Abstract
PCR methodology was developed to identify Salmonella enterica serovars Typhi, Paratyphi A, and Paratyphi B. One multiplex PCR identifies serogroup D, A, and B and Vi-positive strains; another confirms flagellar antigen “d,” “a,” or “b.” Blinded testing of 664 Malian and Chilean Salmonella blood isolates demonstrated 100% sensitivity and specificity.
Identification of the serovars of Salmonella isolated from blood cultures, the lynchpin of enteric fever surveillance, is problematic in developing countries. Classical methods require high-quality O grouping and H typing antisera, reagents that can be difficult to obtain consistently. Accordingly, reference and research laboratories in developing countries, as in industrialized countries, are turning to multiplex PCR methods as a consistent, high-throughput approach to typing etiologic agents (1, 3, 25, 26). We utilized three sequential PCRs to identify the three classical pathogens that cause enteric fever, Salmonella serovars Typhi, Paratyphi A, and Paratyphi B, as an alternative to serotyping. An O grouping multiplex PCR identifies groups A, B, and D, based on described primers (12, 20). An H typing multiplex developed for this work identifies phase 1 H types “a,” “b,” and “d.” A third PCR uses primers described below to identify serovar Paratyphi B biovar Java that ferments d-tartrate (dT) (22). The sequential PCR methodology is robust and amenable to high throughput for use in research and reference laboratories in developing countries.
Classical Salmonella serovar Typhi reference strains expressing H antigen (d) and unusual strains expressing antigen “j” are listed in Table 1, along with reference Salmonella serovar Paratyphi A and B strains and 24 “negative control” strains of other serovars.
TABLE 1.
Twenty-two Salmonella serovar Typhi, Paratyphi A, and Paratyphi B reference strains and 24 “negative control” Salmonella strains consisting of other serovars used for preliminary validation of the multiplex PCR assays
| Strain | Salmonella serovar | O group | Phase 1 H flagellar antigen(s) | Vi capsular antigen status | dT fermentation statusa
|
|
|---|---|---|---|---|---|---|
| Jordan's | Kauffmann's | |||||
| Ty2 | Typhi | D | d | + | NT | NT |
| CVD 908-htrA | Typhi | D | d | + | NT | NT |
| CDC 06-0418 | Typhi | D | j | + | NT | NT |
| CDC 01-0274 | Typhi | D | j | + | NT | NT |
| CDC 2433 | Typhi | D | j | + | NT | NT |
| CDC 96-0344 | Typhi | D | j | + | NT | NT |
| 01-0020 | Paratyphi A | A | a | − | NT | NT |
| 01-0220 | Paratyphi A | A | a | − | NT | NT |
| 02-0209 | Paratyphi A | A | a | − | NT | NT |
| 02-0532 | Paratyphi A | A | a | − | NT | NT |
| 04-0529 | Paratyphi A | A | a | − | NT | NT |
| 04-0571 | Paratyphi A | A | a | − | NT | NT |
| 05-0741 | Paratyphi A | A | a | − | NT | NT |
| 00-0391 | Paratyphi B | B | b | − | − | − |
| 02-0303 | Paratyphi B | B | b | − | − | − |
| 04-0137 | Paratyphi B | B | b | − | − | − |
| 04-0615 | Paratyphi B | B | b | − | − | − |
| 00-0301 | Paratyphi B | B | b | − | + | + |
| 01-0399 | Paratyphi B | B | b | − | + | + |
| 03-0451 | Paratyphi B | B | b | − | + | + |
| 04-0126 | Paratyphi B | B | b | − | + | + |
| 01-0516 | Paratyphi B | B | b | − | + | + |
| CDC 32 | Paratyphi C | C1 | c | − | NT | NT |
| CDC 33 | Paratyphi C | C1 | c | − | NT | NT |
| 06-0868 | Choleraesuis (sensu stricto) | C1 | c | − | NT | NT |
| 06-0894 | Choleraesuis subsp. Kunzendorf | C1 | c | − | NT | NT |
| 06-0707 | Dublin | D | g, p | − | NT | NT |
| State lab 81.23500 | Typhimurium | B | i | − | NT | NT |
| CDC 07-0100 | Braenderup | C1 | e, h | − | NT | NT |
| CDC 07-0222 | Tennessee | C1 | z29 | − | NT | NT |
| CDC 07-0230 | Tennessee | C1 | z29 | − | NT | NT |
| CDC 07-0108 | Orion | E1 | y | − | NT | NT |
| CDC 07-0044 | Newport | C2 | e, h | − | NT | NT |
| CDC 07-0042 | Montevideo | C1 | g, m, s | − | NT | NT |
| CDC 07-0039 | Bardo | C2 | e.h | − | NT | NT |
| CDC 07-0021 | Give | E1 | l, v | − | NT | NT |
| CDC 07-0115 | Meleagridis | E1 | e.h | − | NT | NT |
| CDC 07-0124 | Mbandaka | C1 | z10 | − | NT | NT |
| CDC 07-0217 | Oranienburg | C1 | m, t | − | NT | NT |
| CDC 07-0223 | Virginia | C2 | d | − | NT | NT |
| CDC 07-0001 | Cotham | O28 | i | − | NT | NT |
| CDC 07-0006 | Edinburg | C1 | b | − | NT | NT |
| CDC 07-00014 | Livingstone | C1 | d | − | NT | NT |
| CDC 07-0019 | Inverness | O38 | k | − | NT | NT |
| CDC 07-0094 | Choleraesuis subsp. Kunzendorf | C1 | c | − | NT | NT |
| CVD SE | Enteritidis | D | g, m | − | NT | NT |
NT, not tested.
Two sets of putative Salmonella strains isolated from blood cultures of febrile patients were tested. One set included 443 isolates obtained from blood cultures of 431 febrile patients at l'Hôpital Gabriel Touré in Bamako, Mali, in the course of systematic surveillance for bacteremia and invasive bacterial disease among patients younger than 16 years of age with fever, who were admitted to the hospital or seen in the emergency room (6, 27). Strains initially identified in the clinical microbiology laboratory of the Centre pour le Développement des Vaccins, Bamako, Mali (CVD-Mali), as Salmonella serotype Typhi, Paratyphi A, or Paratyphi B or as Salmonella species were shipped to CVD-Baltimore for bacteriological confirmation. The second set was 34 putative Salmonella serovar Paratyphi A and 189 Salmonella serovar Paratyphi B strains isolated from blood cultures of patients in the course of surveillance for enteric fever in Santiago, Chile (4, 8, 9, 15-18).
The serovars of the vast majority of Malian and Chilean isolates were identified at CVD-Baltimore by agglutination with O grouping (Denka Seiken Co. Ltd, Japan) and H typing antisera (Sifin Institute, Berlin, Germany) (5, 7); remaining isolates were serotyped by the CDC Salmonella Reference Laboratory. CVD-Baltimore and CDC clinical microbiology results were the “gold standard” against which the performance of the multiplex PCR methods was compared to assess their sensitivity, specificity, and positive predictive value.
The 5′ to 3′ sequences for the various primers and the sizes of the expected amplicons are presented in Table 2. To ensure that the absence of PCR products was not a failure in the PCR per se, an internal control (P1 and P2 that amplify oriC) (23) was incorporated into the system (13). The primers in the O serogrouping multiplex have previously been described (12, 20), but heretofore were used in two distinct protocols. We combined these primers into a single multiplex PCR by optimizing primer concentration, annealing temperature, and elongation time.
TABLE 2.
Primers used in the multiplex PCR assays and the expected amplicons
| Primer | Primer sequence (5′ to 3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| O serogrouping | |||
| rfbJ-for | CCAGCACCAGTTCCAACTTGATAC | 662 | 20 |
| rfbJ-rev | GGCTTCCGGCTTTATTGGTAAGCA | ||
| tyv-for | GAGGAAGGGAAATGAAGCTTTT | 614 | 12 |
| tyv-rev | TAGCAAACTGTCTCCCACCATAC | ||
| vi-for | GTTATTCAGCATAAGGAG | 439 | 12 |
| vi-rev | CTTCCATACCACTTTCCG | ||
| prt-for | CTTGCTATGGAAGACATAACGAACC | 256 | 12 |
| prt-rev | CGTCTCCATCAAAAGCTCCATAGA | ||
| H antigen typing | |||
| H-for | ACTCAGGCTTCCCGTAACGC | This study | |
| Ha-rev | GAGGCCAGCACCATCAAGTGC | 423 | This study |
| Hb-rev | GCTTCATACAGACCATCTTTAGTTG | 551 | This study |
| Hd-rev | GGCTAGTATTGTCCTTATCGG | 763 (d) or 502 (j)b | This study |
| dT fermentation | |||
| dT-for | GTAAGGGTAATGGGTTCC | 289 | 22 |
| dT-rev | CACATTATTCGCTCAATGGAG | ||
| Internal controla | |||
| P1 (oriC) | TTATTAGGATCGCGCCAGGC | 163 | 31 |
| P2 (oriC) | AAAGAATAACCGTTGTTCAC |
Internal controls were included in both multiplex mixes and monoplex PCRs.
Letters in parentheses indicate antigens.
The H typing primers were designed by using sequences from GenBank (accession numbers AE014613, X03393, and AY649698) so that the sizes of the DNA fragments would enable facile recognition on 1 to 2% agarose gels. The primers dT-for and dT-rev detect the gene encoding the proficient active enzyme that allows dT fermentation by the Java biovar (22).
Three bacterial colonies were suspended in 100 μl of double-distilled water in 0.5-ml PCR tubes. Tubes were placed in a PCR machine, incubated at 95°C for 10 min, and cooled to 25°C. The cell debris was pelleted by centrifugation at 16,000 × g for 30 s, and 5 μl of clear supernatant was used as the template in a PCR. This constituted “crude DNA.” “Purified DNA” was prepared by hot phenol treatment (19). A dilution of 1:50 was made in double-distilled water, and 5 μl of the diluted DNA was used for the PCRs.
PCR was performed in 1× PCR buffer, 3.5 mM MgCl2, 0.2 mM of deoxynucleoside triphosphates, and 0.2 U of Invitrogen Taq DNA polymerase (final volume of 25 μl) in a Mastercycler (Eppendorf North America, Westbury, NY). Primers were combined at a concentration of 5 μM each (final concentration of 0.2 μM), except for the positive control primers (oriC) that were used at a concentration of 3.5 μM (final concentration of 0.14 μM) in the H mix. For each PCR, 1.0 μl of mix was used per reaction.
The cycling parameters of the PCRs were as follows. The O grouping multiplex PCR consisted of denaturation at 95°C for 2 min, followed by 35 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final step of 72°C for 5 min. The H typing multiplex PCR comprised a denaturation step of 2 min at 95°C, followed by 35 cycles of the following two steps: 95°C for 30 s and 55°C for 15 s. The dT fermentation PCR consisted of a denaturation step of 95°C for 2 min, followed by 35 cycles of 95°C for 30 s and 60°C for 30 s. PCR products were separated on 2% (wt/vol) agarose gels, stained with ethidium bromide and visualized on a UV transilluminator. Generally, for each PCR experiment, phenol-extracted DNA from Salmonella serovar Typhi, Salmonella serovar Paratyphi A, and Salmonella serovar Paratyphi B was used as a positive control. A negative control consisting of no DNA was always included.
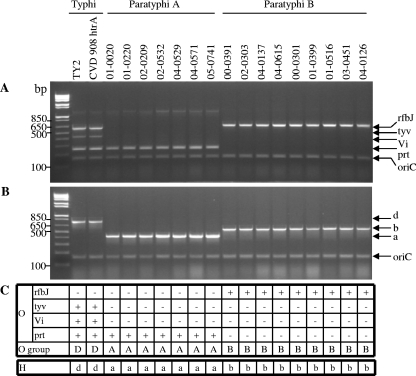
To validate preliminarily the PCR for the identification of enteric fever pathogens, 22 reference strains were analyzed (Table 1). Initially, two classical Salmonella serovar Typhi, seven Salmonella serovar Paratyphi A, four Salmonella serovar Paratyphi B dT nonfermenters (i.e., dT−), and five Salmonella serovar Paratyphi B dT+ strains were tested and the amplicons in Fig. 1 were generated. With the O grouping multiplex, Salmonella serovar Typhi strains produced specific PCR products with the prt primers and with the tyv primers, as expected of group D Salmonella (Fig. 1A and C) and the primers for Vi antigen synthesis, compatible with being Salmonella serovar Typhi. The Salmonella serovar Paratyphi A strains produced a PCR band with the prt, but not the tyv, primers, indicating that the strains are part of group A; the Salmonella serovar Paratyphi B strains yielded a PCR product only with the rfbJ primers, indicative of group B.
FIG. 1.
Validation of the O grouping and H typing multiplex PCRs using 18 reference enteric fever strains. The sample consisted of two Salmonella serovar Typhi, seven Salmonella serovar Paratyphi A, and nine Salmonella serovar Paratyphi B strains. Crude DNA extracts were tested with the O multiplex PCR (A) and H multiplex PCR (B). (C) The interpretation of the PCR products.
When these 18 Salmonella strains were tested with the H antigen multiplex PCR, the two Salmonella serovar Typhi strains yielded a PCR product indicative of flagellar antigen d, the seven Salmonella serovar Paratyphi A isolates yielded a PCR product designating flagellar antigen a, and the nine Salmonella serovar Paratyphi B isolates exhibited a product denoting flagellar antigen b (Fig. 1B and C). Only the Salmonella serovar Paratyphi B strains capable of fermenting dT yielded a positive PCR product in the PCR for dT fermentation (Fig. 2). These initial PCRs demonstrated that results were identical regardless of whether highly purified or crude DNA was used as the template, so crude template DNA was employed for all subsequent PCRs.
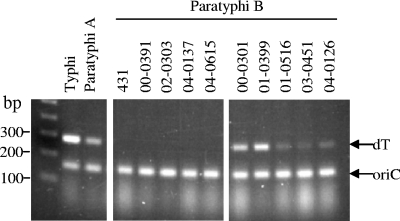
FIG. 2.
Validation of the dT fermentation PCR using nine Salmonella serovar Paratyphi B reference strains. The PCR control strains were Salmonella serovar Typhi CVD 908-htrA, Salmonella serovar Paratyphi A ATCC 9150, and Salmonella serovar Paratyphi B 431. The Salmonella serovar Paratyphi B reference strains included five dT fermenters and four dT nonfermenters.
To preliminarily assess the specificity of the multiplex PCRs, we tested 24 negative control Salmonella strains consisting of serovars other than Typhi, Paratyphi A, or Paratyphi B (Table 1). The multiplex PCR correctly identified Salmonella serotypes Typhimurium as group B, Dublin and Enteritidis as group D, Virginia and Livingstone as flagellar antigen d positive, and Edinburg as flagellar antigen b positive. Since none of these serotypes were positive in both the O group and the H antigen multiplex PCRs, they were clearly not enteric fever serovars.
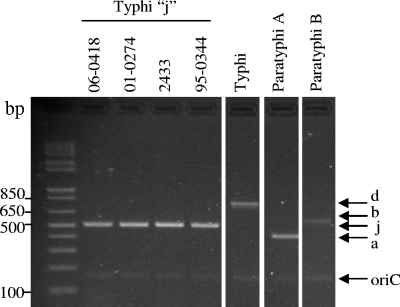
Four reference Salmonella serovar Typhi strains that express j flagellar antigen instead of d antigen were also tested (Table 1); the fliC-j allele results from a 261-bp deletion in fliC-d (10). The H-for and Hd-rev primers bind DNA external to this deletion to amplify a 502-bp fragment rather than the 763-bp d amplicon (Fig. 3). The presence or absence of antigen z66, encoded by fljBz66 on a linear plasmid, has no effect on the amplification of fliC-j (2). The four Salmonella serovar Typhi j strains yielded O multiplex amplicons indistinguishable from those of Salmonella serovar Typhi strains that possess fliC-d.
FIG. 3.
Detection of rare Salmonella serovar Typhi strains that possess fliC-j by the H multiplex PCR. Salmonella serovar Typhi strains 06-0418, 01-0274, and 2433 possess fliC-j and fljBz66. Salmonella serovar Typhi 95-0344 possesses fliC-j only. PCR control strains were Salmonella serovar Typhi CVD 908-htrA, Salmonella serovar Paratyphi A ATCC 9150, and Salmonella serovar Paratyphi B 431.
Systematic blood culture surveillance in Bamako, Mali, yielded 443 Salmonella strains from 431 patients initially identified in a Malian clinical microbiology laboratory as Salmonella serovar Typhi, serovar Paratyphi A, or serovar Paratyphi B or as Salmonella species. The purification of the subcultures in Baltimore revealed that 12 Malian patients harbored two distinct Salmonella serovars in their blood. Classical serotyping revealed that of the 419 Malian patients whose blood cultures yielded a single isolate: 164 were group D Salmonella (101 serovar Typhi, 33 serovar Dublin, and 30 serovar Enteritidis); 246 were group B (210 serovar Typhimurium, 25 serovar Stanleyville, 4 serovar I 4,5,12:i:−, and 7 serovar I 4,5,12:nonmotile strains); six were group C1 (four serovar Paratyphi C, one serovar Choleraesuis subsp. Kunzendorf, and one serovar Virchow); and the remaining three strains were serovars Havana (group O13), Minnesota (group O21), and Poona (group O13). Of the 12 patients whose blood cultures yielded two isolates, 6 harbored serovars Typhi and Typhimurium, 2 had serovars Typhimurium and Dublin, 2 had serovars Typhimurium and Enteritidis, 1 had serovars Dublin and I 4,5,12:nonmotile, and the last had serovars Stanleyville and Dublin. The 443 total strains from these 431 patients are summarized in Table 3.
TABLE 3.
Serovars of 443 Salmonella isolates from blood cultures of 431 febrile patients in Bamako, Mali, and results when tested in blinded fashion with the multiplex PCR assays
| Salmonella serovar | No. of isolates | No. of results by group in by indicated test
|
||||||
|---|---|---|---|---|---|---|---|---|
| O group PCR
|
Phase 1 H flagellar antigen PCR
|
|||||||
| D | Vi | B | A | d | b | a | ||
| Typhi | 107 | 107 | 107 | 0 | 0 | 107 | 0 | 0 |
| Dublin | 37 | 37 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enteritidis | 32 | 32 | 0 | 0 | 0 | 0 | 0 | 0 |
| Serogroup B | ||||||||
| Typhimurium | 220 | 0 | 0 | 220 | 0 | 0 | 0 | 0 |
| I 4,5,12:i:− | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| I 4,5,12:nonmotile | 8 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| Stanleyville | 26 | 0 | 0 | 26 | 0 | 0 | 0 | 0 |
| Serogroup C1 | ||||||||
| Choleraesuis subsp. Kunzendorf | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Paratyphi C | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Virchow | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other serogroups | ||||||||
| Havana | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Minnesota | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Poona | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The 443 Malian strains were coded before being tested blind with the multiplex PCRs to assess the sensitivity and specificity of these molecular diagnostic methods in identifying Salmonella serovar Typhi, serovar Paratyphi A, or serovar Paratyphi B (Table 3). All 107 Salmonella serovar Typhi strains were correctly identified as O group D, Vi positive, and positive for flagellar antigen d (100% sensitivity). The multiplex also detected all 69 of the non-serovar Typhi group D strains (37 serovar Dublin and 32 serovar Enteritidis). The O serogrouping multiplex PCR correctly detected all 258 of the group B strains (220 serovar Typhimurium, 12 serovar I 4,5,12:nonmotile, and 26 serovar Stanleyville strains). Although the precise serovar of these strains could not be determined in the molecular assay, they were clearly not enteric fever organisms. None of the remaining O serogroup strains (none of which were group A, B, or D) were positive in the O serogrouping PCR. The H typing multiplex correctly detected the b allele of the serovar Minnesota strain.
Among the Chilean isolates, 221 were Salmonella, including 26 isolates of Salmonella serovar Paratyphi A, 166 Salmonella serovar Paratyphi B, and 23 Salmonella serovar Typhi. The other six strains were serovars Anatum, Newport, Panama, Reading, Senftenberg, and Typhimurium (Table 4). The multiplex PCRs correctly identified all 26 Salmonella serovar Paratyphi A, 166 presumptive Salmonella serovar Paratyphi B, and 23 Salmonella serovar Typhi strains (Table 4). The O serogrouping PCR also detected the two group B serovars (Reading and Typhimurium) and the group D serovar (Panama), whereas the results for other serogroups (C2, E1, and E4) were all negative. All the Salmonella serovar Paratyphi B strains were of the dT-negative biotype.
TABLE 4.
Serovars of 221 Salmonella isolates from blood cultures of 221 febrile patients in Santiago, Chile, and results when tested in blinded fashion with the multiplex PCR assays
| Salmonella serovar | No. of isolates | O group PCR
|
Phase 1 H antigen PCR
|
Result of PCR to detect functional dT fermentation enzymea | |||||
|---|---|---|---|---|---|---|---|---|---|
| D | Vi | B | A | d | b | a | |||
| Serogroup D | |||||||||
| Typhi | 23 | 23 | 23 | 0 | 0 | 23 | 0 | 0 | NT |
| Panama | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | NT |
| Serogroup A | |||||||||
| Paratyphi A | 26 | 0 | 0 | 0 | 26 | 0 | 0 | 26 | NT |
| Serogroup B | |||||||||
| Paratyphi B | 166 | 0 | 0 | 166 | 0 | 0 | 166 | 0 | 0 |
| Typhimurium | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | NT |
| Reading | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | NT |
| Other serogroups | |||||||||
| Anatum (E1) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NT |
| Newport (C2) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NT |
| Senftenberg (E4) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NT |
NT, not tested.
Among the 664 clinical isolates tested from Mali (n = 443) and Chile (n = 221), there were 130 serovar Typhi, 26 serovar Paratyphi A, and 166 serovar Paratyphi B sensu stricto isolates. The multiplex PCRs proved 100% sensitive in detecting all of these true strains. The O serogrouping PCR was also 100% sensitive in detecting other serovars within groups B and D.
The specificity of the multiplex PCRs for enteric fever bacilli was also 100%, as none of the remaining 342 Salmonella strains belonging to 17 serovars were identified as an enteric fever serovar. The positive predictive value of the multiplex PCR method for detecting enteric fever Salmonella was 100%.
Since our first-step multiplex assay includes both O group and Vi antigen detection, Salmonella serovar Typhi can be presumptively identified at this stage. Although other organisms express Vi antigen, only Salmonella serovar Typhi and a subset of serotype Dublin represent Vi-positive group D strains. Strains positive for one of the three targets in the O grouping multiplex were then tested in the H multiplex. With the exception of some Indonesian strains that express H antigen j, all Salmonella serovar Typhi strains express antigen d (2, 10, 11, 14, 24, 28-30). Other Indonesian strains express “z66” antigen in addition to d or j.
A previously described multiplex PCR Salmonella typing method is not useful for the surveillance of enteric fever (21), since serotypes Paratyphi A and Paratyphi B would not be identified and, without further testing, serotype Paratyphi C cannot be differentiated from serotype Typhi. The fact that highly purified template DNA is not required for our three-tiered multiplex PCR typing system enhances the likelihood of successfully deploying this streamlined technology to laboratories in developing countries in Africa and Asia.
Acknowledgments
This research was supported by grant R01 AI029471 (to M. M. Levine) from the NIAID, NIH, and two grants from the Bill and Melinda Gates Foundation (to M. M. Levine).
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Abdissa, A., D. Asrat, G. Kronvall, B. Shittu, D. Achiko, M. Zeidan, L. K. Yamuah, and A. Aseffa. 2006. High diversity of group A streptococcal emm types among healthy schoolchildren in Ethiopia. Clin. Infect. Dis. 421362-1367. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S., J. Hardy, K. E. Sanderson, M. Quail, I. Goodhead, R. A. Kingsley, J. Parkhill, B. Stocker, and G. Dougan. 2007. A novel linear plasmid mediates flagellar variation in Salmonella Typhi. PLoS Pathog. 3e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkley, J. A., B. S. Lowe, I. Mwangi, T. Williams, E. Bauni, S. Mwarumba, C. Ngetsa, M. P. Slack, S. Njenga, C. A. Hart, K. Maitland, M. English, K. Marsh, and J. A. Scott. 2005. Bacteremia among children admitted to a rural hospital in Kenya. N. Engl. J. Med. 35239-47. [DOI] [PubMed] [Google Scholar]
- 4.Black, R. E., M. M. Levine, C. Ferreccio, M. L. Clements, C. Lanata, J. Rooney, R. Germanier, et al. 1990. Efficacy of one or two doses of Ty21a Salmonella typhi vaccine in enteric-coated capsules in a controlled field trial. Vaccine 881-84. [DOI] [PubMed] [Google Scholar]
- 5.Bopp, C., F. W. Brenner, P. I. Fields, J. G. Wells, and N. A. Strockbine. 2003. Escherichia, Shigella, and Salmonella, p. 654-671. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 6.Campbell, J. D., K. L. Kotloff, S. O. Sow, M. Tapia, M. M. Keita, T. Keita, S. Diallo, J. C. Hormazabal, P. Murray, and M. M. Levine. 2004. Invasive pneumococcal infections among hospitalized children in Bamako, Mali. Pediatr. Infect. Dis. J. 23642-649. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, P., and W. Ewing. 1972. Identification of Enterobacteriaceae. Burgess Publishing Co., Minneapolis, MN.
- 8.Ferreccio, C., M. M. Levine, A. Manterola, G. Rodriguez, I. Rivara, I. Prenzel, R. E. Black, T. Mancuso, and D. Bulas. 1984. Benign bacteremia caused by Salmonella typhi and paratyphi in children younger than 2 years. J. Pediatr. 104899-901. [DOI] [PubMed] [Google Scholar]
- 9.Ferreccio, C., M. M. Levine, H. Rodriguez, and R. Contreras. 1989. Comparative efficacy of two, three, or four doses of Ty21a live oral typhoid vaccine in enteric-coated capsules: a field trial in an endemic area. J. Infect. Dis. 159766-769. [DOI] [PubMed] [Google Scholar]
- 10.Frankel, G., S. M. C. Newton, G. K. Schoolnik, and B. A. D. Stocker. 1989. Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi. EMBO J. 83149-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guinee, P. A., W. H. Jansen, H. M. Maas, M. L. Le, and R. Beaud. 1981. An unusual H antigen (Z66) in strains of Salmonella typhi. Ann. Microbiol. (Paris) 132331-334. [PubMed] [Google Scholar]
- 12.Hirose, K., K. Itoh, H. Nakajima, T. Kurazono, M. Yamaguchi, K. Moriya, T. Ezaki, Y. Kawamura, K. Tamura, and H. Watanabe. 2002. Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. J. Clin. Microbiol. 40633-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoorfar, J., N. Cook, B. Malorny, M. Wagner, D. De Medici, A. Abdulmawjood, and P. Fach. 2003. Making internal amplification control mandatory for diagnostic PCR. J. Clin. Microbiol. 415835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, X., l. Phung, V., S. Dejsirilert, P. Tishyadhigama, Y. Li, H. Liu, K. Hirose, Y. Kawamura, and T. Ezaki. 2004. Cloning and characterization of the gene encoding the z66 antigen of Salmonella enterica serovar Typhi. FEMS Microbiol. Lett. 234239-246. [DOI] [PubMed] [Google Scholar]
- 15.Lanata, C. F., M. M. Levine, C. Ristori, R. E. Black, L. Jimenez, M. Salcedo, J. Garcia, and V. Sotomayor. 1983. Vi serology in detection of chronic Salmonella typhi carriers in an endemic area. Lancet 2441-443. [DOI] [PubMed] [Google Scholar]
- 16.Levine, M. M., R. E. Black, and C. Lanata. 1982. Precise estimation of the numbers of chronic carriers of Salmonella typhi in Santiago, Chile, an endemic area. J. Infect. Dis. 146724-726. [DOI] [PubMed] [Google Scholar]
- 17.Levine, M. M., C. Ferreccio, R. E. Black, R. Germanier, and the Chilean Typhoid Committee. 1987. Large-scale field trial of Ty21a live oral typhoid vaccine in enteric-coated capsule formulation. Lancet 11049-1052. [DOI] [PubMed] [Google Scholar]
- 18.Levine, M. M., C. Ferreccio, S. Cryz, and E. Ortiz. 1990. Comparison of enteric-coated capsules and liquid formulation of Ty21a typhoid vaccine in randomised controlled field trial. Lancet 336891-894. [DOI] [PubMed] [Google Scholar]
- 19.Levy, H., M. Fisher, N. Ariel, Z. Altboum, and D. Kobiler. 2005. Identification of strain specific markers in Bacillus anthracis by random amplification of polymorphic DNA. FEMS Microbiol. Lett. 244199-205. [DOI] [PubMed] [Google Scholar]
- 20.Lim, Y. H., K. Hirose, H. Izumiya, E. Arakawa, H. Takahashi, J. Terajima, K. Itoh, K. Tamura, S. I. Kim, and H. Watanabe. 2003. Multiplex polymerase chain reaction assay for selective detection of Salmonella enterica serovar Typhimurium. Jpn. J. Infect. Dis. 56151-155. [PubMed] [Google Scholar]
- 21.Lin, C. L., C. H. Chiu, C. Chu, Y. C. Huang, T. Y. Lin, and J. T. Ou. 2007. A multiplex polymerase chain reaction method for rapid identification of Citrobacter freundii and Salmonella species, including Salmonella Typhi. J. Microbiol. Immunol. Infect. 40222-226. [PubMed] [Google Scholar]
- 22.Malorny, B., C. Bunge, and R. Helmuth. 2003. Discrimination of d-tartrate-fermenting and -nonfermenting Salmonella enterica subsp. enterica isolates by genotypic and phenotypic methods. J. Clin. Microbiol. 414292-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moshitch, S., L. Doll, B. Z. Rubinfeld, B. A. Stocker, G. K. Schoolnik, Y. Gafni, and G. Frankel. 1992. Mono- and bi-phasic Salmonella typhi: genetic homogeneity and distinguishing characteristics. Mol. Microbiol. 62589-2597. [DOI] [PubMed] [Google Scholar]
- 25.Moyo, S. J., S. Y. Maselle, M. I. Matee, N. Langeland, and H. Mylvaganam. 2007. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect. Dis. 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen, T. V., V. P. Le, H. C. Le, K. N. Gia, and A. Weintraub. 2005. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J. Clin. Microbiol. 43755-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sow, S. O., S. Diallo, J. D. Campbell, M. D. Tapia, T. Keita, M. M. Keita, P. Murray, K. L. Kotloff, and M. M. Levine. 2005. Burden of invasive disease caused by Haemophilus influenzae type b in Bamako, Mali: impetus for routine infant immunization with conjugate vaccine. Pediatr. Infect. Dis. J. 24533-537. [DOI] [PubMed] [Google Scholar]
- 28.Tamura, K., R. Sakazaki, S. Kuramochi, and A. Nakamura. 1988. Occurrence of H-antigen Z66 of R phase in cultures of Salmonella serovar typhi originated from Indonesia. Epidemiol. Infect. 101311-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vieu, J. F., H. Binette, and M. Leherissey. 1986. Absence of the antigen H:z66 in 2355 strains of Salmonella typhi from Madagascar and several countries of tropical Africa. Bull. Soc. Pathol. Exot. Filiales 7922-26. (In French.) [PubMed] [Google Scholar]
- 30.Vieu, J. F., and M. Leherissey. 1988. The antigen H:z66 in 1,000 strains of Salmonella typhi from the Antilles, Central America and South America. Bull. Soc. Pathol. Exot. Filiales 81198-201. (In French.) [PubMed] [Google Scholar]
- 31.Widjojoatmodjo, M. N., A. C. Fluit, R. Torensma, B. H. Keller, and J. Verhoef. 1991. Evaluation of the magnetic immuno PCR assay for rapid detection of Salmonella. Eur. J. Clin. Microbiol. Infect. Dis. 10935-938. [DOI] [PubMed] [Google Scholar]