Abstract
Accurate species determination for anaerobes from blood culture bottles has become increasingly important with the reemergence of anaerobic bacteremia and prevalence of multiple-drug-resistant microorganisms. Our knowledge of the taxonomical diversity of anaerobes that cause bloodstream infections is extremely limited, because identification historically has relied on conventional methods. Over a 5-year period, we profiled anaerobic bacteremia at a large tertiary care hospital with 16S rRNA gene sequencing to gain a better understanding of the taxonomical diversity of the bacteria. Of 316 isolates, 16S rRNA gene sequencing and phylogenetic analysis identified 316 (100%) to the genus or taxonomical group level and 289 (91%) to the species level. Conventional methods identified 279 (88%) to the genus level and 208 (66%) to the species level; 75 (24%) were misidentified at the species level, and 33 (10%) results were inconclusive. High intragenus variability was observed for Bacteroides and Clostridium species, and high intraspecies variability was observed for Bacteroides thetaiotaomicron and Fusobacterium nucleatum. Sequence-based identification has potential benefits in comparison to conventional methods, because it more accurately characterizes anaerobes within taxonomically related clusters and thereby may enable better correlation with specific clinical syndromes and antibiotic resistance patterns.
Anaerobic microorganisms remain an important cause of bloodstream infections and account for 1 to 17% of positive blood cultures in the United States (3, 4, 8, 9, 11). The most commonly isolated pathogens are the Bacteroides fragilis group, other species of Bacteroides, Peptostreptococcus species, and Clostridium species (9). Most laboratories rely on conventional methods for identification of these common microorganisms and use algorithms based on key differential biochemical tests (16). However, DNA target sequencing has emerged as an attractive alternative, because identification is faster, more accurate, and independent of a microorganism's growth characteristics (10, 15, 17, 18). Sequence-based identification has enhanced our knowledge about the taxonomical diversity among anaerobic bacteria and has afforded the opportunity to better define the epidemiology of anaerobe-associated diseases. For example, some anaerobes have been associated with specific clinical syndromes, such as Clostridium sordellii with abortion (1), Clostridium tertium with neutropenia (12), and Fusobacterium necrophorum with hypercoagulability (5). Additionally, national surveys have demonstrated increasing antimicrobial resistance for several anaerobic pathogens (2, 6, 14), and definitive species identification can be extremely useful for guiding selection of empirical antibacterial therapy.
Our knowledge of the taxonomical diversity of anaerobes associated with clinically important bloodstream infections is limited. Most series of anaerobic bacteremia have been based on conventional methods of identification (3, 8, 9, 11), with little or no attention to the genetic diversity within and among genera. Similarly, a systematic approach for identifying the emergence of potentially novel or unusual sequence variants of anaerobes that cause bloodstream infection has not been applied over a 5-year period. We retrospectively studied all anaerobic microorganisms that were recovered from blood cultures at a large, tertiary care hospital. Our aim was to define the spectrum of anaerobes causing bloodstream infections by 16S rRNA gene sequencing and to identify unusual species belonging to taxonomically related groups. Aware that assessing taxonomical diversity relies on representative sequence databases, we specifically used two reference databases and phylogenetic analyses to assess intraspecies, intragenus, and intergenus variability.
MATERIALS AND METHODS
Anaerobic microorganisms recovered from blood cultures between January 2000 and December 2004 at Duke University Hospital, Durham, NC, that were deemed clinically significant (19) were retrospectively identified. During the study period, all three major blood culture systems were used: Bactec 9240 (BD Diagnostics, Sparks, MD), BacT/ALERT (classic and 3D) (bioMérieux, Inc., Durham, NC), and VersaTREK (Trek Diagnostic Systems, Cleveland, OH). Duke University Hospital is a large, 924-bed tertiary and quarternary care facility. Phenotypic identifications were performed by standard laboratory protocols that included a combination of manual biochemical testing, use of the API 20A system (bioMérieux, Marcy l'Etoile, France), and/or use of the Sherlock microbial identification system (MIDI, Inc., Newark, DE).
16S rRNA gene sequencing.
Bacterial DNA was extracted directly from frozen glycerol preparations of bacteria. The tube contents were thawed for 30 min at room temperature, and 50 μl of stock was removed and placed into molecular-grade water to a final volume of 200 μl. DNA was extracted with the QIAmp DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. PCRs were performed in a 20-μl volume containing 1× Taq buffer; 0.25 U of TaKaRa Taq; 3.0 mM MgCl2 (Takara Bio, Inc., Shiga, Japan); 200 μM each dATP, dGTP, and dCTP; 600 μM dUTP (Roche Diagnostics Corporation, Alameda, CA); 0.2 μM of each primer; and 2 μl of template. The primers used for amplification were 5F (5′-TTGGAGAGTTTGATCCTGGCTC-3′) and 1194R (5′-ACGTCATCCCCACCTTCCTC-3′). PCR mixtures were amplified by initial holding at 94°C for 5 min and then 30 cycles of denaturing at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 2 min. The reaction ended with a final extension at 72°C for 2 min and a hold at 4°C. The presence and sizes of amplicons were confirmed by gel electrophoresis. PCR products were purified with ExoSAP-IT reagent (USB Corporation, Cleveland, OH) per the manufacturer's instructions. PCR products were bidirectionally sequenced with the original amplification primer, 5F, and a reverse primer, 810R (5′GGCGTGGACTTCCAGGGTATCT-3′). Sequencing reactions were performed with Big Dye terminator reagents on an ABI Prism 310 or 3730xl instrument (Applied Biosystems, Foster City, CA) by a standard automated sequencer protocol.
Sequence and phylogenetic analyses.
Sequences were analyzed with MicroSeq ID software v2.0 (Applied Biosystems). Sequence-based identifications were determined individually with the MicroSeq 16S rDNA 500 Library v2.0 and SmartGene IDNS-Bacteria (version 3.2.3r8) databases (SmartGene, Inc., Raleigh, NC). SmartGene is a web-based application for sequence comparison with a reference database based on GenBank sequences. Alignments and phylogenetic trees were constructed as previously described (13). Final genus- and species-level identifications were assigned by both phylogenetic analysis and the following general guidelines: ≥99% identity to a reference entry identified a microorganism to the species level, 95.0 to 98.9% identity identified a microorganism to the genus level, and microorganisms with <95% identity to any reference sequence were considered unable to be identified definitively. Multiple species were assigned to isolates when top matches were between 99.0 and 99.9%. When five or more clinical isolates were identified within a group or species, interspecies or intergroup variability was determined by measuring the percent identity and recording the value as the percent difference.
RESULTS
The profile of anaerobes causing clinically significant bacteremia over the 5-year period is delineated in Table 1.
TABLE 1.
Anaerobic bacteremia at Duke University Medical Center, January 2000 to December 2004
| Group and identificationa | No. (%) of isolates |
|---|---|
| Gram-positive isolates | |
| Anaerococcus, Finegoldia, Parvimonas, and | |
| Peptoniphilus spp.b | 21 (7) |
| Clostridium perfringens | 35 (11) |
| Clostridium tertium | 16 (5) |
| Other Clostridium spp.c | 46 (15) |
| Eggerthella spp. | 14 (4) |
| Gram-negative isolates | |
| Alistipes, Porphyromonas, and Prevotella spp.d | 14 (4) |
| Bacteroides fragilis | 73 (23) |
| Bacteroides thetaiotaomicron relatedness group | 22 (7) |
| Other Bacteroides and Parabacteroides spp.e | 41 (13) |
| Fusobacterium spp. | 15 (5) |
| Other anaerobesf | 19 (6) |
| Total | 316 |
Anaerobes identified by 16S rRNA gene sequencing and phylogenetic analysis.
Anaerococcus sp. (n = 7), Finegoldia sp. (n = 3), Parvimonas sp. (n = 3), and Peptoniphilus sp. (n = 8).
Clostridium aerotolerans (n = 1), C. argentinense (n = 1), C. baratii (n = 1), C. bifermentans (n = 4), C. bolteae (n = 2), C. butyricum (n = 1), C. cadaveris (n = 2), C. celerecrescens (n = 1), C. clostridioforme (n = 4), C. colicanis (n = 1), C. hathewayi (n = 4), C. innocuum (n = 3), C. paraputrificum (n = 2), C. ramosum (n = 6), C. septicum (n = 4), C. sporogenes (n = 1), C. subterminale (n = 1), C. symbiosum (n = 3), and Clostridium sp. (n = 4).
Alistipes sp. (n = 2), Porphyromonas sp. (n = 1), Prevotella bivia (n = 6), Prevotella buccae (n = 1), Prevotella denticola (n = 2), Prevotella disiens (n = 1), and Prevotella nigrescens (n = 1).
Bacteroides caccae (n = 3), Bacteroides dorei (n = 4), Bacteroides finegoldii (n = 1), Bacteroides intestinalis (n = 4), Bacteroides ovatus (n = 7), Bacteroides pyogenes (n = 1), Bacteroides splanchnicus (n = 1), Bacteroides uniformis (n = 2), Bacteroides ureolyticus (n = 2), Bacteroides vulgatus (n = 4), Bacteroides sp. (n = 5), Parabacteroides distasonis (n = 6), and Parabacteroides merdae (n = 1).
Actinobaculum sp. (n = 1), Biophila wadsworthia (n = 1), Campylobacter curvus (n = 1), Catabacter hongkongensis (n = 1), Eubacterium limosum (n = 1), Propionibacterium acnes (n = 3), Ruminococcus gnavus (n = 1), Ruminococcus productus (n = 1), Ruminococcus sp. (n = 1), Solobacterium moorei (n = 2), Tissierella praeacuta (n = 2), Veillonella parvula (n = 3), and Veillonella sp. (n = 1).
Sequence-based identification.
Of 316 isolates, 16S rRNA gene sequencing with phylogenetic analysis identified 316 (100%) to the genus or taxonomical group level and 289 (91%) to the species level. Of those identified to the species level, two (0.6%) could not be resolved by sequencing analysis and were assigned to multiple species, Clostridium sporogenes/Clostridium botulinum and Clostridium aerotolerans/Clostridium xylanolyticum.
Identification by conventional methods.
For 316 isolates, conventional methods identified 279 (88%) to the genus level and 208 (66%) to the species level; 75 (24%) were misidentified at the species level, and 33 (10%) of results were inconclusive (Tables 2 and 3).
TABLE 2.
Level of identification for each method (n = 316 isolates)
| Group and genusa (no. of isolates) | No. of isolates identified by:
|
||||
|---|---|---|---|---|---|
| 16S rRNA gene sequencing
|
Conventional methods
|
||||
| Genus level | Species level | Genus level | Species level | Unable to identify or misidentified | |
| Gram-positive isolates | |||||
| Anaerococcus (7) | 7 | 1 | 7 | 0 | 0 |
| Clostridium (97) | 97 | 93 | 93 | 71 | 13b |
| Eggerthella (14) | 14 | 14 | 9 | 7 | 5 |
| Eubacterium (1) | 1 | 1 | 0 | 0 | 1 |
| Finegoldia (3) | 3 | 2 | 2 | 0 | 1 |
| Parvimonas (3) | 3 | 3 | 3 | 0 | 0 |
| Peptoniphilus (8) | 8 | 1 | 0 | 0 | 8 |
| Other (12)c | 12 | 10 | 5 | 1 | 7 |
| Gram-negative isolates | |||||
| Bacteroides (129) | 129 | 124 | 127 | 102 | 26b |
| Fusobacterium (15) | 15 | 15 | 15 | 14 | 0 |
| Parabacteroides (7) | 7 | 7 | 7 | 5 | 2b |
| Porphyromonas (1) | 1 | 1 | 1 | 0 | 0 |
| Prevotella (11) | 11 | 11 | 10 | 8 | 4b |
| Veillonella (4) | 4 | 3 | 0 | 0 | 4 |
| Other (4)d | 4 | 3 | 0 | 0 | 4 |
Genus was defined by sequence-based identification. Conventional identification was considered correct to genus or species level using old nomenclature (e.g., Bacteroides species for Parabacteriodes species).
Some anaerobes were correctly classified to genus level but misidentified to species level.
Actinobaculum (n = 1), Catabacter (n = 1), Propionibacterium (n = 3), Ruminococcus (n = 3), Solobacterium (n = 2), and Tissierella (n = 2).
Alistipes (n = 2), Biophila (n = 1), and Campylobacter (n = 1).
TABLE 3.
Representative misclassifications by conventional methods
| Group | Identification (no. of isolates) by:
|
|
|---|---|---|
| Conventional methods | 16S rRNA gene sequencing | |
| Gram-positive isolates | Clostridium clostridioforme (3) | Clostridium boltae (2), Clostridium hathewayi (1) |
| Clostridium perfringens (2) | Clostridium baratii (1), Clostridium bifermentans (1) | |
| Peptostreptococcus sp. (2) | Veillonella parvula (1), Veillonella sp. (1) | |
| Gram-negative isolates | Parabacteroides distasonis (2) | Bacteroides fragilis (1), Parabacteroides merdae (1) |
| Bacteroides theta/ovatus group (7) | Novel Bacteroides sp. (3), Bacteroides caccae (1), Bacteroides intestinalis (1), Bacteroides uniformis (1), Clostridium hathewayi (1) | |
| Bacteroides uniformis (3) | Bacteroides intestinalis (2), Bacteroides ovatus (2) | |
| Bacteroides vulgatus (8) | Bacteroides caccae (1), Bacteroides dorei (4), Bacteroides finegoldii (1), Bacteroides fragilis (1), Eggerthella lenta (1) | |
| Fusobacterium sp. (3) | Clostridium symbiosum (2), Alistipes finegoldii (1) | |
Analysis of microbial diversity.
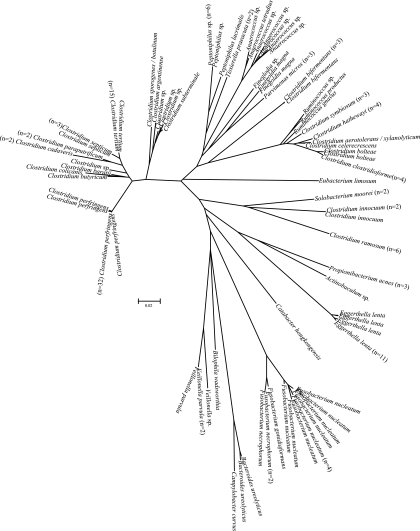
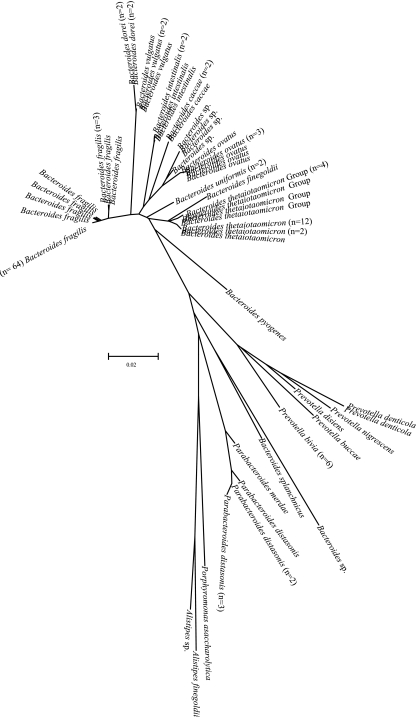
Figures 1 and 2 show radial dendrograms illustrating the genetic diversity of clinical isolates causing bacteremia. Since Clostridium and Bacteroides were the most represented groups, it is not surprising that these genera had the highest intragenus variability, i.e., 27.5% and 15.4%, respectively. Intraspecies variability for the most common species of Clostridium was low: 0.7% for Clostridium perfringens, 0.2% for Clostridium tertium, and 0.0% for Clostridium ramosum. Intraspecies variabilities for B. fragilis, Bacteroides thetaiotaomicron and its relatedness group, and Bacteroides ovatus were 3.3%, 4.0%, and 0.8%, respectively. Intraspecies variabilities of Eggerthella lenta and Fusobacterium nucleatum were 1.7% and 2.7%, respectively. We observed unusual sequence variants that grouped within taxonomical clusters of Anaerococcus sp., Bacteroides thetaiotaomicron, Parabacteroides distasonis, Bacteroides intestinalis, Clostridium subterminale, Eggerthella lenta, and Peptoniphilus sp. For example, we found three isolates with sequences that were distinctly different from reference sequences of Eggerthella lenta and six sequence variants for Bacteroides thetaiotaomicron (Fig. 1 and 2).
FIG. 1.
Neighbor-joining radial dendrogram for all gram-positive anaerobes and a cluster of gram-negative anaerobes. Only unique sequences are illustrated, with numbers of isolates for each unique sequence in parentheses.
FIG. 2.
Neighbor-joining radial dendrogram for gram-negative anaerobes not shown in Fig. 1. Only unique sequences are illustrated, with numbers of isolates for each unique sequence in parentheses.
DISCUSSION
Accurate species determination for anaerobes from blood cultures has become increasingly important, because anaerobic bacteremia with multiple-drug-resistant organisms has emerged as a significant health care problem as there are more patients at risk from immunosuppression and multiple comorbidities (6-9). To our knowledge, this study is the first longitudinal survey of anaerobic bacteremia at a large tertiary care hospital that identified anaerobes by 16S rRNA gene sequencing. We corroborate previous observations that the most common anaerobes that cause bloodstream infection, in decreasing order of frequency, are Bacteroides fragilis, other Bacteroides species, Clostridium species, anaerobic gram-positive cocci, Fusobacterium nucleatum, and Prevotella spp. Unlike prior reports that were limited by conventional methods, we observed with sequence-based identification a significant proportion of bloodstream infections from less common members of the Bacteroides and Clostridium taxonomical groups. We also document the first cases of anaerobic bacteremia from Bacteroides dorei, Bacteroides finegoldii, Parabacteroides merdae, Clostridium argentinense, Clostridium celerecrescens, Clostridium colicanis, Ruminococcus gnavus, and Tissierella praeacuta. Conventional identification misclassified or inconclusively identified approximately 25% of isolates, thereby missing a potential opportunity to define the epidemiology of or susceptibility patterns for these clinically significant anaerobic bloodstream infections. Of importance, conventional methods misclassified the Gram reaction and genera for several isolates and misidentified Parabacteroides distasonis, Bacteroides caccae, and Bacteroides vulgatus, three species known to have resistance to multiple antibacterials (14). Clinical decision-making based on erroneous conventional identifications could adversely affect patient care if a suboptimal empirical antibacterial regimen was selected or if misidentification belied the underlying source of infection.
We acknowledge that many laboratories cannot routinely employ partial 16S rRNA gene sequencing for anaerobic identification due to a lack of technical expertise and to cost. However, over the past several years, various commercial platforms and reference databases have become available for DNA target sequencing, enabling less experienced, nonmolecular bench technologists to determine and analyze DNA sequences. Laboratories should develop algorithms to screen for those isolates that can be adequately identified by conventional methods and should refer only a subset of isolates for 16S rRNA gene sequencing. Additionally, implementation of DNA target sequencing reduces the need for highly experienced personnel, a well-documented diminishing resource, and can result in a labor savings of least one full-time equivalent certified medical technologist (13).
Sequence data are a more valuable tool than identification by conventional methods, because they are objective and can be easily exchanged between different laboratories for comparison. Sequence-based identification enables us to appreciate the degree of heterogeneity within taxa, which can be represented by either high intraspecies variability or unusual sequence variants within taxonomically related clusters. The clinical relevance of reclassifying unusual sequence variants as new species cannot be reliably determined with a single institutional data set. Additionally, phylogeny may vary by the type of DNA target sequenced, with sequences potentially clustering into different groups using 16S rRNA, rpoB, or tuf targets. We propose that investigators maintain viable culture collections of unusual anaerobes and deposit their sequences into public databases, but we caution against the impulse to describe them as unique species. A consensus has not been reached within the microbiology community about drawing finer distinctions between species in a meaningful way, and the concept of species has not been clearly delineated. Instead, we recommend that investigators deposit unusual sequences as “variants within taxonomical relatedness groups,” affording the opportunity to carefully evaluate their taxonomical and clinical significance longitudinally and then determine the need for unique species designations. Improved disease surveillance using DNA target sequencing will provide us with the ability to correlate certain anaerobes with specific clinical syndromes and better understand the development of antibiotic resistance within individual taxonomical groups.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Aldape, M. J., A. E. Bryant, and D. L. Stevens. 2006. Clostridium sordellii infection: epidemiology, clinical findings, and current perspectives on diagnosis and treatment. Clin. Infect. Dis. 431436-1446. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, K. E., D. Ashcraft, K. Cambre, C. L. Pierson, S. G. Jenkins, and J. E. Rosenblatt. 2001. Multicenter survey of the changing in vitro antimicrobial susceptibilities of clinical isolates of Bacteroides fragilis group, Prevotella, Fusobacterium, Porphyromonas, and Peptostreptococcus species. Antimicrob. Agents Chemother. 451238-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blairon, L., Y. De Gheldre, B. Delaere, A. Sonet, A. Bosly, and Y. Glupczynski. 2006. A 62-month retrospective epidemiological survey of anaerobic bacteraemia in a university hospital. Clin. Microbiol. Infect. 12527-532. [DOI] [PubMed] [Google Scholar]
- 4.Diekema, D. J., S. E. Beekmann, K. C. Chapin, K. A. Morel, E. Munson, and G. V. Doern. 2003. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J. Clin. Microbiol. 413655-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forrester, L. J., B. J. Campbell, J. N. Berg, and J. T. Barrett. 1985. Aggregation of platelets by Fusobacterium necrophorum. J. Clin. Microbiol. 22245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hecht, D. W. 2004. Prevalence of antibiotic resistance in anaerobic bacteria: worrisome developments. Clin. Infect. Dis. 3992-97. [DOI] [PubMed] [Google Scholar]
- 7.Hecht, D. W. 2007. Routine anaerobic blood cultures: back where we started? Clin. Infect. Dis. 44901-903. [DOI] [PubMed] [Google Scholar]
- 8.Lark, R. L., S. A. McNeil, K. VanderHyde, A. Noorani, J. Uberti, and C. Chenoweth. 2001. Risk factors for anaerobic bloodstream infections in bone marrow transplant recipients. Clin. Infect. Dis. 33338-343. [DOI] [PubMed] [Google Scholar]
- 9.Lassmann, B., D. R. Gustafson, C. M. Wood, and J. E. Rosenblatt. 2007. Reemergence of anaerobic bacteremia. Clin. Infect. Dis. 44895-900. [DOI] [PubMed] [Google Scholar]
- 10.Lau, S. K. P., P. C. Y. Woo, A. M. Y. Fung, K. Chan, G. K. S. Woo, and K. Yuen. 2004. Anaerobic, non-sporulating, gram-positive bacilli bacteremia characterized by 16S rRNA gene sequencing. J. Med. Microbiol. 531247-1253. [DOI] [PubMed] [Google Scholar]
- 11.Lombardi, D. P., and N. C. Engleberg. 1992. Anaerobic bacteremia: incidence, patient characteristics, and clinical significance. Am. J. Med. 9253-60. [DOI] [PubMed] [Google Scholar]
- 12.Miller, D. L., S. Brazer, D. Murdoch, L. B. Reller, and G. R. Corey. 2001. Significance of Clostridium tertium bacteremia in neutropenic and nonneutropenic patients: review of 32 cases. Clin. Infect. Dis. 32975-978. [DOI] [PubMed] [Google Scholar]
- 13.Simmon, K. E., A. C. Croft, and C. A. Petti. 2006. Application of SmartGene IDNS software to partial 16S rRNA gene sequences for a diverse group of bacteria in a clinical laboratory. J. Clin. Microbiol. 444400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snydman, D. R., N. V. Jacobus, L. A. McDermott, R. Ruthazer, Y. Golan, E. J. C. Goldstein, S. M. Finegold, L. J. Harrell, D. W. Hecht, S. G. Jenkins, C. Pierson, R. Venezia, V. Yu, J. Rihs, and S. L. Gorbach. 2007. National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends in the United States from 1997 to 2004. Antimicrob. Agents Chemother. 511649-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song, Y., C. Liu, M. Bolanos, J. Lee, M. McTeague, and S. M. Finegold. 2005. Evaluation of 16S rRNA sequencing and reevaluation of a short biochemical scheme for identification of clinically significant Bacteroides species. J. Clin. Microbiol. 431531-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song, Y., C. Liu, and S. M. Finegold. 2007. Development of a flow chart for identification of gram-positive anaerobic cocci in the clinical laboratory. J. Clin. Microbiol. 45512-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song, Y., C. Liu, and S. M. Finegold. 2007. Peptoniphilus gorbachii sp. nov., Peptoniphilus olsenii sp. nov., and Anaerococcus murdochii sp. nov. isolated from clinical specimens of human origin. J. Clin. Microbiol. 451746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song, Y., C. Liu, M. McTeague, and S. M. Finegold. 2003. 16S ribosomal DNA sequence-based analysis of clinically significant anaerobic cocci. J. Clin. Microbiol. 411363-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstein, M. P., M. L. Towns, S. M. Quartey, S. Mirrett, L. G. Reimer, G. Parmigiani, and L. B. Reller. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis. 24584-602. [DOI] [PubMed] [Google Scholar]