Abstract
Diarrheagenic Escherichia coli strains are important causes of diarrhea in children from the developing world and are now being recognized as emerging enteropathogens in the developed world. Current methods of detection are too expensive and labor-intensive for routine detection of these organisms to be practical. We developed a real-time fluorescence-based multiplex PCR for the detection of all six of the currently recognized classes of diarrheagenic E. coli. The primers were designed to specifically amplify eight different virulence genes in the same reaction: aggR for enteroaggregative E. coli, stIa/stIb and lt for enterotoxigenic E. coli, eaeA for enteropathogenic E. coli and Shiga toxin-producing E. coli (STEC), stx1 and stx2 for STEC, ipaH for enteroinvasive E. coli, and daaD for diffusely adherent E. coli (DAEC). Eighty-nine of ninety diarrheagenic E. coli and 36/36 nonpathogenic E. coli strains were correctly identified using this approach (specificity, 1.00; sensitivity, 0.99). The single false negative was a DAEC strain. The total time between preparation of DNA from E. coli colonies on agar plates and completion of PCR and melting-curve analysis was less than 90 min. The cost of materials was low. Melting-point analysis of real-time multiplex PCR is a rapid, sensitive, specific, and inexpensive method for detection of diarrheagenic E. coli.
Escherichia coli strains associated with diarrhea have been classified into six groups, based on clinical, epidemiological, and molecular criteria. These E. coli strains are commonly isolated from children with gastroenteritis in the developing world. Recent data suggest that these strains are common in the United States in children less than 5 years of age with acute diarrhea (10, 17). However, diarrheagenic E. coli strains are not routinely sought as stool pathogens in clinical laboratories. Some of these pathogens respond to antimicrobial agents, while for others (e.g., Shiga toxin-producing E. coli [STEC]), antibiotics should be avoided. Because the time frame in which treatment choices must be made is short, there is a need for a rapid, sensitive, and inexpensive detection technique. We have developed a monochromatic, fluorescence-based real-time PCR procedure to simultaneously identify eight virulence genes associated with the six classes of diarrheagenic E. coli. In this assay, the post-PCR products are identified based on melting-point curve analysis. This assay is simple, rapid, inexpensive, and reliable. It is suitable for use in clinical laboratories as well as research facilities.
MATERIALS AND METHODS
Bacterial strains.
One hundred twenty-six E. coli strains (Table 1) were analyzed, including strains representing all six of the currently recognized classes of diarrheagenic E. coli as well as commensal organisms. The prototypical strains, used in laboratories worldwide, included enterotoxigenic E. coli (ETEC) H10407; enteropathogenic E. coli (EPEC) 2348/69; STEC strains H30, HW1, and 86-24; and enteroaggregative E. coli (EAEC) strains O42 and 221. Additional strains were from our previous clinical studies (8, 9, 13, 14) or were provided by Herbert L. DuPont and Zhi Dong Jiang (ETEC and EAEC isolates from ill travelers to Mexico), Mitch Cohen (diffusely adherent E. coli [DAEC] isolates from children with diarrhea in Cincinnati, OH), Guillermo Ruiz-Palacios (EPEC isolates from children in Mexico City), or Thomas Whittam (STEC Center at Michigan State University). The category of each diarrheagenic E. coli strain had previously been determined based on DNA probe, PCR, tissue culture, or toxin assays at the laboratory which provided the strains. The E. coli collection of reference (ECOR) strains studied included all of the human isolates from the ECOR collections (20). These strains, representing the diversity of human E. coli, included each of the major phylogenetic groups (A, B1, B2, and D). They had originally been isolated from healthy individuals or from women with urinary tract infections (ECOR strains 11, 14, 40, 48, 50, 56, 60, 62, 64, 71, and 72). Three of the ECOR strains are known to have virulence genes that have been associated with illness: ECOR 8 is an EAEC strain on the basis of its adherence pattern and its aggregative-adherence-probe positivity (12); ECOR 50 and ECOR 64 are known to be PCR positive for afa and draBC, markers of Dr adhesin-positive strains, including DAEC strains, as well as some urinary tract pathogens (11). Thus, these three strains were included with the pathogens as EAEC, DAEC, and DAEC, respectively, rather than being considered commensals. In total, there were 12 ETEC, 13 EAEC, 7 EPEC, 12 enteroinvasive E. coli (EIEC), 12 DAEC, 34 STEC, and 36 commensal ECOR strains studied.
TABLE 1.
E. coli strains evaluated in the real-time multiplex PCR assay system
| Strains |
|---|
| Nonpathogenic ECOR |
| 1, 2, 4, 5, 6, 9, 10, 11, 12, 13, 14, 15, 24, 28, 35, 36, 38, 39, 40, 41, 42, 43, 48, 49, 51, 53, 54, 55, 56, 59, 60, 61, 62, 63, 71, 72 |
| DAEC |
| 2R57, 3M35, 4W25, 4W42, 4W72j, 5019, 6C61, 6L33, 6V48, 4B78, ECOR 50a, ECOR 64a |
| EIEC |
| 14NLF, 17NLF, 18NLF, 1LF, 20LF, 213, 21LF, 2LF, 40, 5LF, 64, 9LF |
| ETEC |
| H10407, ETEC1, ETEC13, ETEC2, ETEC3, ETEC30, ETEC4, ETEC46, ETEC5, ETEC52, ETEC53, ETEC6 |
| EPEC |
| EPEC56, EPEC19, 2348/69, EPEC7041-3, EPEC7140-3, EPEC7185-3, EPEC7236 |
| EAEC |
| 042, 221, EAEC1, EAEC2, EAEC3, EAEC4, EAEC5, EAEC6, EAEC7, EAEC8, EAEC9, EAEC10, ECOR 8a |
| STEC |
| H30, 1018, 266A1, 304786, 3153-86, 3288-85, 86-24, 91-8026, 91-8120, BA2, E1057, 8607, 1387, 1391, 3067, 4270, 4909, 5992, 7697, 7926, 7931, 7960, 8023, 8101, 8645, 8969, 6315, 8641, 8087, 7936, 6315, 1393, HSC-10, HW1 |
ECOR strains known to be pathogens are listed in the appropriate categories.
DNA isolation.
E. coli strains were subcultured from frozen or peptone stocks onto MacConkey agar plates, utilizing quadrant streaking methods to produce isolated colonies. These strains were then placed in an incubator for culture at 37°C. After overnight incubation, a single colony was carefully removed from the plate by using a sterile toothpick to avoid agar contamination, an important cause of erratic amplification. Crude lysates were prepared and used directly as a template for the PCR. DNA was extracted by boiling a single colony in 50 μl of PCR- or molecular-grade water for 5 min, followed by centrifugation at 14,000 rpm for 10 min. Two microliters of this lysate was used as a template with a 23-μl PCR master mix to make a 25-μl total reaction volume.
Primer design.
The primers were designed to detect eight different virulence genes simultaneously in a single reaction (Table 2). The primers were designed so that amplicons having melting temperatures (Tms) ranging from 77°C to 95°C, with more than 1°C between each peak, would be produced. The primers designed in this study were fashioned in a “bottom up” paradigm. We targeted the amplicon Tm as the first parameter, seeking appropriate primer sequences to amplify unique regions in a given virulence gene that would produce an amplicon of the desired Tm. Sequences of each gene were examined for features such as areas of high or low GC content, size, and identity among reported BLAST sequences for the target gene. These areas were analyzed by an oligonucleotide property calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html) which uses the nearest-neighbor method to predict the amplicon Tm. After selection of areas likely to produce amplicons with the desired Tm, primers were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). All primers were synthesized by Sigma Genosys (Houston, TX). These primers were then sequentially added to the mix to determine their actual Tms as well as to determine whether nonspecific primer amplication occurred in the presence of other oligonucleotide primers. Primer concentrations were then optimized to produce melting curves of similar peak heights (areas under the curve) between products and across a series of dilutions to simulate the various concentrations of template DNA extracted from the crude lysate preparation.
TABLE 2.
Primers for multiplex real-time PCR for diarrheagenic E. coli genesa
| Gene | Orientationb | Primer sequence (5′→3′) | Final concn (μM) | Amplicon size (bp) | Amplicon Tm (mean ± SD) | Source or reference |
|---|---|---|---|---|---|---|
| eaeA | F | ATGCTTAGTGCTGGTTTAGG | 0.56 | 248 | 83.4 ± 0.08 | 29 |
| R | GCCTTCATCATTTCGCTTTC | 0.56 | ||||
| aggR | F | CGAAAAAGAGATTATAAAAATTAAC | 0.44 | 100 | 77.2 ± 0.26 | This study |
| R | GCTTCCTTCTTTTGTGTAT | 0.44 | ||||
| daaD | F | TGAACGGGAGTATAAGGAAGATG | 0.50 | 444 | 93.2 ± 0.38 | This study |
| R | GTCCGCCATCACATCAAAA | 0.50 | ||||
| ipaH | F | GTTCCTTGACCGCCTTTCCGATACCGTC | 0.04 | 619 | 90.9 ± 0.15 | 24 |
| R | GCCGGTCAGCCACCCTCTGAGAGTAC | 0.04 | ||||
| stIa | F | TTTCCCCTCTTTTAGTCAGTCAA | 0.26 | 159 | ||
| stIb | F | TGCTAAACCAGTAGAGTCTTCAAAA | 0.26 | 138 | 80.6 ± 0.23 | This study |
| st | R | GCAGGATTACAACACAATTCACAGCAG | 0.26 | |||
| stx1 | F | CTGGATTTAATGTCGCATAGTG | 0.12 | 150 | 87.0 ± 0.13 | 15 |
| R | AGAACGCCCACTGAGATCATC | 0.12 | ||||
| stx2 | F | GGCACTGTCTGAAACTGCTCC | 0.08 | 255 | 89.1 ± 0.34 | |
| R | TCGCCAGTTATCTGACATTCTG | 0.08 | ||||
| lt | F | TCTCTATGTGCATACGGAGC | 0.36 | 322 | 85.8 ± 0.18 | 26 |
| R | CCATACTGATTGCCGCAAT | 0.36 |
Tm data are based on evaluation of 90 diarrheagenic E. coli strains. EPEC strains are eaeA positive and stx negative; STEC strains are usually eaeA positive and stx1 and/or stx2 positive; EAEC strains are aggR positive; DAEC strains are daaD positive; EIEC strains are ipaH positive; ETEC strains are st and/or lt positive.
F, forward; R, reverse.
PCR conditions.
Initially, we evaluated previously reported multiplex assays (15, 24, 26, 29) to determine whether they would work well in a real-time PCR. Nonspecific amplification or interference with new primers made most of the primers in these assays problematic. We sequentially eliminated primers and eventually were able to use several previously described primers in our system (Table 2). PCR was performed using a PTC-200 thermal cycler and real-time fluorescence monitoring by a Chromo 4 optical detector (MJ Research/Bio-Rad, Hercules, CA). Each multiplex PCR assay was performed with a final reaction volume of 25 μl, containing 0.5 U Phusion polymerase (Finnzyme OY, Espoo, Finland), in high-fidelity Phusion buffer with final concentrations of 200 μM deoxynucleoside triphosphates and 4 mM MgCl2. The primers were used at final concentrations of 0.04 to 0.56 μM (Table 2). Sybr green I (Cambrex Bio Science, Rockland, ME) was diluted as recommended by the manufacturer. The hot-start technique was used to prevent nonspecific amplification. The amplification cycles consisted of incubation at 98°C for 50 s, 60°C for 20 s, 72°C for 30 s, and 75°C for 1 s. After 25 cycles, a melting curve with a ramp speed of 2.5°C/s between 73°C and 95°C was determined with a reading every 0.2°C using fluorescence of Sybr green. Melting peaks were automatically calculated by Opticon Monitor software (Bio-Rad, Hercules, CA), which, after subtracting background fluorescence from a set of water blanks, plotted the negative derivative of fluorescence with respect to temperature (−dF/dT versus T). Representative strains of each diarrheagenic E. coli group were analyzed by agarose gel electrophoresis (2.0% agarose gels) to ensure that no unwanted bands were seen and that the predicted product size was found. To determine machine-to-machine variability, the Tms for the control strains were determined with an iCycler iQ (Bio-Rad, Hercules, CA) as well as with the PTC-200 thermal cycler with the Chromo 4 optical detector (MJ Research/Bio-Rad, Hercules, CA).
RESULTS
We evaluated three enzymes: (i) Phusion (Finnzymes, Finland) (Phusion hot-start DNA polymerase), a Pyrococcus-like enzyme with a processivity-enhancing domain; (ii) Amplitaq gold (Applied Biosystems, Foster City, CA), a recombinant form of Taq DNA polymerase lacking endonuclease and 3′-to-5′ exonuclease activities but having a 5′-to-3′ exonuclease activity (this is provided as an inactive enzyme, requiring heat activation to regenerate polymerase activity); and (iii) Thermus brockianus DNA polymerase from a Dynamo Sybr green quantitative PCR kit (Finnzymes, Finland). Of these, Phusion was the only enzyme that gave reliably reproducible amplification.
After optimization of the primer sequences and concentrations to be used in the multiplex PCR, we arrived at a set of primers that reliably (89/90 strains) amplified the relevant diarrheagenic E. coli genes in strains previously characterized as diarrheagenic E. coli (specificity, 1.00; sensitivity, 0.99). On an individual-gene basis, the sensitivity and specificity were 1.00 for all genes tested except daaD, which was detected in 11/12 strains expected to be positive and not detected in any of the 114 strains expected to lack this gene and be negative (sensitivity, 0.92; specificity, 1.00). The single DAEC strain that was negative in this assay was rechecked for an adherence pattern and confirmed to be diffusely adherent (data not shown). There were no false positives; in no case (0/126) did a peak occur at a melting point inconsistent with the strain genotypes.
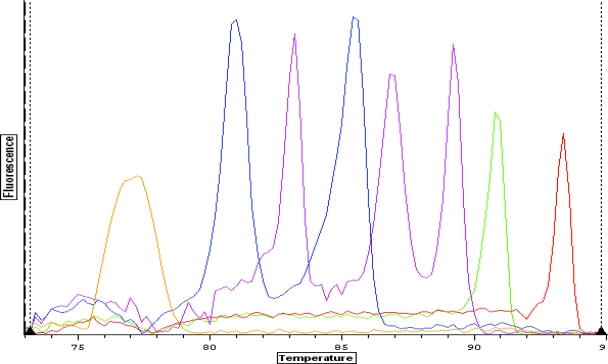
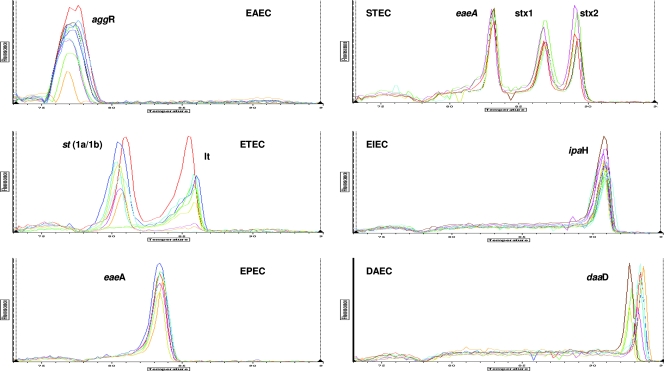
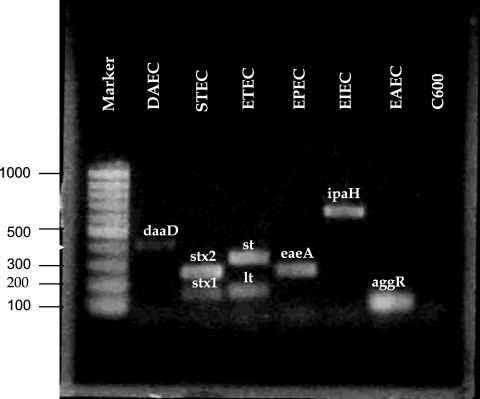
The Tms of the amplicons are spaced such that the identification of multiple samples on a single graph is unambiguous. The spacing between peaks for each gene is shown in Fig. 1. Remarkably little variation in intensity and Tm was observed among the different strains tested (Tm means ± standard deviations are given in Table 2). The amplitudes of the melting curves were quite similar for all strains in each category as well. The individual peaks were symmetric, with the exception of E. coli strains that were lt positive, which frequently gave asymmetric peaks. The individual strains run are shown in Fig. 2, demonstrating the overlap and reproducibility of the assay for specific genes. Figure 2 also shows that DAEC strains give either of two closely spaced peaks well separated from the closest gene (ipaH). The dual closely spaced peaks noted for this gene are due to the fact that daaD has two major variants found in the sequences (n = 30) currently in GenBank. These variants are predicted to produce amplicons of nearly identical sizes, with GC content percentages that are slightly different. For example, the two organisms showing the greatest dissimilarity (only 92% identical) in GenBank, DAEC5 (accession number AY525520.1) and DAEC1 (accession number EU010380.1), would be predicted to produce amplicons of 371 and 372 bases, with GC contents of 60% and 58%, respectively. The predicted Tm difference for these amplicons is 1°C; the actual Tm difference observed for the DAEC peaks is 0.73°C. Analysis with agarose gel confirmed that the amplicons represented on the melting-curve graph were indeed of the correct molecular weights expected based on the primer sequences (Fig. 3).
FIG. 1.
Real-time PCR simultaneously detects eight different diarrheagenic E. coli virulence genes. Data from individual tubes, each containing an EAEC, ETEC, EPEC, STEC, EIEC, or DAEC strain, are shown in a single figure so that the separation between individual amplicon melting curves is illustrated (from left to right: aggR, st, eaeA, lt, stx1, stx2, ipaH, and daaD). The y axis (fluorescence) represents the negative derivative of fluorescence over temperature versus temperature.
FIG. 2.
Melting curves of diarrheagenic E. coli. Curves are superimposed for assays of multiple strains of diarrheagenic E. coli to show the reproducibility of given pathotypes. The y axis (fluorescence) represents the negative derivative of fluorescence over temperature versus temperature.
FIG. 3.
Agarose gel of amplicons from the multiplex real-time PCR. The molecular weight ladder is shown in lane 1; strain identification and amplicons are shown in lanes 2 to 7; nonpathogenic E. coli is shown in lane 8.
Although the studies shown were done with single colonies, we also evaluated results with five colonies. When two different types of diarrheagenic E. coli were present, the assay consistently detected both (data not shown). Thus, the assay could conveniently be used for screening of five colonies simultaneously.
Machine-to-machine variability was determined in 59 separate assays that used representative strains for each E. coli category. The Tm measurements (means ± standard deviations) with the iCycler iQ were as follows: for aggR, 77.1 ± 0.7°C; for st, 81.4 ± 0 0.3°C; for eaeA, 83.9 ± 0.3°C; for lt, 85.9 ± 0.3°C; for stx1, 87.4 ± 0.3°C; for stx2, 89.6 ± 0.3°C; for ipaH, 91.5 ± 0.3°C; and for daaD, 93.8 ± 0.4°C. Thus, the peaks could be easily separated and individual strains distinguished despite small machine-to-machine variations in Tm compared to the Tm values shown in Table 2 for 90 diarrheagenic E. coli strains.
The total time between preparation of DNA from E. coli colonies on agar plates and completion of PCR and melting-curve analysis was less than 90 min. The cost of materials was under $2 per sample analyzed.
DISCUSSION
In recent years, multiplex PCR assays have been developed to detect virulence factors for diarrheagenic E. coli. These assays target common genes for most but not all of the currently recognized classes of diarrheagenic E. coli or require additional steps (4, 5, 6, 15, 25-28, 30). Traditional PCR methods require amplification in a thermocycler followed by product separation by gel electrophoresis (19) or fluorescent capillary electrophoresis (7), time-consuming and laborious processes. However, as shown here, the products of PCR can also be practically detected by using a DNA binding dye, such as Sybr green, in a multiplex format. Real-time PCR offers the advantage of being a faster, more robust assay because it does not require post-PCR procedures to detect amplification products.
The assay that we describe is unique in the number of different genes recognized in a single reaction. Careful selection and testing of primers allowed this approach to be successful. The choice of some individual genes for amplification deserves comment. The EAEC strains were first identified by tissue culture assays, and subsequently, a variety of genes have been associated with this phenotype and with clinical illness. We chose to amplify aggR because it may well be the most reliable indicator of a truly pathogenic EAEC strain (16, 22, 23, 31). For STEC, the toxin genes were chosen because of their central role in hemolytic uremic syndrome and hemorrhagic colitis and because risk of complications is related to toxin type, with Stx2 being more virulent. For EPEC and STEC, eaeA was chosen because of its central role in pathogenesis. The variations in intimin sequence could be problematic, but we were able to design primers that worked well. However, this assay system does not discriminate between typical and atypical EPEC. We were unable to add a ninth gene (bfpA [bundle-forming pilus]) to the mix because there was inadequate space between peaks for one more gene; we had higher-priority genes. Thus, the classification of an eaeA-positive and stx1- and stx2-negative strain based on this assay should be “EPEC” rather than “classic” EPEC, pending the use of separate primers for bfpA or a tissue culture assay. However, a screen for all EPEC strains is appropriate since atypical EPEC strains (lacking bfpA) are also pathogens (1-3, 18, 21). For DAEC, daaD was chosen because the BLAST data indicated that it was better conserved than the other closely linked genes related to the Dr adhesin phenotype and it had a melting point that was useful for this scheme. In several cases, primers that had single-nucleotide mismatches needed to be used as a compromise to best amplify known variants. However, a single internal mismatched base in the primers was well tolerated. For example, all of the E. coli O157:H7 strains reliably amplified eaeA, which in the case of 0157:H7 has a gamma allele of this gene despite the fact that there is a 1-base mismatch in the forward primer that we used for eaeA. Of interest, this assay also allows rapid presumptive identification of nonlactose fermenters as “Shigella/EIEC,” since both Shigella spp. and EIEC have the ipaH gene and are detected with this assay; Salmonella strains were negative because they lack ipaH (data not shown). A weakness of this or any multiplex assay is that important new variants of virulence genes may fail to amplify with the primers described. A second weakness is that we were unable to adapt the assay directly to fresh stool samples.
In testing strains on another real-time PCR machine, we found that Tms were very similar but not identical to those that we describe. The peaks were still well separated so that strains could easily be correctly categorized. However, obviously it is prudent to standardize this assay by using control strains in each laboratory.
The primer sets used in this study were designed with a robust assay in mind. The sequences from the virulence genes that are amplified in this multiplex PCR are from highly conserved regions of the genes, and thus, the assay has a low risk of false negatives related to genotypic variation within the different pathotypes. This assay represents a simple, rapid, sensitive, and inexpensive system for the practical presumptive detection of diarrheagenic E. coli in clinical laboratories. It is practical for use in clinical settings in both developed and many underdeveloped areas.
Acknowledgments
We thank Zhi Dong Jiang, Herbert L. DuPont, Mitchell B. Cohen, and Thomas Whittam for providing strains used in this study.
Theresa Ochoa is supported by PHS-FIC 1K01TW007405, and Tom Cleary is supported by the PHS-NICHD R01-HD051716.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Child Health and Human Development or the National Institutes of Health.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Afset, J. E., G. Bruant, R. Brousseau, J. Harel, E. Anderssen, L. Bevanger, and K. Bergh. 2006. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J. Clin. Microbiol. 443703-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afset, J. E., L. Bevanger, P. Romundstad, and K. Bergh. 2004. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J. Med. Microbiol. 531137-1144. [DOI] [PubMed] [Google Scholar]
- 3.Alikhani, M. Y., A. Mirsalehian, and M. M. Aslani. 2006. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J. Med. Microbiol. 551159-1163. [DOI] [PubMed] [Google Scholar]
- 4.Aranda, K. R., S. H. Fabbricotti, U. Fagundes-Neto, and I. C. Scaletsky. 2007. Single multiplex assay to simultaneously identify enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga toxin-producing Escherichia coli strains in Brazilian children. FEMS Microbiol. Lett. 267145-150. [DOI] [PubMed] [Google Scholar]
- 5.Aranda, K. R., U. Fagundes-Neto, and I. C. A. Scaletsky. 2004. Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J. Clin. Microbiol. 425849-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, C., J. Lüthy, M. Altwegg, and F. Baggi. 2005. Rapid detection of diarrheagenic E. coli by real-time PCR. J. Microbiol. Methods 61335-341. [DOI] [PubMed] [Google Scholar]
- 7.Brandal, L. T., S. Lindstedt, L. Aas, T. Stavnes, J. Lassen, and G. Kapperud. 2007. Octaplex PCR and fluorescence-based capillary electrophoresis for identification of human diarrheagenic Escherichia coli and Shigella spp. J. Microbiol. Methods 68331-341. [DOI] [PubMed] [Google Scholar]
- 8.Cleary, T. G., and B. E. Murray. 1988. Lack of Shiga-like cytotoxin production by enteroinvasive Escherichia coli. J. Clin. Microbiol. 262177-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary, T. G., J. J. Mathewson, E. Faris, and L. K. Pickering. 1985. Shiga-like cytotoxin production by enteropathogenic Escherichia coli serogroups. Infect. Immun. 47335-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, M. B., J. P. Nataro, D. I. Bernstein, J. Hawkins, N. Roberts, and M. A. Staat. 2005. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J. Pediatr. 14654-61. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence associated traits in Escherichia coli. J. Infect. Dis. 18378-88. [DOI] [PubMed] [Google Scholar]
- 12.Lai, X. H., S. Y. Wang, and B. E. Uhlin. 1999. Expression of cytotoxicity by potential pathogens in the standard Escherichia coli collection of reference (ECOR) strains. Microbiology 1453295-3303. [DOI] [PubMed] [Google Scholar]
- 13.Lopez, E. L., M. Diaz, S. Devoto, S. Grinstein, M. Woloj, B. E. Murray, E. Rubeglio, F. Mendilaharzu, M. Turco, M. Vasquez, and T. G. Cleary. 1991. Evidence of infection with organisms producing Shiga-like toxins in household contacts of children with the hemolytic uremic syndrome. Pediatr. Infect. Dis. J. 1020-24. [DOI] [PubMed] [Google Scholar]
- 14.Lopez, E. L., M. Diaz, S. Grinstein, S. Devoto, F. Mendilaharzu, B. E. Murray, S. Ashkenazi, E. Rubeglio, M. Woloj, M. Vasquez, and T. G. Cleary. 1989. Hemolytic uremic syndrome and diarrhea in Argentine children: the role of Shiga-like toxins. J. Infect. Dis. 160469-475. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Saucedo, C., J. F. Cerna, N. Villegas-Sepulveda, R. Thompson, F. R. Velazquez, J. Torres, P. I. Tarr, and T. Estrada-Garcia. 2003. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg Infect. Dis. 9127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyo, S. J., S. Y. Maselle, M. I. Matee, N. Langeland, and H. Mylvaganam. 2007. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect. Dis. 792-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataro, J. P., and J. B. Kaper. 1998. Diarmeagenic Escherichia coli. Clin. Microbiol. Rev. 11142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen, R. N., L. S. Taylor, M. Tauschek, and R. M. Robins-Browne. 2006. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen, T. V., P. Le Van, C. Le Huy, K. N. Gia, and A. Weintraub. 2005. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J. Clin. Microbiol. 43755-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prère, M. F., S. C. Bacrie, O. Baron, and O. Fayet. 2006. Bacterial etiology of diarrhoea in young children: high prevalence of enteropathogenic Escherichia coli (EPEC) not belonging to the classical EPEC serogroups. Pathol. Biol. (Paris) 54600-602. [DOI] [PubMed] [Google Scholar]
- 22.Rüttler, M. E., C. S. Yanzón, M. J. Cuitiño, N. F. Renna, M. A. Pizarro, and A. M. Ortiz. 2006. Evaluation of a multiplex PCR method to detect enteroaggregative Escherichia coli. Biocell 30301-308. [PubMed] [Google Scholar]
- 23.Sarantuya, J., J. Nishi, N. Wakimoto, S. Erdene, J. P. Nataro, J. Sheikh, M. Iwashita, K. Manago, K. Tokuda, M. Yoshinaga, K. Miyata, and Y. Kawano. 2004. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J. Clin. Microbiol. 42133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sethabutr, O., M. Venkatesan, G. S. Murphy, B. Eampokalap, C. W. Hoge, and P. Echeverria. 1993. Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J. Infect. Dis. 167458-461. [DOI] [PubMed] [Google Scholar]
- 25.Sharma, V. K., E. A. Dean-Nystrom, and T. A. Casey. 1999. Semi-automated fluorogenic PCR assays (TaqMan) for rapid detection of Escherichia coli 0157:H7 and other Shiga toxigenic E. coli. Mol. Cell. Probes 13291-302. [DOI] [PubMed] [Google Scholar]
- 26.Toma, C., Y. Lu, N. Higa, N. Nakasone, I. Chinen, A. Baschkier, M. Rivas, and M. Iwanaga. 2003. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J. Clin. Microbiol. 412669-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidal, M., E. Kruger, C. Durán, R. Lagos, M. Levine, V. Prado, C. Toro, and R. Vidal. 2005. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J. Clin. Microbiol. 435362-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidal, R., M. Vidal, R. Lagos, M. Levine, and V. Prado. 2004. Multiplex PCR for diagnosis of enteric infections associated with diarrheagenic Escherichia coli. J. Clin. Microbiol. 421787-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, G., C. G. Clark, and F. G. Rodgers. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the 0157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 403613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watterworth, L., E. Topp, H. Schraft, and K. T. Leung. 2005. Multiplex PCR-DNA probe assay for the detection of pathogenic Escherichia coli. J. Microbiol. Methods 6093-105. [DOI] [PubMed] [Google Scholar]
- 31.Zamboni, A., S. H. Fabbricotti, U. Fagundes-Neto, and I. C. Scaletsky. 2004. Enteroaggregative Escherichia coli virulence factors are found to be associated with infantile diarrhea in Brazil. J. Clin. Microbiol. 421058-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]