Abstract
Despite shortcomings, cultures of blood and sterile sites remain the “gold standard” for diagnosing systemic candidiasis. Alternative diagnostic markers, including antibody detection, have been developed, but none are widely accepted. In this study, we used an enzyme-linked immunosorbent assay to measure serum antibody responses against 15 recombinant Candida albicans antigens among 60 patients with systemic candidiasis due to various Candida spp. and 24 uninfected controls. Mean immunoglobulin G (IgG) responses against all 15 antigens were significantly higher among patients with systemic candidiasis than among controls, whereas IgM responses were higher against only seven antigens. Using discriminant analysis that included IgG responses against the 15 antigens, we derived a mathematical prediction model that identified patients with systemic candidiasis with an error rate of 3.7%, a sensitivity of 96.6%, and a specificity of 95.6%. Furthermore, a prediction model using a subset of four antigens (SET1, ENO1, PGK1-2, and MUC1-2) identified through backward elimination and canonical correlation analyses performed as accurately as the full panel. Using the simplified model, we predicted systemic candidiasis in a separate test sample of 32 patients and controls with 100% sensitivity and 87.5% specificity. We also demonstrated that IgG titers against each of the four antigens included in the prediction model were significantly higher in convalescent-phase sera than in paired acute-phase sera. Taken together, our findings suggest that IgG responses against a panel of candidal antigens might represent an accurate and early marker of systemic candidiasis, a hypothesis that should be tested in future trials.
Candida spp. are major causes of systemic infections among hospitalized patients in the developed world. Candidemia, for example, is the fourth most common bloodstream infection in the United States and is associated with attributable mortality as high as 40 to 50% despite treatment with antifungal agents (4, 9). The management of systemic candidiasis is complicated by the limitations of current diagnostic tests. Blood cultures, generally taken as the “gold standard” for the diagnosis of candidemia, are negative in up to 50% of autopsy-proven cases of disseminated candidiasis (1). Moreover, blood cultures often become positive late in the course of disease (5, 18), which delays appropriate therapy. Such delays in the institution of an antifungal agent are associated with significantly increased mortality rates among patients with candidemia (8, 17). Cultures of deep-tissue sites are limited by uncertainties over sensitivity and the necessity for invasive sampling procedures. Because of these shortcomings, there is great interest in non-culture-based diagnostic tests.
Investigators have assessed a wide range of potential diagnostic markers including the detection of candidal nucleic acids, metabolites such as d-arabinitol, cell wall components such as β-d-glucan, and antigens such as secreted aspartyl proteinase (SAP), enolase (ENO1), and mannan (5). Despite reports of reasonable diagnostic yields for each of these tests, none have been broadly validated or accepted in widespread clinical practice. There has been less enthusiasm for antibody detection as a diagnostic strategy because of concerns regarding false-negative tests in immunocompromised patients (26). Nevertheless, recent studies detecting antibodies against SAP (19, 20), ENO1 (13, 16), mannan (30, 33, 36), a 52-kDa metalloprotein (6), hyphal wall protein 1 (HWP1) (14), and a Candida albicans germ tube antigen (CGTA) (26) reported sensitivities and specificities that are consistent with those of other diagnostic markers, even among highly immunocompromised hosts like stem cell transplant and liver transplant recipients (11, 12, 21, 31, 34). Moreover, various combinations of an antibody test with an antigen test have been shown to be superior to either test alone in diagnosing systemic candidiasis (23, 28, 29). Clearly, much work in developing diagnostic markers remains to be done; antibody detection strategies merit exploration as part of these endeavors.
In ongoing work in our laboratories, we have used a human antibody-based screening strategy to identify C. albicans genes that encode immunogenic proteins including previously uncharacterized virulence factors (2, 3, 22, 27). In the present project, we have chosen 12 proteins of diverse function and cellular localization to study as targets for antibody detection assays. These proteins are classified into four groups: classic cell wall proteins (Bgl2 and MUC1), glycolytic enzymes localized to the cell wall (ENO1, FBA1, GAP1, and PGK1), intracellular proteins localized to the cell wall (NOT5 and MET6), and intracellular proteins likely not localized to the cell wall (CAR1, RBT4, SET1, and IPF9162) (2, 15, 16, 24, 27, 33). Our objectives were to determine if serum antibody responses against any purified recombinant antigens could reliably distinguish patients with systemic candidiasis from uninfected controls. We also sought to derive a predictive model for systemic candidiasis that considered antibody responses against multiple antigens.
MATERIALS AND METHODS
Definitions.
Outcomes were classified as systemic candidiasis or controls. Systemic candidiasis was defined as the recovery of Candida spp. from blood or a sterile site. Controls were defined as patients hospitalized at the Shands Teaching Hospital at the University of Florida (UF) (STH-UF) who did not have any clinical or microbiological evidence of systemic Candida infection. Predictor variables were the antibodies against specific antigens.
Collection of sera.
Patients at STH-UF were identified on the day blood or sterile-site cultures were positive for Candida spp. Controls were identified by the Infectious Diseases Consultation Service at STH-UF. Sera were collected in accordance with procedures approved by the UF Institutional Review Board, frozen, and stored at −70°C in the repository at the UF Mycology Research Unit. For patients with candidiasis, sera were obtained from the earliest possible date on or after the date that the first positive cultures were drawn. In all cases, this was within 7 days of the first positive culture (acute-phase sera). For eight patients with candidiasis, sera were also recovered 4 to 12 weeks after the date on which the first positive cultures were drawn (convalescent-phase sera).
Enzyme-linked immunosorbent assay (ELISA).
Antibody titers were evaluated for a set of 12 proteins that were identified using in vivo-induced antigen technology (MET6, SET1, GAP1, ENO1, NOT5, BGL2, FBA1, MUC1, CAR1, RBT4, IPF9162, and PGK1) (2, 3, 27). Whole or partial DNA sequences of the genes encoding the proteins were amplified by PCR using the primers listed in Table 1. Two fragments of MET6, PGK1, and MUC1 were amplified, resulting in a total of 15 DNA sequences. The resulting PCR products were cloned into plasmid pET30 using an EK/LIC cloning kit (EMD Biosciences, Inc.). All inserts were confirmed by DNA sequencing. Each plasmid was transformed into Escherichia coli BL21(DE3) cells (Novagen). Expression of the recombinant antigens was induced by isopropyl-β-d-thiogalactopyranoside (IPTG). The recombinant antigens were purified from cell-free supernatants by chromatography on Ni2+-nitrilotriacetic acid-agarose as previously described (3). Briefly, E. coli cell pellets were resuspended in BugBuster (Novagene) with Benzonase nuclease and rLysozyme and incubated at room temperature for 30 min with gentle shaking. Samples were then separated into a soluble fraction or an insoluble pellet by centrifugation at 10,000 × g for 10 min. The location of the recombinant protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). If soluble, the supernatant fraction was filtered through a 0.45-μm filter (Millipore) and passed through a His-Bind column, followed by washing the column with binding buffer and then with wash buffer (both from EMD Biosciences, Inc.). The peptide of interest was then eluted with 60 to 100 mM imidazole buffer. The His6-tagged recombinant proteins were confirmed by 15% SDS-PAGE, which showed a single band of the expected size, and by Western blot analysis with anti-His monoclonal antibody (Invitrogen). The insoluble proteins were pelleted by centrifugation and then resuspended in 1:10-diluted BugBuster reagent (in deionized water). The suspension was centrifuged at 5,000 × g for 15 min at 4°C to collect the inclusion bodies. These inclusion bodies were then resuspended in 5 ml of 1:10-diluted BugBuster, which then underwent centrifugation as described above. The resultant pellet was again resuspended in diluted BugBuster and centrifuged at 16,000 × g for 15 min at 4°C. The final pellet of purified inclusion bodies was resuspended in 1× binding buffer including 6 M urea and incubated on ice for 1 h to completely dissolve the protein. The insoluble material was removed by centrifugation at 16,000 × g for 30 min; the purification process of the supernatant was performed as discussed above.
TABLE 1.
Primers used for cloning of antigens
| Antigen | Sense primer (5′→3′) | Antisense primer (3′→5′) | Length (amino acids) of antigen (positions) |
|---|---|---|---|
| MET6-1 | 5′-GACGACGACAAGATGGTTCAATCTTCCGTCTTAGGT | 5′-GAGGAGAAGCCCGGTTAAGAAGATTCGGATCTAGC | 400 (1-400) |
| MET6-2 | 5′-GACGACGACAAGATGATACCAACGATCCAAAG | 5′-GAGGAGAAGCCCGGTTAGTATTTAGCTCTGAATTC | 367 (401-768) |
| NOT5 | 5′-GACGACGACAAGATGACTCATGCACCAGCAGCAGTGT | 5′-GAGGAGAAGCCCGGTTAAACAATTCGAGAAATGGTATCAC | 71 (401-474) |
| SET1 | 5′-GACGACGACAAGATGTCATACAATAACAGAAGCGGA | 5′-GAGGAGAAGCCCGGTTATTGAGGTAATTGCTTATATGG | 208 (1-208) |
| RBT4 | 5′-GACGACGACAAGATGAAGTTTTCTCAAGTTGCCACTACTGCTGCTGCCATT | 5′-GAGGAGAAGCCCGGTAATAACACCAGAGTTCTGTAAAAGTCGGTA | 359 (entire gene) |
| IPF9162 | 5′-GACGACGACAAGATGAAAAAAAGGTTAGTTTTGTTTGATGATTCTGATGAT | 5′-GAGGAGAAGCCCGGTAATTTATCAATTTACATATAGTGCTCAAAATGGACCTGTCAA | 272 (entire gene) |
| CAR1 | 5′-GACGACGACAAGATGTCATCAATTCAATATAAATATCATCCAGACAA | 5′-GAGGAGAAGCCCGGTAATGTTATTTCAAACTGGGTTACGTGTAGAT | 318 (entire gene) |
| GAP1 | 5′-GACGACGACAAGATGGCTATTAAAATTGGTATTAAC | 5′-GAGGAGAAGCCCGGTTAAGCAGAAGCTTTAGCAAC | 336 (entire gene) |
| ENO1 | 5′-GACGACGACAAGATGTCTTACGCCACTAAAATCCAC | 5′-GAGGAGAAGCCCGGTTACAATTGAGAAGCCTTTTGGAA | 441 (entire gene) |
| BGL2 | 5′-GACGACGACAAGATGCAAATCAAATTCTTGACTACT | 5′-GAGGAGAAGCCCGGTTAGTTGAATTTACAGTCAATTGA | 309 (entire gene) |
| FBA1 | 5′-GACGACGACAAGATGGCTCCTCCAGCAGTTTTAAGT | 5′-GAGGAGAAGCCCGGTTACAATTGTCCTTTGGTGTGGAA | 360 (entire gene) |
| MUC1-1 | 5′-GACGACGACAAGATGTCATTTTGGGACAACAACAA | 5′-GAGGAGAAGCCCGGTTAGGTTGAGTTATTGGTTAAAA | 306 (1-306) |
| MUC1-2 | 5′-GACGACGACAAGATGGAGTATATCGCATCTTGGTGT | 5′-GAGGAGAAGCCCGGTTATTCATGTGGCATTGCTCGATA | 153 (735-888) |
| PGK1-1 | 5′-GACGACGACAAGATGTCATTATCTAACAAATTATCA | 5′-GAGGAGAAGCCCGGTTAACCACCACCAACAATC | 238 (1-238) |
| PGK1-2 | 5′-GACGACGACAAGATGGCCTTCACTTTCAAGAAA | 5′-GAGGAGAAGCCCGGTTAGTTTTTGTTGGAAAGAGC | 180 (239-438) |
Ninety-six-well flat-bottom microtiter plates were coated overnight at 4°C with purified recombinant antigens in carbonate buffer (pH 9.6) at a concentration of 0.5 μg per well (3). The plates were washed in phosphate-buffered saline with 0.1% Tween-20 (PBS-T), blocked with 0.25% gelatin in PBS-T for 1 h at 37°C, and washed again with PBS-T. Serially diluted serum specimens were added to each well, and the plates were incubated for 1 h at 37°C. The plates were washed, and peroxidase-conjugated goat anti-human immunoglobulin M (IgM) (1:5,000 dilution) or IgG (1:25,000 dilution) in PBS-T was added. After 1 h of incubation at 37°C, the plates were washed and developed with o-phenylenediamine in citrate buffer and 5% hydrogen peroxidase. The developing solution was stopped with 1 M phosphoric acid. The optical densities were determined using a spectrophotometer at 450 nm. Background was defined in wells coated with the protein to which the secondary antibody was added but the primary antibody was not. The reactive titer was defined as the inverse of the greatest dilution at which the optical density was twofold greater than background. All serum samples were tested in duplicate. In addition to wells lacking the primary antibody, wells that were not coated with antigen were further included as negative controls.
Statistical analyses.
The antibody titers for individual antigens were first log2 transformed to approximate normal distribution prior to data analysis. Means and standard errors for each predictor were calculated for each outcome variable. The differences in log2 antibody titers (IgM or IgG) were assessed using unpaired t tests with Welch correction; statistical significance was set at 0.05. The sensitivity and specificity were calculated for IgM or IgG against individual predictors. The optimal cutoff was determined by receiver operating characteristic analyses. Multicolinearity among the predictor variables was assessed using colinearity diagnostics in SAS PROC REG.
Potentially significant predictor variables for the discriminant model were identified by backward elimination analysis using the STEPDISC procedure and by canonical correlation analysis using the CANCORR procedure in SAS/STAT. In the backward elimination analysis, the predictor variables chosen to leave the model were based on the significance level of an F test from an analysis of covariance. In the canonical analysis, standardized canonical coefficients, which reflect the relative contribution of each predictor variable to the power of discriminating between the two outcomes, were generated; the variables with highest absolute values were included in the discriminant model.
The DISCRIM procedure in SAS/STAT was used to identify the smallest subset of predictor variables that best discriminated the two outcomes. The performance of this discriminant analysis was evaluated by estimating the error rate (probability of misclassification of outcome). Finally, linear regression analysis using PROC REG in SAS was performed to generate the predicted function for the best set of predictors that were identified. The prediction score takes the form of y = α + β1x1 + β2x2 + … + βnxn, where α is the constant where the regression line intercepts the y axis and β is a regression coefficient.
To compare IgG responses in acute and convalescent-phase sera from eight patients with candidiasis, the titers against the recombinant antigens of interest were log2 transformed, and means and standard errors were calculated. The differences in log2 antibody titers were assessed using the Student t test; statistical significance was set at 0.05.
RESULTS
Study population.
Between January 2004 and December 2006, we collected sera from 68 patients with systemic candidiasis, including 66 patients with candidemia and 2 patients with deep-seated candidiasis (one with Candida peritonitis related to chronic peritoneal dialysis and one with biopsy-proven Candida pneumonia). Patients were subclassified as newborn infants (<6 months old), immunocompromised hosts, and burn victims as well as by portal of entry. Descriptive data for the patients are provided in Table 2. The median time from the date of positive culture to serum collection was 2 days. Seven patients died, and four of the survivors had infections that persisted for over 2 weeks. We also collected sera from 24 hospitalized patients who had no evidence of candidiasis as controls.
TABLE 2.
Descriptive data of patients with systemic candidiasis
| Characteristic | Value |
|---|---|
| Median (range) age (yr) | 51.5 (3 days-81 yr) |
| No. of patients | |
| <6 mo old | 8 |
| Immunocompromised | 12a |
| Burn victims | 8 |
| With portal of entry of: | |
| Catheter | 33 |
| Abdominal | 21 |
| Wound | 6 |
| With complication of: | |
| Endocarditis | 2 |
| Mediastinitis | 2 |
| Vascular graft infection | 2 |
| With Candida sp. infection by: | |
| C. albicans | 28b |
| C. glabrata | 19 |
| C. parapsilosis | 10c |
| C. tropicalis | 5 |
| C. krusei | 4 |
| C. lusitaniae | 1 |
| Unspeciated | 1 |
Three bone marrow transplant recipients, five solid-organ transplant recipients, one recipient of both a bone marrow transplant and a solid-organ transplant, two patients with hematologic malignancies receiving chemotherapy, and one patient with systemic lupus on high-dose steroid.
Three patients were coinfected with C. glabrata, C. parapsilosis, and C. tropicalis; they are not included under these species.
One patient was coinfected with Candida guilliermondii.
Expression and purification of antigens.
We selected 12 proteins for inclusion in the study. SET1 and NOT5 were previously shown by our laboratories to contribute to virulence and to elicit higher antibody responses among patients with systemic candidiasis than among uninfected controls (2, 3, 27). Six proteins were identified by our laboratories as being immunogenic and were shown by others to elicit higher antibody responses among patients with systemic candidiasis than among uninfected controls (ENO1, BGL2, PGK1, GAP1, FBA1, and MET6). Finally, four proteins were found to be immunogenic exclusively by our laboratories (CAR1, MUC1, RBT4, and IPF11897). Fifteen antigens from the 12 proteins were expressed as His6-tagged polypeptides in E. coli cells and purified from cell-free supernatants by chromatography on Ni2+-nitrilotriacetic acid-agarose. Seven of the purified recombinant antigens were full-length proteins (Table 1). The remaining antigens could not be efficiently expressed as full-length proteins and were instead purified as smaller fragments. The purified antigens appeared as single bands of expected sizes by SDS-PAGE and were detected by probing with anti-His antibodies (data not shown).
Antibody responses against recombinant antigens in sera of patients with systemic candidiasis and uninfected controls.
Sera from the 68 patients with systemic candidiasis and the uninfected controls were tested against the 15 recombinant antigens using ELISA. Sera from two patients (one with systemic candidiasis and one control) were sufficient to test against only 13 antigens (all except PGK1-1 and PGK1-2). As anticipated, IgM and IgG titers against each of the recombinant antigens in sera of eight newborn infants were consistently indistinguishable from background; these sera were excluded from further data analysis.
IgM and IgG data for the 60 patients (excluding newborn infants) with systemic candidiasis and 24 controls are presented in Table 3. Overall, IgG responses were better than IgM responses in differentiating patients with systemic candidiasis from controls. The mean log2 IgG titers were significantly higher for patients with systemic candidiasis than for controls against all 15 antigens. The mean log2 IgM titers, on the other hand, were significantly higher for patients with systemic candidiasis against only seven antigens: ENO1 and PGK1-1 (glycolytic enzymes localized to the cell wall), NOT5 (an intracellular protein localized to the cell wall), and SET1, RBT4, IPF9162, and CAR1 (intracellular proteins not localized to the cell wall). In addition, whereas IgG titers against specific antigens were detectable in sera of 85 to 98.3% of patients with systemic candidiasis, IgM titers were detectable in only 10 to 68.3% of patients.
TABLE 3.
Summary of IgM and IgG data against specific antigens among patients with systemic candidiasis versus controls
| Predictor variable | Mean log2 titer ± SE of sera from:
|
P valuea | % of patients with systemic candidiasis with detectable titers (no. with detectable titers/total no. of patients) | |
|---|---|---|---|---|
| Patients with systemic candidiasis | Controls | |||
| IgM titers | ||||
| MET6-1 | 4.57 ± 0.28 | 4.93 ± 0.48 | NS | 25 (15/60) |
| MET6-2 | 4.16 ± 0.23 | 4.80 ± 0.44 | NS | 18.3 (11/60) |
| NOT5 | 6.52 ± 0.33 | 4.35 ± 0.42 | 0.0003 | 63.3 (38/60) |
| SET1 | 6.71 ± 0.34 | 4.65 ± 0.49 | 0.001 | 65 (39/60) |
| RBT4 | 5.37 ± 0.32 | 4.12 ± 0.38 | 0.03 | 41.7 (25/60) |
| IPF9162 | 6.96 ± 0.34 | 4.30 ± 0.26 | <0.0001 | 68.3 (41/60) |
| CAR1 | 5.41 ± 0.31 | 4.22 ± 0.37 | 0.03 | 43.3 (26/60) |
| GAP1 | 3.75 ± 0.17 | 4.22 ± 0.37 | NS | 10 (6/60) |
| ENO1 | 5.62 ± 0.33 | 4.30 ± 0.41 | 0.02 | 46.7 (28/60) |
| BGL2 | 4.40 ± 0.26 | 4.62 ± 0.43 | NS | 23.3 (14/60) |
| FBA1 | 5.16 ± 0.32 | 4.26 ± 0.38 | NS | 36.7 (22/60) |
| MUC1-1 | 3.90 ± 0.19 | 4.48 ± 0.42 | NS | 13.3 (8/60) |
| MUC1-2 | 4.29 ± 0.25 | 4.13 ± 0.38 | NS | 20 (12/60) |
| PGK1-1 | 5.57 ± 0.34 | 3.56 ± 0.23 | 0.0005 | 44.1 (26/59)* |
| PGK1-2 | 4.12 ± 0.23 | 3.97 ± 0.36 | NS | 20.3 (12/59)* |
| IgG titers | ||||
| MET6-1 | 8.67 ± 0.28 | 6.28 ± 0.50 | 0.0002 | 93.3 (56/60) |
| MET6-2 | 9.60 ± 0.21 | 8.57 ± 0.48 | 0.02 | 98.3 (59/60) |
| NOT5 | 8.18 ± 0.37 | 3.74 ± 0.23 | <0.0001 | 85 (51/60) |
| SET1 | 9.16 ± 0.22 | 4.51 ± 0.36 | <0.0001 | 98.3 (59/60) |
| RBT4 | 9.42 ± 0.27 | 4.78 ± 0.40 | <0.0001 | 98.3 (59/60) |
| IPF9162 | 8.92 ± 0.22 | 5.14 ± 0.39 | <0.0001 | 96.7 (58/60) |
| CAR1 | 8.40 ± 0.28 | 4.28 ± 0.35 | <0.0001 | 90 (54/60) |
| GAP1 | 8.38 ± 0.29 | 3.74 ± 0.23 | <0.0001 | 91.7 (55/60) |
| ENO1 | 9.07 ± 0.26 | 4.46 ± 0.38 | <0.0001 | 98.3 (59/60) |
| BGL2 | 8.69 ± 0.27 | 4.32 ± 0.36 | <0.0001 | 95 (57/60) |
| FBA1 | 8.49 ± 0.29 | 3.60 ± 0.19 | <0.0001 | 93.3 (56/60) |
| MUC1-1 | 8.72 ± 0.23 | 5.86 ± 0.56 | <0.0001 | 96.7 (58/60) |
| MUC1-2 | 8.69 ± 0.24 | 7.02 ± 0.52 | 0.001 | 95 (57/60) |
| PGK1-1 | 7.93 ± 0.27 | 2.20 ± 0.46 | 0.01 | 91.5 (54/59)b |
| PGK1-2 | 7.49 ± 0.26 | 6.52 ± 0.43 | 0.05 | 89.8 (53/59)b |
NS, not significant.
One patient with systemic candidiasis and one control did not have sufficient sera to test against PGK1-1 and PGK1-2.
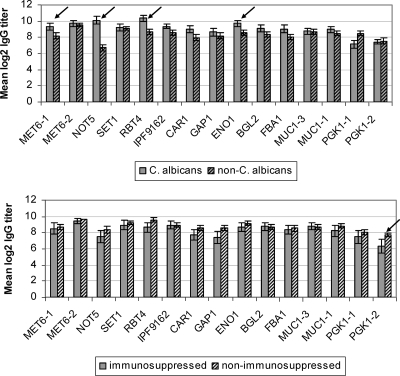
Mean IgG titers against MET6-1, NOT5, RBT4, and ENO1 were significantly higher for patients infected with C. albicans than for patients infected with non-C. albicans spp. (Fig. 1). In addition, higher percentages of patients infected with C. albicans than non-C. albicans spp. had detectable IgG titers against MET6-1 (76.5% [20/26] versus 55.9% [18/34], respectively [P = 0.07]) and NOT5 (96.2% [25/26] versus 76.5% [26/34], respectively [P = 0.06]). For the remaining antigens, there were no significant differences in the means of log2 IgG titers (Fig. 1) or the percentages of patients with detectable IgG titers among those infected with C. albicans versus those infected with non-C. albicans spp. (data not shown).
FIG. 1.
Data on IgG titers from patients with systemic candidiasis stratified by Candida spp. (top) and immunosuppression (bottom). Arrows indicate a significant difference (P values of <0.05) in IgG titers between groups.
Among patients with systemic candidiasis, IgG titers against 14 antigens did not significantly differ between immunocompromised and immunocompetent hosts (Fig. 1); immunocompetent hosts demonstrated higher IgG titers only against PGK1-2. IgG titers did not differ between burn victims and other patients or based on portal of entry (data not shown).
Identification of antibody responses that discriminated patients with systemic candidiasis from controls.
For each of the antigens, we assigned cutoff antibody titers that best discriminated patients from controls using receiver operating characteristic analyses (data not shown). The sensitivities and specificities of IgM and IgG antibody responses in identifying patients with systemic candidiasis are presented in Table 4.
TABLE 4.
Performance of IgM and IgG antibody titers against specific antigens in predicting systemic candidiasisa
| Antigen | IgM test
|
IgG test
|
|||
|---|---|---|---|---|---|
| % Sensitivity (no. of positive samples/total no. of samples) | % Specificity (no. of positive samples/total no. of samples) | % Sensitivity (no. of positive samples/total no. of samples) | % Specificity (no. of positive samples/total no. of samples) | % Positive predictive value (no. of positive samples/total no. of samples) | |
| MET6-1 | 25 (15/60) | 66.7 (16/24) | 65 (39/60) | 83.3 (20/24) | 90.7 (39/43) |
| MET6-2 | 18.3 (11/60) | 66.7 (16/24) | 60 (36/60) | 54.2 (13/24) | 76.6 (36/47) |
| NOT5 | 63.3 (38/60) | 79.2 (19/24) | 85 (51/60) | 87.5 (21/24) | 94.4 (51/54) |
| SET1 | 65 (39/60) | 75 (18/24) | 98.3 (59/60) | 66.7 (16/24) | 88.1 (59/67) |
| RBT4 | 41.7 (25/60) | 83.3 (20/24) | 98.3 (59/60) | 62.5 (15/24) | 86.8 (59/68) |
| IPF9162 | 68.3 (41/60) | 79.2 (19/24) | 96.7 (58/60) | 50 (12/24) | 82.9 (58/70) |
| CAR1 | 43.3 (26/60) | 79.2 (19/24) | 90 (54/60) | 75 (18/24) | 90 (54/60) |
| GAP1 | 10 (6/60) | 79.2 (19/24) | 91.7 (55/60) | 87.5 (21/24) | 94.8 (55/58) |
| ENO1 | 46.7 (28/60) | 79.2 (19/24) | 98.3 (59/60) | 70.8 (17/24) | 89.4 (59/66) |
| BGL2 | 23.3 (14/60) | 70.8 (17/24) | 95 (57/60) | 75 (18/24) | 90.5 (57/63) |
| FBA1 | 36.7 (22/60) | 79.2 (19/24) | 93.3 (56/60) | 91.7 (22/24) | 96.6 (56/58) |
| MUC1-1 | 13.3 (8/60) | 75 (18/24) | 96.7 (58/60) | 50 (12/24) | 82.9 (58/70) |
| MUC1-2 | 20 (12/60) | 83.3 (20/24) | 86.7 (52/60) | 54.2 (13/24) | 82.5 (52/63) |
| PGK1-1 | 44.1 (26/59)* | 95.6 (22/23)* | 72.9 (43/59)* | 56.5 (13/23)* | 81.1 (43/53)* |
| PGK1-2 | 20.3 (12/59)* | 87.0 (20/23)* | 62.7 (37/59)* | 52.2 (12/23)* | 52.2 (37/48)* |
* indicates that one patient with systemic candidiasis and one control did not have sufficient sera to test against PGK1-1 and PGK1-2.
Having shown that IgG titers against specific antigens identified systemic candidiasis with reasonable sensitivity and specificity, we developed a predictive model that considered IgG responses against a panel of antigens. As a first step, we assessed for colinearity by determining the tolerance of each of the 15 predictor variables (i.e., antibodies to specific antigens) on the others. After ruling out colinearity, we included all 15 antigens in backwards elimination and canonical correlation analyses to identify variables that best predicted systemic candidiasis. The two methods demonstrated excellent overall agreement in identifying predictors likely to be significant (data not shown). Based on these results, a panel of seven predictors (SET1, MUC1-2, FBA1, PGK1-1, PGK1-2, BGL2, and ENO1) was selected for discriminant analysis.
Discriminant analysis of the full set of 15 predictors yielded an outcome classification error rate of 3.7% (3/82) (Table 5). Among the 58 patients and 24 controls for whom sera were adequate to test all antigens, only 3 were classified incorrectly: 1 patient with systemic candidiasis was predicted to be a control, and 2 controls were predicted to have systemic candidiasis. The sensitivity and specificity of the panel of 15 predictors were 96.6% (57/59) and 95.6% (22/23), respectively. Of note, discriminant analysis of the panel of seven predictors yielded results identical to those for the full set of 15 predictors.
TABLE 5.
Performance of IgG against panels of recombinant antigensa
| Model | % Error rate (no. of positive samples/total no. of samples) | % Sensitivity (no. of positive samples/total no. of samples) | % Specificity (no. of positive samples/total no. of samples) |
|---|---|---|---|
| Full model (with 15 predictors) | 3.7 (3/82)* | 96.6 (57/59)* | 95.6 (22/23)* |
| SET1, ENO1, FBA1, PGK1-1, PGK1-2, MUC1-2, BGL21 | 3.7 (3/82)* | 96.6 (57/59)* | 95.6 (22/23)* |
| SET1, ENO1, PGK1-2, MUC1-2 | 3.7 (3/82)* | 96.6 (57/59)* | 95.6 (22/23)* |
| SET1, ENO1, MUC1-2 | 4.8 (4/84) | 95.0 (57/60) | 95.8 (23/24) |
* indicates that one patient with systemic candidiasis and one control did not have sufficient sera to test against PGK1-1 and PGK1-2.
Using classification tables generated for smaller subsets of predictors, we identified a panel of four predictors (SET1, ENO1, MUC1-2, and PGK1-2) that performed as well as the larger panels. Further decreasing the number of predictors to three (SET1, ENO1, and MUC1-2) increased the error rate to 4.8% (4/84) and decreased the sensitivity to 95% (57/60).
Using regression analysis, we confirmed that the panel of 4 predictors performed as well as the 15 predictors in identifying systemic candidiasis. The analysis of the four predictors yielded an R2 value of 0.69, which was not significantly different from the R2 value of 0.73 for the full model (F = 0.98; P = 0.47).
Testing a prediction model for systemic candidiasis.
Based on our data, the equation that best predicted systemic candidiasis was as follows: prediction score = (0.10 × SET1 + 0.07 × ENO1 − 0.04 × MUC1-2 − 0.02 × PGK1-2 − 0.12), where SET1, ENO1, MUC1-2, and PGK1-2 indicate the log2 specific antibody titers in individual patients. A score of >0.5 for a given patient was predictive of systemic candidiasis.
To assess the validity of this model, we performed ELISAs against the 15 recombinant antigens using additional sera that had been collected from 16 patients with systemic candidiasis within 2 days of the positive blood cultures and from 16 uninfected controls. The panel of four predictors yielded sensitivity and specificity of 100% (16/16) and 87.5% (14/16), respectively. The only classification errors were two controls who were predicted to have systemic candidiasis.
IgG responses against recombinant SET1, ENO1, MUC1-2, and PGK1-2 in acute- and convalescent-phase sera of patients with systemic candidiasis.
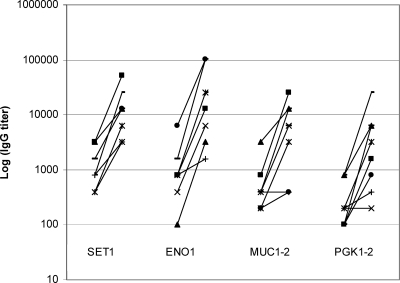
Finally, we performed ELISA using acute- and convalescent-phase sera from eight patients against the recombinant antigens included in the prediction model. The mean log2 IgG titer was significantly higher in convalescent-phase sera in than acute-phase sera against all four recombinant antigens (Table 6). Moreover, 100% (8/8), 87.5% (7/8), 62.5% (5/8), and 75% (6/8) of convalescent-phase sera exhibited at least fourfold increases in IgG titers against SET1, ENO1, MUC1-2, and PGK 1-2, respectively (Fig. 2).
TABLE 6.
IgG responses against SET1, ENO1, MUC1-2, and PGK1-2 in acute- and convalescent-phase sera of eight patients with systemic candidiasis
| IgG titer | Mean log2 titer ± SE
|
P value | |
|---|---|---|---|
| Acute-phase sera | Convalescent-phase sera | ||
| SET1 | 9.77 ± 0.46 | 13.39 ± 0.49 | 0.0004 |
| ENO1 | 9.64 ± 0.60 | 13.89 ± 0.77 | 0.0007 |
| MUC1-2 | 8.89 ± 0.45 | 12.02 ± 0.80 | 0.004 |
| PGK1-2 | 7.77 ± 0.44 | 11.02 ± 0.82 | 0.004 |
FIG. 2.
IgG titers against SET1, ENO1, MUC1-2, and PGK1-2 in acute- and convalescent-phase sera of eight patients with systemic candidiasis. For each recombinant protein, titers in acute-phase sera are shown at left and convalescent-phase sera are shown at right. Convalescent-phase sera were recovered 4 to 12 weeks after the first blood culture was positive for Candida spp.
DISCUSSION
The most striking finding of this study was that serum IgG responses against selected Candida antigens were accurate and early markers of systemic candidiasis. We measured IgG titers against 15 recombinant C. albicans antigens by ELISA and derived a prediction model that identified patients with systemic candidiasis with an error rate of only 3.7%, a sensitivity of 96.6%, and a specificity of 95.6%. The performance of the prediction model was superior to that of antibody detection against any individual antigen. Using backward elimination and canonical correlation analyses, we identified a subset of four antigens that performed as accurately as the full panel. We further confirmed the validity of the four-antigen panel by testing sera that were different from those used to derive the prediction model. Given the limitations of current diagnostic tests, measuring antibody responses against a panel such as ours might represent a significant advance in the diagnosis of systematic candidiasis.
Our data refute three of the major concerns about the limitations of antibody detection as a diagnostic tool. First, we demonstrated that patients with systemic candidiasis exhibited significant IgG titers against a wide range of antigens at the time that the diagnosis was made by conventional culture-based methods. This observation corroborates a number of reports documenting significant IgG titers against individual proteins like ENO1, HWP1, mannan, and CGTA before or at the time of the first positive blood culture (13, 14, 23, 36). Such findings are consistent with the fact that blood cultures are often not positive until relatively late in the course of disease (8, 17, 18). It is also possible that invasive diseases like candidemia are preceded in at least some patients by low-level systemic exposure to Candida spp., perhaps reflecting “leakage” from mucosal sites of colonization. Alternatively, the relatively early and potent IgG responses could represent amnestic responses against tissue invasion by a commensal organism to which the host has already been exposed. Along these lines, IgG responses were superior to those of IgM against each of the 15 antigens in discriminating patients with systemic candidiasis from controls. Previous studies of IgM responses yielded conflicting results, with results superior to IgG against blastospore cytoplasm and mannan in one study and inferior against hyphal cell wall proteins in another (10, 32). In our study, it is interesting that IgM titers against the four intracellular proteins that are not localized to the cell wall among patients with systemic candidiasis were significantly higher than those among controls, which suggests that these antigens might be less visible to the humoral immune system during commensal growth.
Second, we showed that antibody responses against recombinant C. albicans antigens diagnosed patients infected with C. albicans and non-C. albicans spp. Indeed, the prediction model was accurate regardless of the infecting species, a finding that corroborates data from previous studies. IgG responses against C. albicans ENO1, for example, have been reported to be effective in identifying patients with candidemia caused by diverse Candida spp. (14). Interestingly, a recent study showed that the detection of antibodies against HWP1, a protein produced exclusively during hyphal growth by C. albicans, is also useful among patients infected with non-C. albicans spp. (13), suggesting that epitopes within nonconserved proteins might elicit cross-reactive responses. Finally, our limited data on immunocompromised hosts suggest that the IgG responses were not attenuated in these patients. Again, these results are consistent with data from previous reports (11, 12, 21, 31, 34).
The sensitivities and specificities that we observed using individual proteins are within the ranges previously reported for antibody detection against a variety of antigens. Studies of IgG responses to ENO1 yielded sensitivities of 50 to 92% and specificities of 78 to 95% (14, 15, 16, 26, 33), and similar performances have been reported for tests against SAP (20, 35), HWP1 (14), a 52-kDa metalloprotein (6), mannan (28, 36), and CGTA (26). In attempts to overcome the diagnostic limitations of existing antibody tests, investigators have combined antibody responses against individual proteins with tests of antigen and/or metabolite detection. In general, these strategies have improved the sensitivity and specificity of individual tests as well as resulted in earlier diagnoses of systemic candidiasis (7, 25, 28-30, 36). It is quite possible, therefore, that a prediction model such as ours, based on antibody responses to multiple antigens, will ultimately be of the greatest utility in combination with cultures and other diagnostic markers.
It is important that antibody responses in this study were measured against recombinant antigens. As such, we cannot make definitive conclusions about humoral responses against native Candida proteins. Nevertheless, we demonstrated that mean IgG titers against each of the recombinant antigens included in our prediction model were significantly higher in convalescent-phase sera than in paired acute-phase sera from eight patients. These data suggest that the elevated IgG responses reflected responses to the causative Candida isolate.
In conclusion, we have clearly shown that a prediction model based on IgG responses against a panel of recombinant proteins is a potentially powerful tool for diagnosing systemic candidiasis. Our findings support future studies to validate and define the role of antibody detection in the diagnosis of systemic candidiasis. Moreover, it will be useful to assess other applications of antibody testing against panels of candidal antigens, such as tracking responses to antifungal therapy and identifying high-risk patients who could benefit from preventive or preemptive treatment.
Acknowledgments
This study was supported by the University of Florida Mycology Research Unit (NIH PO1 AI061537-01 to M.H.N., C.J.C., and J.R.W.) and by the Medical Research Service of the Department of Veterans Affairs.
Experiments were conducted in the laboratories of M.H.N. and C.J.C. at the North Florida/South Georgia Veterans Health System.
There is no conflict of interest.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Berenguer, J., M. Buck, F. Witebsky, F. Stock, P. A. Pizzo, and T. J. Walsh. 1993. Lysis-centrifugation blood cultures in the detection of tissue-proven invasive candidiasis. Disseminated versus single-organ infection. Diagn. Microbiol. Infect. Dis. 17103-109. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, S., C. J. Clancy, M. A. Checkley, M. Handfield, J. D. Hillman, A. Progulske-Fox, A. S. Lewin, P. L. Fidel, and M. H. Nguyen. 2003. Identification of Candida albicans genes induced during thrush offers insight into pathogenesis. Mol. Microbiol. 481275-1288. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, S., C. J. Clancy, M. A. Checkley, Z. Zhang, K. L. Wozniak, K. R Seshan, H. Y. Jia, P. Fidel, Jr., G. Cole, and M. H. Nguyen. 2005. The role of Candida albicans NOT5 in virulence depends upon diverse host factors in vivo. Infect. Immun. 737190-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29239-244. [DOI] [PubMed] [Google Scholar]
- 5.Ellepola, A. N., and C. J. Morrison. 2005. Laboratory diagnosis of invasive candidiasis. J. Microbiol. 4365-84. [PubMed] [Google Scholar]
- 6.El Moudni, B., M.-H. Rodier, G. Daniault, and J. L. Jacquemin. 1998. Improved immunodiagnosis of human candidiasis by an enzyme-linked immunosorbent assay using a Candida albicans 52-kilodalton metallopeptidase. Clin. Diagn. Lab. Immunol. 5823-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher, J. F., R. C. Trincher, J. F. Agel, T. B. Buxton, C. A. Walker, D. H. Johnson, R. E. Cormier, W. H. Chew, and J. P. Rissing. 1985. Disseminated candidiasis: a comparison of two immunologic techniques in the diagnosis. Am. J. Med. Sci. 290135-142. [DOI] [PubMed] [Google Scholar]
- 8.Garey, K. W., M. Rege, M. P. Pai, D. E. Mingo, K. J. Suda, R. S Turpin, and D. T. Bearden. 2006. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin. Infect. Dis. 4325-31. [DOI] [PubMed] [Google Scholar]
- 9.Gudlaugsson, O., S. Gillespie, K. Lee, J. Vande Berg, J. Hu, S. Messer, L. Herwaldt, M. Pfaller, and D. Diekema. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 371172-1177. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez, J., C. Maroto, G. Piedrola, E. Martin, and J. A. Perez. 1993. Circulating Candida antigens and antibodies: useful markers of candidemia. J. Clin. Microbiol. 312550-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoppe, J. E., D. Friess, and D. Niethammer. 1995. Orointestinal yeast colonization of paediatric oncologic patients during antifungal prophylaxis: results of quantitative culture and Candida serology and comparison of three polyenes. Mycoses 3841-49. [DOI] [PubMed] [Google Scholar]
- 12.Klingspor. L., G. Stintzing, and J. Tollemar. 1995. Deep Candida infection in child liver transplant recipients: serological diagnosis and incidence. Acta Paediatr. 84424-428. [DOI] [PubMed] [Google Scholar]
- 13.Lain, A., M. D. Moragues, J. C. Ruiz, J. Mendoza, A. Camacho, A. Del Palacio, and J. Ponton. 2007. Evaluation of a novel enzyme-linked immunosorbent assay to detect immunoglobulin G antibody to enolase for serodiagnosis of invasive candidiasis. Clin. Vaccine Immunol. 14318-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lain, A., N. Elguezabal, S. Brena, J. C. Garcia-Ruiz, A. Del Palacio, M. D. Moragues, and J. Ponton. 2007. Diagnosis of invasive candidiasis by enzyme-linked immunosorbent assay using the N-terminal fragment of Candida albicans hyphal wall protein 1. BMC Microbiol. 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsutake, K., S. Kohno, T. Miyazaki, H. Miyazaki, S. Maesaki, and H. Koga. 1994. Detection of Candida enolase antibody in patients with candidiasis. J. Clin. Lab. Anal. 8207-210. [DOI] [PubMed] [Google Scholar]
- 16.Mitsutake, K., T. Miyazaki, T. Tashiro, Y. Yamamoto, H. Kakeya, T. Otsubo, S. Kawamura, M. A. Hossain, T. Noda, Y. Hirakata, and S. Kohno. 1996. Enolase antigen, mannan antigen, Cand-Tec antigen, and β-glucan in patients with candidemia. J. Clin. Microbiol. 341918-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrell, M., V. J. Fraser, and M. H. Kollef. 2005. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 493640-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris, A. J., T. C. Byrne, J. F. Madden, and L. B. Reller. 1996. Duration of incubation of fungal cultures. J. Clin. Microbiol. 341583-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison, C. J., S. F. Hurst, and E. Reiss. 2003. Competitive binding inhibition enzyme-linked immunosorbent assay that uses the secreted aspartyl proteinase of Candida albicans as an antigenic marker for diagnosis of disseminated candidiasis. Clin. Diagn. Lab. Immunol. 10835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Na, B. K., and C. Y. Song. 1999. Use of monoclonal antibody in diagnosis of candidiasis caused by Candida albicans: detection of circulating aspartyl proteinase antigen. Clin. Diagn. Lab. Immunol. 6924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro, D., E. Monzonis, J. L. Lopez-Ribot, P. Sepulveda, M. Casanova, J. M. Nogueira, and J. P. Martinez. 1993. Diagnosis of systemic candidiasis by enzyme immunoassay detection of specific antibodies to mycelial phase cell wall and cytoplasmic candidal antigens. Eur. J. Clin. Microbiol. Infect. Dis. 12839-846. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, M. H., S. Cheng, and C. J. Clancy. 2004. Assessment of Candida albicans genes expressed during infections as a tool to understand pathogenesis. Med. Mycol. 42293-304. [DOI] [PubMed] [Google Scholar]
- 23.Pazos, C., M. D. Moragues, G. Quindos, J. Ponton, and A. del Palacio. 2006. Diagnostic potential of (1,3)-beta-D-glucan and anti-Candida albicans germ tube antibodies for the diagnosis and therapeutic monitoring of invasive candidiasis in neutropenic adult patients. Rev. Iberoam. Micol. 23209-215. [DOI] [PubMed] [Google Scholar]
- 24.Pitarch, A., A. Jimenez, C. Nombela, and C. Gil. 2006. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol. Cell. Proteomics 579-96. [DOI] [PubMed] [Google Scholar]
- 25.Platenkamp, G. J., A. M. Van Duin, J. C. Porsius, H. J. Schouten, P. E. Zondervan, and M. F. Michel. 1987. Diagnosis of invasive candidiasis in patients with and without signs of immune deficiency: a comparison of six detection methods in human serum. J. Clin. Pathol. 401162-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quindos, G., M. D. Moragues, and J. Ponton. 2004. Is there a role for antibody testing in the diagnosis of invasive candidiasis? Rev. Iberoam. Micol. 2110-14. [PubMed] [Google Scholar]
- 27.Raman, S. B., M. H. Nguyen, Z. Zhang, S. Cheng, H. Y. Jia, N. Weisner, K. Iczkowski, and C. J. Clancy. 2006. Candida albicans SET1 encodes a histone 3 lysine 4 methyltransferase that contributes to the pathogenesis of invasive candidiasis. Mol. Microbiol. 60697-709. [DOI] [PubMed] [Google Scholar]
- 28.Sendid, B., T. Jouault, R. Coudriau, D. Camus, F. Odds, M. Tabouret, and D. Poulain. 2004. Increased sensitivity of mannanemia detection tests by joint detection of α- and β-linked oligomannosides during experimental and human systemic candidiasis. J. Clin. Microbiol. 42164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sendid, B., J. L. Poirot, M. Tabouret, A. Bonnin, D. Caillot, D. Camus, and D. Poulain. 2002. Combined detection of mannanaemia and antimannan antibodies as a strategy for the diagnosis of systemic infection caused by pathogenic Candida species. J. Med. Microbiol. 51433-442. [DOI] [PubMed] [Google Scholar]
- 30.Sendid, B., M. Tabouret, J. L. Poirot, D. Mathieu, J. Fruit, and D. Poulain. 1999. New enzyme immunoassays for sensitive detection of circulating Candida albicans mannan and antimannan antibodies: useful combined test for diagnosis of systemic candidiasis. J. Clin. Microbiol. 371510-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tollemar, J., K. Holmberg, O. Ringden, and B. Lonnqvist. 1989. Surveillance tests for the diagnosis of invasive fungal infections in bone marrow transplant recipients. Scand. J. Infect. Dis. 21205-212. [DOI] [PubMed] [Google Scholar]
- 32.Torres-Rodríguez, J. M., N. Madrenys-Brunet, J. Nolla-Salas, A. Carceller, and C. Tur. 1997. Candiduria in non-neutropenic critically-ill surgical patients. Detection of IgA, IgG and IgM antibodies to Candida albicans by germ tube immunofluorescence. Mycoses 40439-444. [DOI] [PubMed] [Google Scholar]
- 33.van Deventer, A. J., W. H. Goessens, J. H. van Zeijl, J. W. Mouton, M. F. Michel, and H. A. Verbrugh. 1996. Kinetics of anti-mannan antibodies useful in confirming invasive candidiasis in immunocompromised patients. Microbiol. Immunol. 40125-131. [DOI] [PubMed] [Google Scholar]
- 34.Villalba, R., A. Gonzalez, M. J. Linares, M. Casal, and A. Torres. 1993. Detection of antibodies to Candida albicans germ tube as a possible aid in diagnosing systemic candidiasis in bone marrow transplant patients. Eur. J. Clin. Microbiol. Infect. Dis. 12347-349. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Q., Q. P. Su, G. Y. Wang, D. Z. Wen, Y. H. Zhang, H. Z. Bao, and L. Wang. 2007. SAP production of hybrid phage displaying secreted aspartyl proteinase epitope of Candida albicans and its application for the diagnosis of disseminated candidiasis. Mycoses 50165-171. [DOI] [PubMed] [Google Scholar]
- 36.Yera, H., B. Sendid, N. Francois, D. Camus, and D. Poulain. 2001. Contribution of serological tests and blood culture to the early diagnosis of systemic candidiasis. Eur. J. Clin. Microbiol. Infect. Dis. 20864-870. [DOI] [PubMed] [Google Scholar]