Abstract
Pseudomonas fluorescens F113 is motile by means of type b flagella. Analysis of the region encoding the synthesis of the flagellar filament has shown a transcriptional organization different from that of type a flagella. Additionally to the promoters driving fliC, fliD, and fleQ expression, we have found promoters upstream of the flaG gene and the fliST operon. These promoters were functional in vivo. Both promoters have been mapped and appear to be dependent on the vegetative sigma factor and independent of FleQ, the master regulator of flagellum synthesis.
Pseudomonads are ubiquitous bacteria that can adapt to different lifestyles, with species that are either saprophytic or pathogenic for plants and animals. Motility is an important trait for pseudomonads, and it has been shown to be involved in rhizosphere colonization (7, 13, 14, 18), biofilm formation (9, 20, 27), and pathogenesis in plant (10, 22) and animal (4) models. Flagellar biosynthesis in pseudomonads is regulated in a hierarchical way, with the transcriptional activator fleQ on top of the regulatory cascade (2, 8). Besides, the alternative sigma factors encoded by fliA and rpoN are required for flagellum assembly (25, 26). The regulation of flagellar biosynthesis has been analyzed using DNA microarrays and Pseudomonas aeruginosa PAK (12), which contains type a flagella (24). Other pseudomonads, like Pseudomonas fluorescens F113 or P. aeruginosa PAO1, contain type b flagella and are characterized by different syntenies in the region encoding the formation of the flagellar filament (7, 12, 24). In type b strains, this region contains fliC, encoding type b flagellin; flaG, encoding a protein of unknown function implicated in filament length (7); fliD, encoding the flagellar-cap protein (3); fliS, encoding a FliC chaperone required for flagellin export and assembly (7); fliT, encoding a protein of unknown function required for full motility and rhizosphere colonization in P. fluorescens F113 (7); fleQ (2, 8), encoding the master regulatory protein; and fleSR, encoding a two-component system required for flagellar biosynthesis (23).
Analysis of the transcriptional organization of a region implicated in the synthesis of the flagellar filament in P. fluorescens F113.
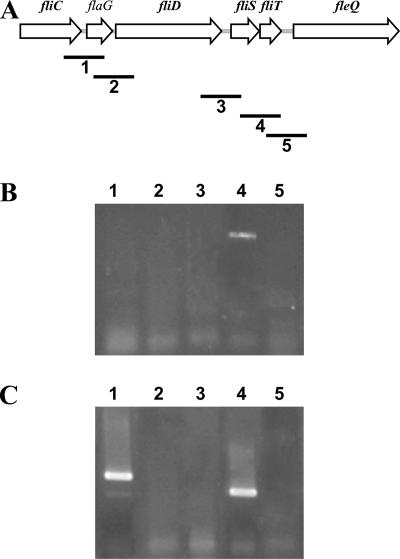
In order to establish the transcriptional organization of the fliC-fleQ region of Pseudomonas fluorescens F113, the upstream regions of the fliC, flaG, fliS, fliT, and fleQ genes were amplified and cloned into the reporter vector pMP220 to form transcriptional fusions with a promoterless lacZ gene. These constructs were introduced into strain F113 by triparental mating, and β-galactosidase activity was tested (19). Substantial activity was shown for the regions upstream of all genes (125 to 350 Miller units), except the region upstream of the fliT gene (<5 Miller units) (see Fig. S1 in the supplemental material). Previous studies of other pseudomonads have shown the existence of promoters upstream of the fliC, fliD, and fleQ genes. To our knowledge, no promoters have been found upstream of the flaG or fliS gene. These results indicated a monocistronic organization for the fliC, flaG, and fliD operons and the existence of a bicistronic fliST operon. To confirm this organization, primers for the coamplification of adjacent genes in this region were designed (Fig. 1A) from the genomic sequences of strain F113 and P. aeruginosa PAO1. PCR products of the expected sizes were obtained for all primer pairs from genomic DNA from strains F113 and PAO1 (data not shown). Total RNA from F113 and PAO1 was subjected to reverse transcription (RT)-PCR using these primers. In the case of F113 (Fig. 1B), only the fliS-fliT primer pair yielded an RT-PCR product, confirming the transcriptional organization defined by lacZ fusions. However, in the case of P. aeruginosa PAO1 (Fig. 1C), RT-PCR products were obtained with the fliC-flaG and fliS-fliT pairs, indicating that in P. aeruginosa, fliC, flaG, and fliST are transcribed as bicistronic operons.
FIG. 1.
RT-PCR determination of putative promoters in the flagellar-synthesis region. (A) Physical map of the region in type b pseudomonads. Numbered bars indicate the expected fragments amplified with each primer pair. Each fragment partially spans two adjacent genes. (B) Results from RT-PCR amplification from P. fluorescens F113 RNA. (C) Results from RT-PCR amplification from P. aeruginosa PAO1 RNA. DNA controls from both strains yielded the expected amplified fragments from all the primer pairs. Total RNA was extracted according to the instructions of the manufacturer (Roche Boehringer Mannheim) from P. fluorescens strains grown at 28°C in LB medium (5). RT-PCRs were carried out by using a one-step RT-PCR kit from Amersham Biosciences according to manufacturer's specified running conditions and by using a melting temperature of 59°C. Primers used are indicated in Table S1 in the supplemental material. The presence of DNA contamination was discarded by running a PCR under the conditions indicated above for the RNA samples. Strains, plasmids, and primers used are described in Table S1 in the supplemental material.
Analysis of the flaG and fliST promoters of Pseudomonas fluorescens F113.
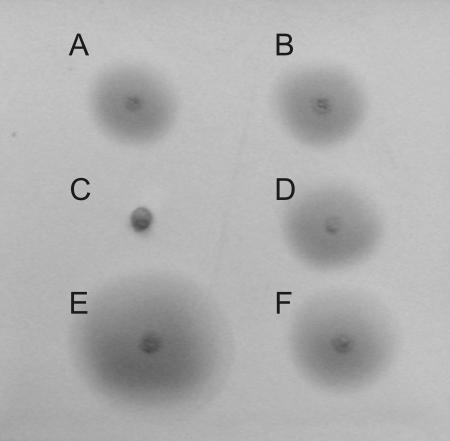
An analysis of the transcriptional organization of the flagellum synthesis region of P. fluorescens F113 revealed the presence of two previously unknown promoters controlling the transcription of the flaG gene and the fliST operon. In order to confirm the functionality in vivo of these two promoters, we used a complementation assay of flaG and fliS F113 mutants. Genomic DNA fragments containing each of the flaG and fliS genes with their putative promoter regions (as identified in the lacZ fusion assays) were cloned into the pFAJ1700 vector, which contains transcriptional terminators flanking both sides of the insertion site. Figure 2 shows the results of the complementation assay. Introduction of a cloned fliS gene under the control of its putative promoter restores wild-type motility to the nonmotile fliS mutant (7), demonstrating in vivo the functionality of the fliS upstream region as a promoter. Similarly, the introduction of a cloned flaG gene under the control of its own putative promoter reverts the hypermotility phenotype of a flaG mutant (7) to the wild-type level of motility, demonstrating that the upstream region of the flaG gene also contains a functional promoter.
FIG. 2.
In vivo determination of flaG and fliST promoters in P. fluorescens F113. The flaG and fliS genes with their upstream regions were cloned into the pFAJ1700 vector, which contains transcriptional terminators flanking both sides of the insertion site. The constructs were used to complement the swimming motility phenotypes of flaG and fliS mutants, after introduction of the constructs by triparental mating. Swimming assays of organisms on LB medium were performed as previously described (7). (A) F113; (B) F113(pFAJ1700); (C) F113 fliS mutant; (D) F113 fliS mutant (pfliS); (E) F113 flaG mutant; (F) F113 flaG mutant (pflaG). Strains, plasmids, and primers used are described in Table S1 in the supplemental material.
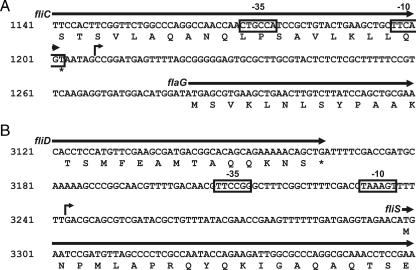
In order to analyze the structure of the newly described promoters, rapid amplification of 5′ cDNA ends (5′RACE) was performed. As shown in Fig. 3A, a single transcription start site was detected for the flaG transcript in the intergenic fliC-flaG region, 75 bp upstream of the translation start codon. Upstream of this transcriptional start site, a putative region containing −10 and −35 sites was observed. This region lies within the fliC coding sequence. Figure 3B shows the fliST promoter region. 5′RACE detected a single transcription start site in the fliD-fliS intergenic region, 55 bp upstream of the fliS translational start codon. A putative −10/−35 region was also observed for this promoter. The existence of these boxes, together with the lack of putative fliA and rpoN boxes, suggests that these two promoters use the housekeeping σ70 factor for their expression.
FIG. 3.
Physical map and sequences of the flaG and fliST promoters in P. fluorescens F113 as determined by 5′RACE. 5′RACE was performed using the RACE second-generation kit (Roche Applied Science). Primers used in each amplification are specified in Table S1 in the supplemental material. cDNA containing the transcription initiation site was subcloned into pDRIVE (Qiagen), and at least three clones were sequenced for the mapping of each promoter using universal primers. Putative −10 and −35 regions are boxed. Arrows indicate the determined transcription start site. The numbering corresponds to the sequence AF399739, deposited in GenBank. Strains, plasmids, and primers used are described in Table S1 in the supplemental material.
Regulatory analysis of the flaG and fliST operons in Pseudomonas fluorescens.
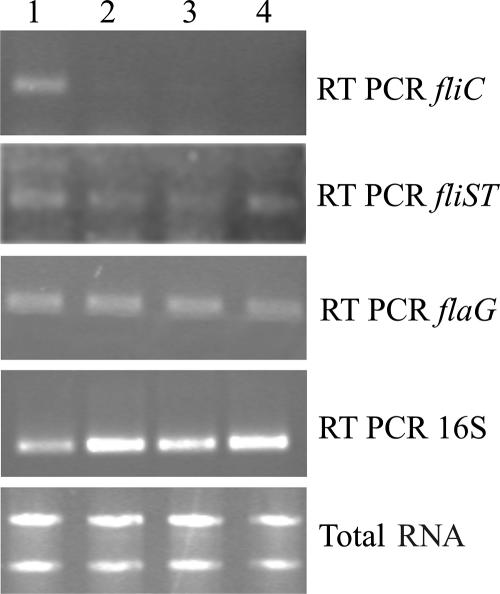
To analyze the regulation of the two operons, we tested their expression in different genetic backgrounds by RT-PCR. Expression was analyzed in the wild-type strain and isogenic strains containing mutations in the master regulatory gene fleQ and in the genes encoding the alternative sigma factors required for flagellar biosynthesis, fliA and rpoN. The expression of the fliC gene, encoding flagellin, was also monitored in the same backgrounds. As shown in Fig. 4, fliC expression is strictly dependent on fleQ, fliA, and rpoN. The requirement of these genes for fliC expression has already been shown for P. aeruginosa (12, 25, 26). Conversely, the expression of flaG and fliST was observed in all backgrounds, indicating that the expression of these genes does not require FleQ or the alternative sigma factor. This result and the presence of putative −10 and −35 boxes in the promoter regions strongly indicate that these promoters are dependent on the vegetative sigma factor. However, some effect of these regulatory proteins on their transcriptional regulation cannot be excluded. Since fleQ is also σ70 dependent in P. aeruginosa (11), it is likely that in P. fluorescens, flaG and fliST are at the same hierarchical level as fleQ. Previous studies of P. aeruginosa PAK, a type a strain, indicated that fliC and flaG (fleL) were cotranscribed (12). Here we have shown that this is also true for the b-type strain P. aeruginosa PAO1 but not for P. fluorescens F113. The lack of amplification by RT-PCR of a band spanning both genes and the in vivo complementation of a flaG mutant by a DNA fragment containing the flaG promoter clearly show that in P. fluorescens F113, flaG expression is totally independent of fliC expression. Similarly, in P. aeruginosa PAK14, fliS and fliT (fleP) seemed to be cotranscribed from a FleQ-dependent promoter upstream of fliD (12). Here, we present evidence that this is not the case in b-type strains. The results presented here show that flagellar biosynthesis is regulated differently in different Pseudomonas strains and even in strains containing the same type of flagella. Furthermore, genes implicated in important functions of flagellar synthesis, such as the regulation of flagellar length and flagellin secretion, are outside the regulatory four-tiered circuit and appear to depend on housekeeping regulation. The regulation of flagellar genes outside the typical hierarchy is uncommon but has been observed for other bacteria. In Salmonella enterica serovar Typhimurium, expression of the flk gene, encoding a regulator of flagellar-gene expression (1), was shown to be independent of the master regulatory factors FlhDC and did not require the σ28 factor (16). In Caulobacter crescentus, an operon containing the flgBC and fliE genes has been shown to be regulated outside of the flagellar regulatory hierarchy (6). Finally, in several spirochetes, such as Borrelia burgdorferi (15), Leptospira borgpetersenii (17), and Spirochaeta aurantia (21), important genes implicated in flagellum synthesis and encoding flagellins and hook-associated proteins require a vegetative σ70 factor.
FIG. 4.
Expression analysis of the flaG and fliST promoters. Total RNA (10 ng) from F113 (lane 1), the F113 fleQ mutant (lane 2), the F113 fliA mutant (lane 3), and the F113 rpoN mutant (lane 4) were subjected to RT-PCR with primers to amplify the flaG and fliST fragments. 16S RNA RT-PCR was used as a control. RT-PCR conditions were as described for Fig. 1. The strains, plasmids, and primers used are described in Table S1 in the supplemental material.
Supplementary Material
Acknowledgments
This work was funded by Spanish Ministry of Science and Education grant BIO2006-08596 and by the Research Program MICROAMBIENTE-CM from Comunidad de Madrid. M. Redondo-Nieto is the recipient of a Juan de la Cierva contract from the Spanish Ministry of Science and Education. Ana Navazo is the recipient of an FPI fellowship from the Spanish Ministry of Science and Education. Emma Barahona is the recipient of a predoctoral contract from Comunidad de Madrid.
Footnotes
Published ahead of print on 28 March 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aldridge, P., J. E. Karlinsey, E. Becker, F. F. Chevance, and K. T. Hughes. 2006. Flk prevents premature secretion of the anti-sigma factor FlgM into the periplasm. Mol. Microbiol. 60630-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1795574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 661000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora, S. K., A. N. Neely, B. Blair, S. Lory, and R. Ramphal. 2005. Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 734395-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani, G. 1951. Studies on lysogenesis. 1. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, C. H., and J. W. Gober. 2001. Temporal regulation of genes encoding the flagellar proximal rod in Caulobacter crescentus. J. Bacteriol. 183725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capdevila, S., F. M. Martínez-Granero, M. Sánchez-Contreras, R. Rivilla, and M. Martín. 2004. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 1503889-3897. [DOI] [PubMed] [Google Scholar]
- 8.Casaz, P., A. Happel, J. Keithan, D. L. Read, S. R. Strain, and S. B. Levy. 2001. The Pseudomonas fluorescens transcription activator AdnA is required for adhesion and motility. Microbiology 147355-361. [DOI] [PubMed] [Google Scholar]
- 9.Choy, W. K., L. Zhou, C. K. C. Syn, L. H. Zhang, and S. Swarup. 2004. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol. 1867221-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czelleng, A., Z. Bozso, P. G. Ott, E. Besenyei, G. J. Varga, A. Szatmari, L. Kiraly, and Z. Klement. 2006. Identification of virulence-associated genes of Pseudomonas viridiflava activated during infection by use of a novel IVET promoter probing plasmid. Curr. Microbiol. 52282-286. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta, N., E. P. Ferrell, K. J. Kanack, S. E. H. West, and R. Ramphal. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 1845240-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 13.de Weert, S., L. C. Dekkers, I. Kuiper, G. V. Bloemberg, and B. J. J. Lugtenberg. 2004. Generation of enhanced competitive root-tip-colonizing Pseudomonas bacteria through accelerated evolution. J. Bacteriol. 1863153-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Weger, L. A., C. I. M. van der Vlugt, A. H. M. Wijfjes, P. A. H. M. Bakker, B. Schippers, and B. Lugtenberg. 1987. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 1692769-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge, Y., I. G. Old, I. S. Girons, and N. W. Charon. 1997. The flgK motility operon of Borrelia burgdorferi is initiated by a sigma 70-like promoter. Microbiology 1431681-1690. [DOI] [PubMed] [Google Scholar]
- 16.Karlinsey, J. E., A. J. Pease, M. E. Winkler, J. L. Bailey, and K. T. Hughes. 1997. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J. Bacteriol. 1792389-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, M., H. Dan, and Y. Li. 2004. Identification of a second flagellin gene and functional characterization of a sigma70-like promoter upstream of a Leptospira borgpetersenii flaB gene. Curr. Microbiol. 48145-152. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-Granero, F., R. Rivilla, and M. Martin. 2006. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 723429-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 21.Parales, J., Jr., and E. P. Greenberg. 1993. Analysis of the Spirochaeta aurantia flaA gene and transcript. FEMS Microbiol. Lett. 106245-251. [DOI] [PubMed] [Google Scholar]
- 22.Quiñones, B., G. Dulla, and S. E. Lindow. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant-Microbe Interact. 18682-693. [DOI] [PubMed] [Google Scholar]
- 23.Ritchings, B. W., E. C. Almira, S. Lory, and R. Ramphal. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 634868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spangenberg, C., T. Heuer, C. Burger, and B. Tummler. 1996. Genetic diversity of flagellins of Pseudomonas aeruginosa. FEBS Lett. 396213-217. [DOI] [PubMed] [Google Scholar]
- 25.Starnbach, M. N., and S. Lory. 1992. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6459-469. [DOI] [PubMed] [Google Scholar]
- 26.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toutain, C. M., N. C. Caizza, M. E. Zegans, and G. A. O'Toole. 2007. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res. Microbiol. 158471-477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.