Abstract
High-throughput epidemiological typing systems that provide phylogenetic and genotypic information are beneficial for tracking bacterial pathogens in the field. The incidence of Salmonella enterica serovar Typhi infection in Indonesia is high and is associated with atypical phenotypic traits such as expression of the j and the z66 flagellum antigens. Utilizing a high-throughput genotyping platform to investigate known nucleotide polymorphisms dispersed around the genome, we determined the haplotypes of 140 serovar Typhi isolates associated with Indonesia. We identified nine distinct serovar Typhi haplotypes circulating in Indonesia for more than 30 years, with eight of these present in a single Jakarta suburb within a 2-year period. One dominant haplotype, H59, is associated with j and z66 flagellum expression, representing a potential pathotype unique to Indonesia. Phylogenetic analysis suggests that H59 z66+, j+ isolates emerged relatively recently in terms of the origin of serovar Typhi and are geographically restricted. These data demonstrate the potential of high-throughput genotyping platforms for analyzing serovar Typhi populations in the field. The study also provides insight into the evolution of serovar Typhi and demonstrates the value of a molecular epidemiological technique that is exchangeable, that is internet friendly, and that has global utility.
Salmonella enterica serovar Typhi remains a major public health problem in many developing countries with approximately 22 million cases of typhoid fever reported annually (3, 22). After primary culture, serovar Typhi is identified using serotyping, and strains are subsequently differentiated using phage typing, pulsed-field gel electrophoresis (30), or assays based on PCR including IS200 fingerprinting (31), RAPD [random(ly) amplified polymorphic DNA]-PCR (26), variable number tandem repeat scanning (18), and amplified fragment length polymorphisms (20). All of these techniques have the potential to distinguish between different isolates, but they have limited or no ability to identify phylogenetic relationships, evolutionary trends, or genotypic characteristics. Techniques such as RAPD-PCR, for example, have considerable inherent experimental variability, and data are not readily comparable between laboratories.
Approaches involving direct interrogation of DNA sequence are extremely reproducible and can also supply phylogenetic information. For example, multilocus sequence typing (MLST) involves the comparative sequencing of DNA fragments from different bacterial genes and facilitates the calculation of evolutionary relationships based on sequence diversity (5). MLST has proved highly discriminative at the serovar level within S. enterica (33), but this approach identified few sequence types in serovar Typhi, indicating that this is a monophyletic, recently emerged human pathogen, which is perhaps only 10,000 to 50,000 years old (15, 24).
Recently, Roumagnac et al. (24) used denaturing high-performance liquid chromatography and DNA sequencing of 199 gene fragments to identify rare single nucleotide polymorphisms (SNPs) that defined specific haplotypes illustrative of the evolutionary history of serovar Typhi. SNP typing unequivocally assigns individual serovar Typhi strains to a particular haplotype and can simultaneously gather information about traits such as fluoroquinolone resistance (24, 36). This is because SNP analysis, which has been used extensively to analyze human DNA, can be performed using equipment that interrogates thousands of nucleotide changes simultaneously in a single DNA sample (10).
SNP typing can also potentially be exploited to analyze serovar Typhi populations circulating in a particular geographical region (36). Typhoid fever has a particularly high incidence (810/100,000) and mortality rate in Indonesia (27), where cases are sometimes associated with unusual clinical manifestations, including neurological complications (11). In addition, serovar Typhi isolated in Indonesia can express uncommon flagellum antigens that are not routinely found elsewhere, such as H:j (j) and H:z66 (z66). The j antigen, like the d antigen, is encoded by the chromosomal gene fliC, but the j fliC allele harbors a 251-bp deletion in the variable region of the gene, thus changing the dominant antigenic epitope on the flagellum (6). Expression of j antigen has been associated with a milder clinical presentation of typhoid fever (8). The z66 antigen was first described in 1981, and the genetic determinants for expression were recently shown to be located on a novel linear plasmid pBSSB1 (1, 9, 13). Thus, z66+ serovar Typhi harbor two different flagellin genes, although z66 expression is dominant due to the action of a repressor protein encoded on pBSSB1 that targets the chromosomal fliC gene (2).
It has been hypothesized that serovar Typhi strains from Indonesia have a higher level of genetic diversity than isolates from many other typhoid endemic regions (19). Furthermore, because z66+ strains harbor two flagellin genes they were postulated to be ancestral to global serovar Typhi (7). However, this conclusion was not supported by SNP typing of 105 strains, according to which, all seven z66+ strains were restricted to a local branch (24). In the present study we have used a high-throughput platform to assign 140 Indonesian serovar Typhi isolates to particular haplotypes. Furthermore, we have mapped the origin of some of these haplotyped isolates within a particular geographical area and have associated haplotypes with a particular flagellum antigen type.
MATERIALS AND METHODS
Bacterial strains.
The strains used in the present study were isolated either in Indonesia, in the vicinity of Indonesia, or from tourists returning to their native countries who had contracted typhoid in Indonesia; they are from five different sources: (i) Institut Pasteur, Paris, France, 17 strains including 11 controls strains that were described by Roumagnac et al. (24); (ii) The Salmonella Genetic Stock Centre, Calgary, Alberta, Canada, 27 strains exhibiting an E+40 I-CeuI pattern from Kothapalli et al. (16); (iii) The Sanger Institute, Cambridge, United Kingdom, 11 strains, including two z66+ strains from Baker et al. (1) and two strains whose genome has been sequenced (4, 21); (iv) Leiden University Medical Centre, Leiden, The Netherlands, 84 strains from Vollaard et al. (35); and (v) Microbiological Diagnostic Unit-Public Health Laboratory, The University of Melbourne, Melbourne, Victoria, Australia, 22 strains isolated from travelers returning from Indonesia. Detailed descriptions of the strains used and the data produced are provided in Table S1 in the supplemental material.
Serovar Typhi genotyping assay.
Primers for the study were specifically designed to target the previously described single nucleotide base changes (24) using MassARRAY software and were supplied by Sigma Genosys (Haverhill, United Kingdom). Genotyping was performed by using Sequenom homogenous mass extend (Sequenom, Inc., San Diego, CA). Multiplexed assays of 1 to 10 SNPs per multiplex were designed by using Sequenom Assay Design v3.0.2.0. Genomic DNA was prepared predominantly using the Wizard Genomic DNA purification kit (Promega) and was diluted to a final concentration of 4 ng/μl. The genomic DNA underwent a locus-specific PCR before being treated with shrimp alkaline phosphatase to remove unincorporated nucleotides. The Sequenom homogenous mass extend reaction was then performed according to the manufacturer's specifications. Oligonucleotide primers anneal adjacent to the SNP of interest, and a mixture of terminator nucleotides allow extension through the SNP site, thus creating allele specific extension products each with unique mass. Extension products were then desalted through the addition of an anion-exchange resin, before their weights were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry. In total, 84 DNA fragments were analyzed for calculation of the genotype for each individual bacterial isolate. Genotypes were assigned and viewed by using the SpectroCALLER and SpectroACQUIRE softwares (Sequenom), respectively. The specific haplotypes for all strains were assigned on the basis of the previously described haplotypes (24). A control strain E00-7866 (superscript c in Fig. 1) originating from Morocco was originally designated H45 but was redesignated H46 on the basis of our data. The specific polymorphisms in E00-7866 were verified by DNA sequencing, and this strain was found to be correctly assigned to H46. Furthermore, two Indonesian serovar Typhi controls (403Ty and 404Ty) were assigned to H59 by our analysis, rather than to H5 and H2, respectively, as previously described (24). It was subsequently shown that the SNPs differentiating 403Ty H5 and 404Ty H2 from H59 were introduced during laboratory manipulations and were therefore not investigated.
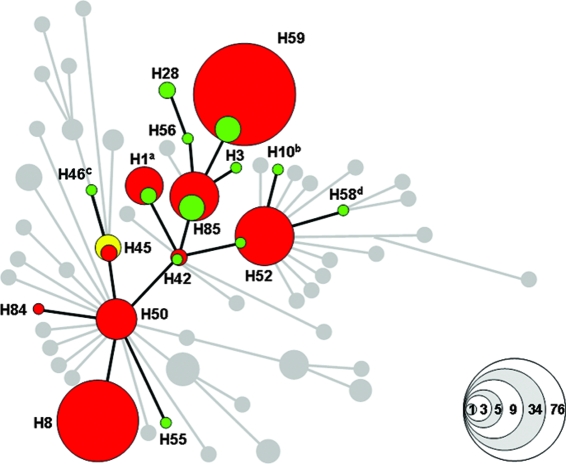
FIG. 1.
Minimal spanning tree based upon SNPs, demonstrating the relationship of the isolates used in the present study. This illustration is modified from Fig. 1 of Roumagnac et al. (24) and demonstrates the relationship of the 161 strains used here. The 18 serovar Typhi control strains with known haplotypes are labeled green and include CT18 (superscript a), Ty2 (superscript b), E00-7866 (superscript c), and AG3 (superscript d). The 140 experimental serovar Typhi strains are distinguished by red clusters, and the three serovar Paratyphi A isolates are colored yellow. The specific haplotype for each cluster is labeled (e.g., H45), and undetected haplotypes are gray. The size of each cluster corresponds to the number of strains identified in each group; this is demonstrated by the key in the bottom right corner.
PCR for flagellin gene variation.
The fliC gene and the fljB(z66) gene were amplified with primers specific for each locus. Amplification of the fliC gene was performed with the primers fliC_F (TTAACGCAGTAAAGAGAG) and fliC_R (ATGGCACAAGTCATTAATAC) and produced a 1,521-bp product for the d allele and a 1,273-bp product for the j allele (6). Amplification of the fljB(z66) gene was performed as previously described (1) using z66Flag_F (ATGGCACAAGTCATCAATAC) and z66Flag_R (TTAACGCAGCAGAGACAGTAC). Control PCR amplicons from the aroC gene were produced for all experimental isolates using the primers aroCfor (CCTGGCACCTCGCGCTATAC) and aroCrev (CCACACACGGATCGTGGCG) (10). PCR was performed in a 25-μl volume using PCR Supermix Taq polymerase (Invitrogen) and cycled on an MJ Research thermal cycler; the products were analyzed on a 1% agarose gel, and the sizes were estimated by comparison to the migration of Hyperladder I (Bioline).
RESULTS
Utility of a high-throughput genotyping method for the analysis of serovar Typhi.
We examined 143 serovar Typhi strains isolated between 1975 and 2005 from typhoid cases originating in or around Indonesia or from travelers returning from Indonesia. These isolates were subjected to conventional H and O typing and many were found to express the antigenic flagella variants j and z66. We have previously described a scheme based on denaturing high-performance liquid chromatography analysis that facilitates the assignment of serovar Typhi isolates to individual haplotypes that can be placed on a phylogenetic tree illustrative of the evolution of this serovar (24). We included 18 strains that had been previously haplotyped (24) as controls; therefore, the total number of isolates analyzed was 161. SNPs are frequently used in human genotyping as markers for regions of genome variation and as reference points to map genetic associations. Consequently, we selected the Sequenom high-throughput genotyping platform that is routinely used for analyzing mammalian DNA for SNP typing of serovar Typhi. The design of the Sequenom serovar Typhi SNP assay permitted the simultaneous detection of 84 SNPs in the serovar Typhi isolates, and the full data set is provided in Table S1 in the supplemental material.
The minimal spanning tree assembled from the raw data is shown in Fig. 1 and is adapted from Roumagnac et al. (24). Sequenom analysis of the 161 serovar Typhi isolates correctly assigned all control strains to the predicted haplotype (green circles; Fig. 1). The novel Indonesian serovar Typhi isolates were assigned to nine different haplotypes (red circles in Fig. 1), five of which also contained control isolates (green circles within red circles in Fig. 1). Significantly, the remaining 41 haplotypes reported previously in a global serovar Typhi collection, including three haplotypes described in Indonesia—H14, H58, and H70 (24), were undetected (gray circles in Fig. 1). Therefore, only a limited selection of the known haplotypes in the serovar Typhi population are circulating in this region.
The assay was additionally validated by assigning three experimental isolates to the ancestral haplotype, H45 (yellow circle in Fig. 1). These isolates were originally designated serovar Typhi, but when reserotyped they were found to be serovar Paratyphi A (588, 618, and 2585; see Table S1 in the supplemental material). All of the SNPs in the assay are specific for serovar Typhi; therefore, analysis of any other Salmonella serovars with this SNP assay would assign them to the ancestral haplotype H45.
Haplotypes H59 and H8 accounted for 53% (73) and 24% (34), respectively, of the serovar Typhi isolates recovered, and all haplotypes, except H84, included more than one isolate. Serovar Typhi haplotypes H42, H45, H50, H52, and H85 have been described previously as originating from geographically dispersed locations including Asia, Africa, and South America, providing evidence for the global dissemination of some strains (24). In contrast, H59 appears to be restricted to Indonesia and may be a recently evolved variant. H59 strains were identified in all five collections (see Materials and Methods) of experimental strains and were consistently isolated over a 30-year period (see Table S1 in the supplemental material). In addition, H42, H50, and H85 were also identified in different collections, with H85 representatives being isolated as far apart as 1987 and 2003 (see Table S1 in the supplemental material). This supports our previous observations that serovar Typhi strains can persist in a particular geographical region for periods extending several decades (24).
Serovar Typhi strains of haplotype H58 are frequently isolated in many parts of Southeast Asia and are associated with resistance to nalidixic acid, which may be a direct consequence of the common use of fluoroquinolones (17). We were unable to identify any H58 strains within these experimental Indonesian strains (superscript d in Fig. 1), although one fluoroquinolone-resistant strain belonging to H58 has been detected from a French traveler returning from Indonesia in 2003 (24). Importantly, our assay also interrogated the DNA for mutations in DNA gyrase, which leads to fluoroquinolone resistance (34). No such mutations in DNA gyrase were detected in this experimental population of serovar Typhi. This result indicates that there has been no recent clonal expansion of H58 in Indonesia, regardless of the fact that fluoroquinolone-resistant H58 strains may have been introduced in this country from near neighbors, such as Vietnam, where such strains are common. These data concur with those of other studies demonstrating the antimicrobial susceptibility of serovar Typhi and other enteric bacteria in Indonesia (14, 32) and may indicate a different epidemiological dynamic of the disease in this country.
Multiple serovar Typhi haplotypes are circulating in a local region.
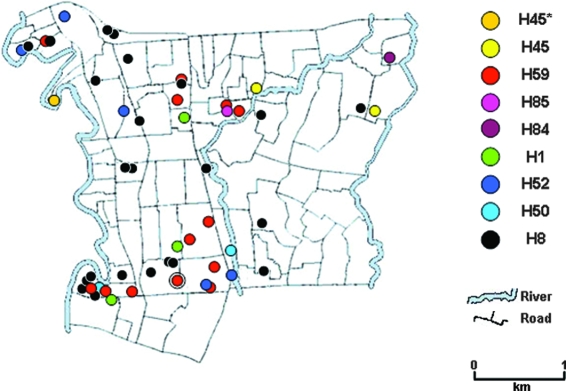
Serovar Typhi described in collection 4 (see Table S1 in the supplemental material) were isolated between June 2001 and February 2003 in the Jatinegara district in Eastern Jakarta in a study by Vollaard et al. (35). To understand the circulation of serovar Typhi strains in this local region, we attempted to retrospectively correlate data, locating the residence of the individual typhoid patients with a specific serovar Typhi haplotype. A map demonstrating the distribution of individual serovar Typhi strains isolated within the ∼10.5-km2 Jatinegara district is shown in Fig. 2. We were able to pinpoint the serovar Typhi haplotype from all of the residence of the patients for which we had residential mapping data (54 of 84 cases in total). We identified eight circulating haplotypes in the Jatinegara district, providing unequivocally evidence that there are independent clusters of typhoid transmission involving distinct hapotypes in this small area. Four serovar Typhi isolates of identical haplotype (H59) were isolated in the same household (red circle within a black circle), suggesting direct horizontal transmission. The methodology presented here indicates that haplotyping data may prove to be a powerful tool to assist the tracking of serovar Typhi within a community by stratifying the bacterial population in a reliable and reproducible manner. A prospective study combining mapping information and haplotyping data would give insight into the mechanisms of transmission and long-term carriage within a population where typhoid is endemic.
FIG. 2.
Specific locations of serovar Typhi haplotypes isolated in Jatinegara, Jakarta. A map of the Jatinegara area in Eastern Jakarta highlighted to demonstrate the association of the household of the typhoid patient with the haplotype of the serovar Typhi bacteria isolated from the patient. Each specific haplotype corresponds to a colored circle as depicted on the right. H45* demonstrates the location of a household from which serovar Paratyphi A was isolated. The red circle enclosed by a black circle depicts the location of four individual typhoid patients from which H59 serovar Typhi was isolated.
Flagellin gene variation.
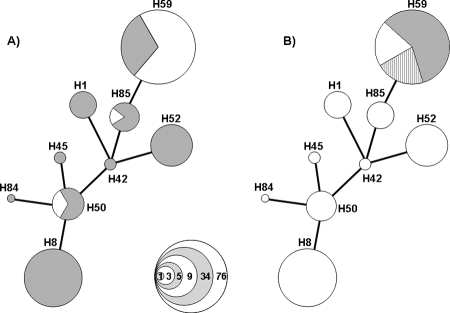
Serovar Typhi Indonesian isolates can express h, j, or z66 flagellum antigens. Consequently, we used PCR analysis of the fliC gene and the fljB(z66) gene using DNA extracted from all 140 experimental strains to determine which flagellin type they encoded. The primers were designed so that the fliC d flagellin allele would generate a DNA fragment of 1,521 bp compared to one of 1,273 bp for the j antigen (data not shown). The globally ubiquitous d fliC allele amplicon was detected in 85 of 140 (61%) of serovar Typhi and was dominant in all haplotypes except for the Indonesian specific H59 group (gray in Fig. 3A). The smaller fliC j allele, which was amplified in the remaining 55 isolates, was dominant in group H59 and was additionally identified in two other haplotypes (white in Fig. 3A).
FIG. 3.
Association of the fliC and fljB(z66) flagellin genes with specific haplotypes. (A) Diagram displaying the association of fliC allele with haplotype of 140 serovar Typhi strains. The strains with harbor the fliC gene which directs expression of the d antigen are colored gray the strains which harbor the fliC gene which directs expression of the j antigen are colored white. The specific haplotypes are labeled in association with Fig. 1, and the sizes of the clusters correspond to the number of strains located within each group; this is demonstrated by the key in the bottom right corner. (B) Diagram displaying the association of fljB(z66) gene with haplotype H59 in 140 serovar Typhi strains. The groups of strains lacking the fljB(z66) gene are white, and the strains harboring the fljB(z66) gene are shaded gray. The strains harboring the fljB(z66) gene are further subdivided in to groups carrying the fliC j allele (solid gray) or the fliC d allele (broken gray). The specific haplotypes are labeled in association with Fig. 1, and the sizes of the clusters correspond to the number of strains located within each group, as Fig. 3A.
Similarly, presence of the fljB(z66) gene on the pBSSB1 linear plasmid was confirmed by amplification of a 1,479-bp product specific to this region (data not shown). A 1,479-bp amplicon particular for the fljB(z66) gene was generated in 59 of 140 isolates, all of which fall into the H59 haplotype (gray shading in Fig. 3B). In addition, when the fljB(z66) data are combined with the fliC data the majority of strains (43 of 59) that can express the z66 antigen harbored a fliC j allele on the chromosome. Haplotype H59 contains four different subgroups based upon flagellin genes: fliC d, fliC j, fljB(z66)+fliC d, and fljB(z66)+fliC j, with the latter group being the dominant one. Thus, while the j allele was restricted to a limited number of haplotypes including H59, z66 was found only in H59 isolates and may be associated with the j allele.
DISCUSSION
Serovar Typhi is a young, genetically monomorphic bacterial pathogen that has not been in existence long enough to generate extensive sequence polymorphisms (15, 23-25). Here, we have investigated the population structure of serovar Typhi associated with Indonesia by using SNP typing and flagellin-based PCR analysis. In the collections we identified nine haplotypes composed of two dominant and seven less common haplotypes. Our data show that in a relatively small area (∼10.5 km2) there are several different haplotypes of serovar Typhi circulating within the local population, suggesting the existence of independent transmission clusters. These data are in agreement with other studies, which report multiple strain types circulating within a specific location (28-30). Furthermore, we identified bacteria of the same haplotype isolated over a 30-year period, providing further support for a vital role of persistent carriage and/or prolonged dissemination of serovar Typhi in the environment.
Typhoid fever remains a significant public health problem in many parts of Southeast Asia. Dissemination of serovar Typhi is of particular importance considering the wave of multiple drug-resistant strains that is currently spreading across Asia and the movement of people associated with current rapid economic growth. There is a pressing need for more effective epidemiological scrutiny of this organism, enabling better understanding of the nature of spread, which will ultimately facilitate control policies. Molecular methods appear to be the most robust, but such methods require standardization for the communication and analysis of the resulting data. The SNP-based approach is reproducible, results are applicable in any setting, and the data can be integrated into datasets produced in multiple laboratories using a variety of SNP detection technologies. This is in contrast to other schemes that detect strain differences but produce variable results in different laboratory settings. In addition, such schemes typically provide little or no information on phylogeny or phenotype and are unable to identify clonal expansion in a region.
The high-throughput system utilized here has been used to perform MLST on a number of Neisseria meningitidis reference strains (12). We have demonstrated the effectiveness of the same platform for detecting haplotypes in a genetically monomorphic gram-negative bacterial pathogen. Further flexibility can be anticipated when additional sensitivity is incorporated after more SNP variation is discovered.
Although the system has benefits, high-throughput SNP typing is currently not suitable for routine use in laboratories without specific infrastructure or financial support to run such assays. However, SNP detection technology has been driven in recent years by the need for massive throughput (currently >1 million SNPs for human genotyping studies). Microbial genotyping is much less demanding, with studies such as this providing epidemiologically informative data by assaying fewer than 100 SNP loci. With current high-throughput SNP detection technology, running costs for bacterial genotyping assays should be feasible for clinical laboratories, although setup costs may be prohibitive in some settings. The development of novel medium-throughput SNP detection technologies will be an important step toward the adoption of SNP-based microbial genotyping in clinical laboratories worldwide.
A practical solution for short-term development would be to house SNP typing facilities in regional reference laboratories linked via the internet through a global database. Simple “in the field” assays can also be established to detect a subset of discriminatory SNPs using a targeted approach. For example, MLST is performed routinely in many laboratories, while PCR analysis of nine gene fragments would have detected all of the haplotypes circulating in the present study. The resulting data would provide information about population structure in a particular region. Indeed, we are currently developing such a method based on PCR that can distinguish many of the major serovar Typhi haplotypes, which could potentially be universally adopted in clinical and research laboratories alike. However, it is important to note that the validity of this kind of simplified genotyping scheme depends on prior knowledge of the underlying phylogeny provided by comprehensive SNP typing studies.
It was originally hypothesized that z66+ serovar Typhi isolates from Indonesia were the precursor to global serovar Typhi, and it was assumed that the possession of a second phase of the flagella antigen was an ancestral state which has subsequently been lost (19). Conversely, an “out of Africa” hypothesis was proposed as the global source of Typhi (25), which would predict that the z66 gene was acquired later, possibly by horizontal gene transfer. We now know that z66 is present in only a single haplotype, and that the z66 flagellin gene [fljB(z66)] and fliC repressor [fljA(z66)] are located on a plasmid, indicating a relatively recent origin (1). The present study demonstrates that the acquisition of the pBSSB1 linear plasmid permitting the expression of z66 antigen most likely occurred only once and does not readily transfer to other genotypes. Furthermore, we suggest that pBSSB1 was horizontally transferred into an serovar Typhi strain carrying the d fliC allele, and over time the gene became truncated in some strains to form the j flagellin epitope. Expression of the z66 antigen is the default in serovar Typhi stains harboring the fljB(z66) locus, and fliC is effectively silenced (2). The truncated j allele can be found in three haplotypes, i.e., H50, H59, and H85, suggesting either that deletion has occurred spontaneously on several independent occasions or that homologous recombination has taken place between different haplotypes. However, it seems that strains harboring fljB(z66) preferentially possess the j fliC chromosomal allele.
In conclusion, our results demonstrate the adaptation of a global serovar Typhi genotyping scheme for a single country and ultimately a localized typhoid endemic region. The methodology offers a high level of sensitivity, thus allowing interrogation of phenotypes or pinpointing the locality of specific strains. The technique can in principle be used to define serovar Typhi circulating globally and is also potentially applicable to other bacterial pathogens. Our data prove that a number of distinct haplotypes of serovar Typhi can circulate in relatively small geographical area. However, somewhat paradoxically, we found that H58 strains that are currently circulating in other parts of South Asia and are associated with multiple drug resistance were not detected in Indonesia. Furthermore, strains harboring the z66 antigen are associated with the Indonesian archipelago, there is no evidence for the spread of these organisms to other countries where typhoid is endemic. It is likely that understanding the “Indonesian exception” would aid our understanding of the global epidemiology of typhoid fever. By combining haplotyping with phenotyping we were able to gain considerable insight into the population structure of serovar Typhi circulating in this region. Surveillance of strains using these methods combined with assessment of social and medical practices over a prolonged period will add vital information on how serovar Typhi is evolving and spreading in the human population.
Supplementary Material
Acknowledgments
We thank Soegianto Ali and Jonathan Hardy for supplying strains and information that have assisted with this study.
This study was funded by The Wellcome Trust, London, United Kingdom. K.E.S. was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada. M.A. is a Principal Investigator of the Scientific Foundation of Ireland (grant 05/FE1/B882) and the Max Planck Society for the Advancement of Science.
Footnotes
Published ahead of print on 5 March 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Baker, S., J. Hardy, K. E. Sanderson, M. Quail, I. Goodhead, R. A. Kingsley, J. Parkhill, B. Stocker, and G. Dougan. 2007. A novel linear plasmid mediates flagellar variation in Salmonella Typhi. PLoS Pathog. 3e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S., K. Holt, S. Whitehead, L. Goodhead, T. Perkins, B. Stocker, J. Hardy, and G. Dougan. 2007. A linear plasmid truncation induces unidirectional flagellar phase change in H:z66 positive Salmonella Typhi. Mol. Microbiol. 661207-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crump, J. A., S. P. Luby, and E. D. Mintz. 2004. The global burden of typhoid fever. Bull. W. H. O. 82346-353. [PMC free article] [PubMed] [Google Scholar]
- 4.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 1852330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7482-487. [DOI] [PubMed] [Google Scholar]
- 6.Frankel, G., S. M. Newton, G. K. Schoolnik, and B. A. Stocker. 1989. Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi. EMBO J. 83149-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel, G., S. M. Newton, G. K. Schoolnik, and B. A. Stocker. 1989. Unique sequences in region VI of the flagellin gene of Salmonella typhi. Mol. Microbiol. 31379-1383. [DOI] [PubMed] [Google Scholar]
- 8.Grossman, D. A., N. D. Witham, D. H. Burr, M. Lesmana, F. A. Rubin, G. K. Schoolnik, and J. Parsonnet. 1995. Flagellar serotypes of Salmonella typhi in Indonesia: relationships among motility, invasiveness, and clinical illness. J. Infect. Dis. 171212-216. [DOI] [PubMed] [Google Scholar]
- 9.Guinee, P. A., W. H. Jansen, H. M. Maas, L. Le Minor, and R. Beaud. 1981. An unusual H antigen (Z66) in strains of Salmonella typhi. Ann. Microbiol. 132331-334. [PubMed] [Google Scholar]
- 10.Higgins, G. S., D. P. Little, and H. Koster. 1997. Competitive oligonucleotide single-base extension combined with mass spectrometric detection for mutation screening. BioTechniques 23710-714. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman, S. L., N. H. Punjabi, S. Kumala, M. A. Moechtar, S. P. Pulungsih, A. R. Rivai, R. C. Rockhill, T. E. Woodward, and A. A. Loedin. 1984. Reduction of mortality in chloramphenicol-treated severe typhoid fever by high-dose dexamethasone. N. Engl. J. Med. 31082-88. [DOI] [PubMed] [Google Scholar]
- 12.Honisch, C., Y. Chen, C. Mortimer, C. Arnold, O. Schmidt, D. van den Boom, C. R. Cantor, H. N. Shah, and S. E. Gharbia. 2007. Automated comparative sequence analysis by base-specific cleavage and mass spectrometry for nucleic acid-based microbial typing. Proc. Natl. Acad. Sci. USA 10410649-10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, X., V. Phung le, S. Dejsirilert, P. Tishyadhigama, Y. Li, H. Liu, K. Hirose, Y. Kawamura, and T. Ezaki. 2004. Cloning and characterization of the gene encoding the z66 antigen of Salmonella enterica serovar Typhi. FEMS Microbiol. Lett. 234239-246. [DOI] [PubMed] [Google Scholar]
- 14.Isbandrio, B. B., M. H. Gasem, W. M. Dolmans, and J. A. Hoogkamp-Korstanje. 1994. Comparative activities of three quinolones and seven comparison standard drugs against Salmonella typhi from Indonesia. J. Antimicrob. Chemother. 331055-1056. [DOI] [PubMed] [Google Scholar]
- 15.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 239-45. [DOI] [PubMed] [Google Scholar]
- 16.Kothapalli, S., S. Nair, S. Alokam, T. Pang, R. Khakhria, D. Woodward, W. Johnson, B. A. Stocker, K. E. Sanderson, and S. L. Liu. 2005. Diversity of genome structure in Salmonella enterica serovar Typhi populations. J. Bacteriol. 1872638-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le, T. A., L. Fabre, P. Roumagnac, P. A. Grimont, M. R. Scavizzi, and F. X. Weill. 2007. Clonal expansion and microevolution of quinolone-resistant Salmonella enterica serotype Typhi, Vietnam, 1996-2004. J. Clin. Microbiol. 453485-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Y., M. A. Lee, E. E. Ooi, Y. Mavis, A. L. Tan, and H. H. Quek. 2003. Molecular typing of Salmonella enterica serovar Typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 414388-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moshitch, S., L. Doll, B. Z. Rubinfeld, B. A. Stocker, G. K. Schoolnik, Y. Gafni, and G. Frankel. 1992. Mono- and bi-phasic Salmonella typhi: genetic homogeneity and distinguishing characteristics. Mol. Microbiol. 62589-2597. [DOI] [PubMed] [Google Scholar]
- 20.Nair, S., E. Schreiber, K. L. Thong, T. Pang, and M. Altwegg. 2000. Genotypic characterization of Salmonella typhi by amplified fragment length polymorphism fingerprinting provides increased discrimination as compared to pulsed-field gel electrophoresis and ribotyping. J. Microbiol. Methods 4135-43. [DOI] [PubMed] [Google Scholar]
- 21.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug-resistant Salmonella enterica serovar Typhi CT18. Nature 413848-852. [DOI] [PubMed] [Google Scholar]
- 22.Parry, C. M. 2004. Typhoid fever. Curr. Infect. Dis. Rep. 627-33. [DOI] [PubMed] [Google Scholar]
- 23.Reeves, M. W., G. M. Evins, A. A. Heiba, B. D. Plikaytis, and J. J. Farmer III. 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roumagnac, P., F. X. Weill, C. Dolecek, S. Baker, S. Brisse, N. T. Chinh, T. A. Le, C. J. Acosta, J. Farrar, G. Dougan, and M. Achtman. 2006. Evolutionary history of Salmonella typhi. Science 3141301-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selander, R. K., P. Beltran, N. H. Smith, R. Helmuth, F. A. Rubin, D. J. Kopecko, K. Ferris, B. D. Tall, A. Cravioto, and J. M. Musser. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 582262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shangkuan, Y. H., and H. C. Lin. 1998. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella typhi and other Salmonella species. J. Appl. Microbiol. 85693-702. [DOI] [PubMed] [Google Scholar]
- 27.Simanjuntak, C. H., F. P. Paleologo, N. H. Punjabi, R. Darmowigoto, Soeprawoto, H. Totosudirjo, P. Haryanto, E. Suprijanto, N. D. Witham, and S. L. Hoffman. 1991. Oral immunisation against typhoid fever in Indonesia with Ty21a vaccine. Lancet 3381055-1059. [DOI] [PubMed] [Google Scholar]
- 28.Thong, K. L., Y. M. Cheong, S. Puthucheary, C. L. Koh, and T. Pang. 1994. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J. Clin. Microbiol. 321135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thong, K. L., Y. L. Goh, R. M. Yasin, M. G. Lau, M. Passey, G. Winston, M. Yoannes, T. Pang, and J. C. Reeder. 2002. Increasing genetic diversity of Salmonella enterica serovar Typhi isolates from Papua New Guinea over the period from 1992 to 1999. J. Clin. Microbiol. 404156-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thong, K. L., S. Puthucheary, R. M. Yassin, P. Sudarmono, M. Padmidewi, E. Soewandojo, I. Handojo, S. Sarasombath, and T. Pang. 1995. Analysis of Salmonella typhi isolates from Southeast Asia by pulsed-field gel electrophoresis. J. Clin. Microbiol. 331938-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Threlfall, E. J., E. Torre, L. R. Ward, A. Davalos-Perez, B. Rowe, and I. Gibert. 1994. Insertion sequence IS200 fingerprinting of Salmonella typhi: an assessment of epidemiological applicability. Epidemiol. Infect. 112253-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tjaniadi, P., M. Lesmana, D. Subekti, N. Machpud, S. Komalarini, W. Santoso, C. H. Simanjuntak, N. Punjabi, J. R. Campbell, W. K. Alexander, H. J. Beecham III, A. L. Corwin, and B. A. Oyofo. 2003. Antimicrobial resistance of bacterial pathogens associated with diarrheal patients in Indonesia. Am. J. Trop. Med. Hyg. 68666-670. [PubMed] [Google Scholar]
- 33.Torpdahl, M., M. N. Skov, D. Sandvang, and D. L. Baggesen. 2005. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis, and amplified fragment length polymorphism. J. Microbiol. Methods 63173-184. [DOI] [PubMed] [Google Scholar]
- 34.Turner, A. K., S. Nair, and J. Wain. 2006. The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J. Antimicrob. Chemother. 58733-740. [DOI] [PubMed] [Google Scholar]
- 35.Vollaard, A. M., S. Ali, H. A. van Asten, S. Widjaja, L. G. Visser, C. Surjadi, and J. T. van Dissel. 2004. Risk factors for typhoid and paratyphoid fever in Jakarta, Indonesia. JAMA 2912607-2615. [DOI] [PubMed] [Google Scholar]
- 36.Weill, F. X., H. H. Tran, P. Roumagnac, L. Fabre, N. B. Minh, T. L. Stavnes, J. Lassen, G. Bjune, P. A. Grimont, and P. J. Guerin. 2007. Clonal reconquest of antibiotic-susceptible Salmonella enterica serotype Typhi in Son La Province, Vietnam. Am. J. Trop. Med. Hyg. 761174-1181. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.