2005. Biosynthesis of hexahydroxyperylenequinone melanin via oxidative aryl coupling by cytochrome P-450 in Streptomyces griseus. J. Bacteriol. 1878149-8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funa, N., Y. Ohnishi, I. Fujii, M. Shibuya, Y. Ebizuka, and S. Horinouchi. 1999. A new pathway for polyketide synthesis in microorganisms. Nature 400897-899. [DOI] [PubMed] [Google Scholar]
- 13a.Funabashi, M., N. Funa, and S. Horinouchi. Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem., in press. [DOI] [PubMed]
- 14.Goshi, K., T. Uchida, A. Lezhava, M. Yamasaki, K. Hiratsu, H. Shinkawa, and H. Kinashi. 2002. Cloning and analysis of the telomere and terminal inverted repeat of the linear chromosome of Streptomyces griseus. J. Bacteriol. 1843411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higashi, T., Y. Iwasaki, Y. Ohnishi, and S. Horinouchi. 2007. A-factor and phosphate depletion signals are transmitted to the grixazone biosynthesis genes via the pathway-specific transcriptional activator GriR. J. Bacteriol. 1893515-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horinouchi, S. 2007. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem. 71283-299. [DOI] [PubMed] [Google Scholar]
- 17.Horinouchi, S., and T. Beppu. 2007. Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B 83277-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, C.-H., H.-H. Tsai, Y.-G. Tsay, Y.-N. Chien, S.-L. Wang, M.-Y. Cheng, C.-H. Ke, and C. W. Chen. 2007. The telomere system of the Streptomyces linear plasmid SCP1 represents a novel class. Mol. Microbiol. 631710-1718. [DOI] [PubMed] [Google Scholar]
- 19.Huang, C.-H., Y.-S. Lin, Y.-L. Yang, S.-W. Huang, and C. W. Chen. 1998. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 28905-916. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Ômura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21526-531. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 10114925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakimowicz, D., J. Majka, W. Messer, C. Speck, M. Fernandez, M. C. Martin, J. Sanchez, F. Schauwecker, U. Keller, H. Schrempf, and J. Zakrzewska-Czerwińska. 1998. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology 1441281-1290. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, J., X. He, and D. E. Cane. 2007. Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat. Chem. Biol. 3711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegräbe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 1045-25. [DOI] [PubMed] [Google Scholar]
- 25.Khokhlov, A. S., I. I. Tovarova, L. N. Borisova, S. A. Pliner, L. N. Shevchenko, E. Y. Kornitskaia, N. S. Ivkina, and I. A. Rapoport. 1967. The A-factor, responsible for streptomycin biosynthesis by mutant strains of Actinomyces streptomycini. Dokl. Akad. Nauk SSSR 177232-235. [PubMed] [Google Scholar]
- 25a.Komatsu, M., M. Tsuda, S. Ômura, H. Oikawa, and H. Ikeda. 2008. 2-Methylisoborneol: identification and functional analysis of genes controlling biosynthesis. Proc. Natl. Acad. Sci. USA (in press). [DOI] [PMC free article] [PubMed]
- 26.Lee, H.-S., Y. Ohnishi, and S. Horinouchi. 2001. A σB-like factor responsible for carotenoid biosynthesis in Streptomyces griseus. J. Mol. Microbiol. Biotechnol. 395-101. [PubMed] [Google Scholar]
- 27.Lezhava, A., T. Mizukami, T. Kajitani, D. Kameoka, M. Redenbach, H. Shinkawa, O. Nimi, and H. Kinashi. 1995. Physical map of the linear chromosome of Streptomyces griseus. J. Bacteriol. 1776492-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lykidis, A., K. Mavromatis, N. Ivanova, I. Anderson, M. Land, G. DiBartolo, M. Martinez, A. Lapidus, S. Lucas, A. Copeland, P. Richardson, D. B. Wilson, and N. Kyrpides. 2007. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 1892477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manabe, Y. C., C. L. Hatem, A. K. Kesavan, J. Durack, and J. R. Murphy. 2005. Both Corynebacterium diphtheriae DtxR(E175K) and Mycobacterium tuberculosis IdeR(D177K) are dominant positive repressors of IdeR-regulated genes in M. tuberculosis. Infect. Immun. 735988-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLeod, M. P., R. L. Warren, W. W. L. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. M. Jones, R. Holt, F. S. L. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 10315582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22881-893. [DOI] [PubMed] [Google Scholar]
- 32.Normand, P., P. Lapierre, L. S. Tisa, J. P. Gogarten, N. Alloisio, E. Bagnarol, C. A. Bassi, A. M. Berry, D. M. Bickhart, N. Choisne, A. Couloux, B. Cournoyer, S. Cruveiller, V. Daubin, N. Demange, M. P. Francino, E. Goltsman, Y. Huang, O. R. Kopp, L. Labarre, A. Lapidus, C. Lavire, J. Marechal, M. Martinez, J. E. Mastronunzio, B. C. Mullin, J. Niemann, P. Pujic, T. Rawnsley, Z. Rouy, C. Schenowitz, A. Sellstedt, F. Tavares, J. P. Tomkins, D. Vallenet, C. Valverde, L. G. Wall, Y. Wang, C. Medigue, and D. R. Benson. 2007. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohnishi, Y., H. Yamazaki, J.-Y. Kato, A. Tomono, and S. Horinouchi. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69431-439. [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34102-111. [DOI] [PubMed] [Google Scholar]
- 35.Oliynyk, M., M. Samborskyy, J. B. Lester, T. Mironenko, N. Scott, S. Dickens, S. F. Haydock, and P. F. Leadlay. 2007. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 25447-453. [DOI] [PubMed] [Google Scholar]
- 36.Ômura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 9812215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poralla, K., G. Muth, and T. Härtner. 2000. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 18993-95. [DOI] [PubMed] [Google Scholar]
- 38.Shirling, E. B., and D. Gottlieb. 1972. Cooperative description of type strains of Streptomyces V. Additional descriptions. Int. J. Syst. Bacteriol. 22268-394. [Google Scholar]
- 39.Udwary, D. W., L. Zeigler, R. N. Asolkar, V. Singan, A. Lapidus, W. Fenical, P. R. Jensen, and B. S. Moore. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA 10410376-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Keulen, G., D. A. Hopwood, L. Dijkhuizen, and R. G. Sawers. 2005. Gas vesicles in actinomycetes: old buoys in novel habitats? Trends Microbiol. 13350-354. [DOI] [PubMed] [Google Scholar]
- 41.Waksman, S. A. 1953. Streptomycin: background, isolation, properties, and utilization. Science 118259-266. [DOI] [PubMed] [Google Scholar]
- 42.Walsby, A. E., and P. G. Dunton. 2006. Gas vesicles in actinomycetes? Trends Microbiol. 1499-100. [DOI] [PubMed] [Google Scholar]
- 43.Willey, J. M., and W. A. van der Donk. 2007. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61477-501. [DOI] [PubMed] [Google Scholar]
- 44.Yamanaka, K., H. Oikawa, H.-O. Ogawa, K. Hosono, F. Shinmachi, H. Takano, S. Sakuda, T. Beppu, and K. Ueda. 2005. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 1512899-2905. [DOI] [PubMed] [Google Scholar]
- 45.Zazopoulos, E., K. Huang, A. Staffa, W. Liu, B. O. Bachmann, K. Nonaka, J. Ahlert, J. S. Thorson, B. Shen, and C. M. Farnet. 2003. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat. Biotechnol. 21187-190. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, R., Y. Yang, P. Fang, C. Jiang, L. Xu, Y. Zhu, M. Shen, H. Xia, J. Zhao, T. Chen, and Z. Qin. 2006. Diversity of telomere palindromic sequences and replication genes among Streptomyces linear plasmids. Appl. Environ. Microbiol. 725728-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.