Abstract
Aelurostrongylus abstrusus (Nematoda, Strongylida, Metastrongyloidea) is a cosmopolitan parasite of cats and causes severe respiratory distress. Information on the biology and epidemiology of feline aelurostrongylosis is fragmentary, mainly due to the limits inherent in the classical diagnosis. In the present work, a two-step nested PCR based on the use of genetic markers in the second internal transcribed spacer (ITS2) of ribosomal DNA was established for A. abstrusus in different biological samples. Characterization of the ITS2 (321 bp of length) revealed a G+C content of 39.5%. To exploit the sequence difference between the ITS2 of A. abstrusus and those of other common feline endoparasites, specific primers were designed and tested by PCR for their specificities and sensitivities. The PCR assay was validated on a panel of fecal (i.e., feces, flotation supernatant, and Baermann sediment) and pharyngeal swab samples from cats with known histories of lungworm infections, and it showed a specificity of 100% and a sensitivity of up to 96.6%. Also, the nested PCR was able to identify cats that were actually infected but that tested negative by the classical diagnostic methods. This PCR method was shown to be a powerful tool for the molecular diagnosis of feline aelurostrongylosis, overcoming the constraints of the classical diagnosis. The implications of such a molecular tool for further bioepidemiological studies of both intermediate and definitive hosts have been discussed.
Adults of the lungworm Aelurostrongylus abstrusus (Nematoda, Strongylida, Metastrongyloidea) live in the respiratory bronchioles and alveolar ducts of domestic cats and other felids. After mating, the females produce eggs that embryonate and hatch within the pulmonary ducts and alveoli. The first-stage larvae (L1s) pass up the bronchial escalator and are swallowed and released via the feces into the environment (2, 4). L1s are then ingested by slugs and snails that act as intermediate hosts (14, 17), while rodents, frogs, lizards, snakes, and birds can be paratenic hosts (2, 4). The felid definitive hosts become infected by ingesting a mollusk and/or a paratenic host (2).
The disease in cats is clinically relevant, as the parasite's eggs and larvae induce damage of the lung parenchyma (4, 23), inducing pulmonary effects ranging from minimal respiratory signs (e.g., occasional cough) to bronchopneumonia, open-mouthed abdominal breathing, intense and chronic coughing, sneezing, mucopurulent discharge, dyspnea, and hydrothorax; also, the disease can be fatal due to respiratory failure (13, 20, 26).
Infection by A. abstrusus has a worldwide distribution and in the past few years has been increasingly reported in America (10, 11) and Europe (7, 12, 18, 21), with a prevalence of up to about 20% in areas of endemicity (15).
Although the infection appears to be emerging and severe, there are still significant gaps in the knowledge of cat aelurostrongylosis, mainly due to the difficulties in its diagnosis in living animals. Indeed, the disease poses a significant diagnostic challenge, since the clinical presentation is hard to differentiate from those of other feline respiratory diseases. Furthermore, inapparent or subclinical infections may occur, thus impairing a reliable diagnosis by parasitological and collateral imaging approaches. More specifically, many cases of infection are not diagnosed, since cat feces are not routinely examined by the Baermann migration method, which is considered the gold standard for the diagnosis of infections, although its sensitivity is 90% or less (31). Other coprological methods, i.e., direct fecal smear and fecal flotation, are less accurate due to the irregularity of larval presence and sample size. Furthermore, the flotation approaches show inherent limits, depending on the time of flotation and on the dehydration of the larvae with certain salt solutions (23).
The characterization of the internal transcribed spacer 1 (ITS1) and ITS2 of nuclear ribosomal DNA (rDNA) recently defined the basis for the molecular identification of a huge range of parasites of veterinary importance, including lungworms (6). Moreover, highly sensitive two-step PCR-based assays using genetic markers within the ITSs have allowed researchers to overcome some constraints of the conventional diagnosis for a range of parasitic nematodes, e.g., human nodule worm and hookworm infections (22, 29), filariasis (9), and equine habronemiasis (27).
Given the limits of the classical diagnosis of aelurostrongylosis, there is a significant interest in molecular studies of A. abstrusus instrumentally leading to a PCR-based detection of the pathogen. Therefore, the aim of the present study was to validate a molecular assay for the diagnosis of cat aelurostrongylosis in different biological samples by using genetic markers within the ribosomal ITS2 of A. abstrusus.
MATERIALS AND METHODS
Characterization of the A. abstrusus ITS2.
Single fecal samples were collected from 30 cats previously diagnosed as infected by A. abstrusus, and L1s were sedimented, concentrated, and isolated with the Baermann migration technique (8). Twenty-five of the 30 cats were diagnosed also with a mixed infection caused by other endoparasites, i.e., 11 cats were infected by ascarids, 4 by hookworms, 4 by ascarids and hookworms, 3 by tapeworms, 2 by coccidia, and 1 by ascarids and tapeworms.
Genomic DNA was individually extracted from each of the 30 A. abstrusus larval samples by using a commercial kit (i.e., Qiagen stool minikit; Qiagen Gmbh, Germany). The ITS2 plus 5.8S and 28S rRNA flanking regions of A. abstrusus were individually PCR amplified from all DNA extracts. PCR mixtures were prepared in 50-μl reaction mixtures containing 100 pmol of each of the two rDNA conserved oligonucleotide primers, NC1 (5′ACGTCTGGTTCAGGGTTGTT3′; designed within the ribosomal 5.8S flanking region of Caenorhabditis elegans; GenBank accession number X03680) (forward) and NC2 (5′TTAGTTTCTTTTCCTCCGCT3′; designed within the ribosomal 28S flanking region of C. elegans, GenBank accession number X03680) (reverse) (6), 5 μl of template, and 25 μl of Red Taq ready mix (Sigma-Aldrich, St. Louis, MO). Reactions were performed on an Applied Biosystems 2700 thermocycler as follows: 94°C for 7 min and 40 cycles at 94°C for 45 s, 50°C for 45 s, and 72°C for 45 s, followed by a final extension for 10 min at 72°C.
Amplicons were electrophoresed in a 1.6% agarose gel, stained with ethidium bromide (10 mg/ml), and photographed using a documentation system (Gel Doc 2000; Bio-Rad, Hercules, CA). All PCR products were further purified by using Ultrafree-DA columns (Millipore, Billerica, MA) and then sequenced using the Taq DyeDeoxy Terminator cycle sequencing kit (version 2; Applied Biosystems, Foster City, CA) in the automated sequencer ABI Prism 377. The ITS2 sequences obtained were determined in both directions, and the electropherograms were verified by eye with the software Chromas Lite (version 2.01). The sequences were aligned using the computer software program DAMBE (version 4.5.55) (32), and the 5′ and 3′ ends were determined by comparisons with those of C. elegans (GenBank accession number X03680) and other Metastrongyloidea nematodes. In particular, the ITS2 sequences obtained were compared with those of the rDNA of other metastrongylid nematodes available in the GenBank by using the nucleotide-nucleotide BLAST (1). Then the A. abstrusus ITS2 sequences were further compared with one another, and the software MEGA (version 3.1) (16) was used to calculate the nucleotide pairwise distance with the Kimura two-parameter model among the A. abstrusus ITS2 sequence and the same sequences of other Metastrongyloidea.
Establishing a diagnostic PCR assay for the specific detection of A. abstrusus rDNA.
The primer set AabFor (forward 5′GTAACAACGATATTGGTACTATG3′; ITS2 oligonucleotidic residues 62 to 84) and AabRev (reverse 5′GAACTCCTTCACGTGCTACTCG3′; ITS2 oligonucleotidic residues 273 to 294) was designed by following general criteria (24) for the consensus ITS2 sequences of A. abstrusus in regions without intraspecific polymorphisms which also displayed the greatest interspecific difference with the three most common parasitic nematodes of cats, i.e., Toxocara cati (GenBank accession number Y09493), Ancylostoma tubaeforme (GenBank accession number AJ001592), and Uncinaria stenocephala (GenBank accession number AF194145).
The ability of a nested-PCR protocol to achieve specific amplification of an ITS2 internal segment (i.e., 233 bp) from A. abstrusus was evaluated with a set of biological samples (i.e., direct fecal sample, flotation supernatant sample, Baermann sediment sample, and pharyngeal swab) collected from each of the 30 cats affected by aelurostrongylosis (see the previous section). In particular, primer sets NC1-NC2 and AabFor-AabRev were used in the first and second rounds, respectively. Genomic DNA was extracted from samples of fecal origin, as described above, and from pharyngeal swabs, as described previously (28). Common endoparasites of cats collected by the authors or provided by colleagues (see Acknowledgments) and from cat blood and lung tissue (Table 1) were subjected to DNA extraction by the Qiagen DNeasy tissue kit (Qiagen Gmbh, Germany). The protocol of the nested PCR was optimized by consequent trials (i.e., different titrations of primer concentrations and various annealing times, temperatures, and cycle numbers) to achieve the highest number of PCR amplicons from each kind of biological sample. The optimal conditions of the PCR mixtures of the first round were the same as those for the protocol described above for the amplification of the ITS2 from A. abstrusus L1s. The optimized mixture conditions of the second round were as follows: 50-μl reaction mixture containing 200 pmol of each AabFor and AabRev primer, 3 μl of a 1/20 dilution of each NC1-NC2 amplicon as the template, and 25 μl of Red Taq ready mix (Sigma-Aldrich, St. Louis, MO). The cycling protocol used in both rounds was the same as that used for the ITS2 characterization, and all AabFor-AabRev amplicons produced in the second round were electrophoresed and sequenced as described above.
TABLE 1.
DNA samples used to verify the specificity of the nested PCR assay for the specific identification of A. abstrusus rDNAa
| Sample | Stage |
|---|---|
| Cystoisospora felis | O |
| Cystoisospora rivolta | O |
| Giardia spp. | C |
| Mesocestoides lineatus | Ssa |
| Dipylidium caninum | Ssa |
| Taenia taeniaeformis | Ssa |
| Ancylostoma tubaeforme | Sa |
| Uncinaria stenocephala | Sa |
| Ollulanus tricuspis | Sa |
| Oslerus rostratus | Sa |
| Toxocara cati | Sa |
| Toxascaris leonina | Sa |
| Eucoleus aerophilus | Sa |
| Feline blood | |
| Feline lung tissue |
O, concentrated oocysts of fecal origin; C, concentrated cysts of fecal origin; Ssa, single segments from adult specimens; Sa, single adult specimen.
The presence of inhibition in the PCRs that did not produce any amplicon detectable on agarose gels was verified by spiking the DNA of A. abstrusus into DNA extracts (27).
Application of nested PCR in a clinical setting.
To verify the applicability of nested PCR for diagnosis of aelurostrongylosis, individual fecal samples and pharyngeal swabs were collected from 50 cats that were admitted to private veterinary clinics; the cats were suspected of having lungworm infections based on histories of respiratory signs and anamneses, i.e., all 50 animals (younger than 1.5 years) were free roaming and came from an area of endemicity for A. abstrusus in central Italy.
All fecal samples were subjected to routine coprological diagnosis, i.e., direct smear, flotation procedures by using both a sugar and a zinc sulfate solution, with specific gravities of 1,200 and of 1,350, respectively, and the Baermann technique.
The direct smear was performed by using ∼0.5 g of feces mixed with a drop of saline solution on a microscope slide to produce a thin layer that was examined under a microscope (Axioskop 40; Zeiss, Oberkochen, Germany) at a magnification of ×20. Approximately 3 g of feces was added to 20 ml of flotation solution and centrifuged at 600 × g for 5 min. A supernatant aliquot of ∼100 μl was aspirated with a Pasteur pipette, transferred to a glass slide, and examined using a light microscope at a magnification of ×20. The Baermann technique was performed by using 5 to 10 g from each sample (8).
All parasites retrieved at the coprological examination were identified according to morphological keys, and A. abstrusus L1s were recognized by their characteristic notched and S-shaped caudal ends (23, 25).
Genomic DNA was extracted from the 50 pharyngeal swabs as described previously (28), and all the DNA extracts were subjected to the nested-PCR assay specific for the 233-bp-long fragment internal to the A. abstrusus ITS2 (described above). A positive control (larval A. abstrusus DNA) and a negative control (sterile water) were included in every run, and all amplicons produced in the second round were electrophoresed and sequenced as described above. All the molecular procedures were performed in separate rooms (i.e., DNA preparation and pre-PCR steps were carried out in a room different than that used for post-PCR manipulation) to avoid PCR contamination. All procedures were validated twice.
Nucleotide sequence accession number.
Nucleotide sequence data for ITS2 of the ribosomal DNA plus partial flanking regions of the 5.8S and 28S ribosomal rRNA genes of A. abstrusus have been registered in the GenBank database under accession number EU034168.
RESULTS
The primer set NC1-NC2 yielded amplicons of ∼500 bp from A. abstrusus L1s recovered from the 30 infected cats. ITS2 sequences showed no variation in length (321 bp), a G+C content of 39.5%, and two polymorphic nucleotidic positions (i.e., two C→T transitions at ITS2 residues 183 and 219), with an intraspecific difference of 0.6%.
The ITS2 sequence of A. abstrusus showed consistency with ribosomal sequences of 20 other metastrongylid nematodes, with a nucleotide pairwise distance ranging from 0.631 (A. abstrusus versus Umingmakstrongylus pallikuukensis) to 1.823 (A. abstrusus versus Torynurus convolutus) (Table 2).
TABLE 2.
Distance matrix showing the nucleotide pairwise distane calculated by MEGA 3.1 among ITS2 sequences
| Nematode | A. abstrusus | A. tubaeforme | T. cati | U. stenocephala | Meta-strongylus salmi | Meta-strongylus pudendotectus | Meta-strongylus elongatus | Meta-strongylus confusus | Meta-strongylus asymmetricus | Hovor-konema variegatum | Angio-strongylus cantonensis | Angio-strongylus vasorum | Angio-strongylus costaricensis | Muellerius capillaris | Umingmak-strongylus pallikuukensis | Elapho-strongylus rangiferi | Vare-strongylus alpena | Parelapho-strongylus odocoilei | Oto-strongylus circumlitus | Torynurus convolutus | Parelapho-strongylus andersoni | Parelapho-strongylus tenuiss | Elapho-strongylus alces |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. tubaeforme | 0.943 | ||||||||||||||||||||||
| T. cati | 0.817 | 1.730 | |||||||||||||||||||||
| U. stenocephala | 0.934 | 0.153 | 1.806 | ||||||||||||||||||||
| Metastrongylus salmi | 0.719 | 0.864 | 1.184 | 0.776 | |||||||||||||||||||
| Metastrongylus pudendotectus | 0.787 | 1.174 | 1.196 | 1.107 | 0.594 | ||||||||||||||||||
| Metastrongylus elongatus | 0.660 | 0.936 | 1.198 | 0.859 | 0.297 | 0.351 | |||||||||||||||||
| Metastrongylus confusus | 1.208 | 1.660 | 1.833 | 1.487 | 1.074 | 1.046 | 0.872 | ||||||||||||||||
| Metastrongylus asymmetricus | 1.110 | 1.620 | 1.694 | 1.566 | 1.083 | 1.030 | 0.858 | 0.105 | |||||||||||||||
| Hovorkonema variegatum | 1.450 | 0.633 | 1.680 | 0.614 | 0.709 | 1.419 | 1.008 | 1.347 | 1.535 | ||||||||||||||
| Angiostrongylus cantonensis | 1.442 | 2.375 | 1.912 | 1.757 | 1.792 | 1.274 | 1.303 | 1.121 | 1.180 | 1.824 | |||||||||||||
| Angiostrongylus vasorum | 1.289 | 1.471 | 1.832 | 1.383 | 1.040 | 1.137 | 1.078 | 1.318 | 1.402 | 1.360 | 0.666 | ||||||||||||
| Angiostrongylus costaricensis | 1.365 | 1.520 | 2.499 | 1.406 | 1.001 | 1.071 | 1.017 | 1.328 | 1.342 | 1.699 | 0.770 | 0.307 | |||||||||||
| Muellerius capillaris | 0.725 | 0.870 | 1.364 | 0.799 | 0.612 | 0.708 | 0.631 | 1.128 | 1.223 | 0.793 | 1.455 | 1.369 | 1.323 | ||||||||||
| Umingmakstrongylus pallikuukensis | 0.631 | 1.081 | 1.324 | 0.999 | 0.425 | 0.674 | 0.467 | 1.036 | 1.109 | 0.659 | 1.696 | 1.170 | 1.308 | 0.353 | |||||||||
| Elaphostrongylus rangiferi | 0.774 | 0.936 | 1.351 | 0.828 | 0.554 | 0.726 | 0.604 | 0.816 | 0.864 | 0.724 | 1.021 | 1.001 | 1.050 | 0.589 | 0.397 | ||||||||
| Varestrongylus alpena | 0.933 | 1.088 | 1.690 | 0.941 | 0.634 | 0.950 | 0.663 | 1.368 | 1.414 | 0.862 | 1.648 | 1.148 | 1.496 | 0.581 | 0.445 | 0.540 | |||||||
| Parelaphostrongylus odocoilei | 0.759 | 0.970 | 1.151 | 0.892 | 0.474 | 0.647 | 0.598 | 0.790 | 0.869 | 0.665 | 0.874 | 0.880 | 1.004 | 0.597 | 0.384 | 0.137 | 0.502 | ||||||
| Otostrongylus circumlitus | 1.227 | 1.191 | 1.652 | 1.236 | 1.129 | 1.133 | 1.065 | 1.350 | 1.367 | 1.531 | 1.377 | 1.520 | 1.629 | 1.215 | 1.048 | 0.979 | 1.219 | 1.019 | |||||
| Torynurus convolutus | 1.823 | 1.628 | 1.634 | 1.742 | 1.634 | 1.521 | 1.595 | 1.233 | 1.075 | 1.446 | 1.523 | 1.685 | 1.303 | 1.432 | 1.594 | 1.038 | 1.520 | 1.058 | 2.036 | ||||
| Parelaphostrongylus andersoni | 0.708 | 0.852 | 1.126 | 0.784 | 0.451 | 0.634 | 0.559 | 0.760 | 0.833 | 0.637 | 0.845 | 0.838 | 0.961 | 0.556 | 0.387 | 0.120 | 0.494 | 0.013 | 0.967 | 1.054 | |||
| Parelaphostrongylus tenuiss | 0.744 | 0.932 | 1.167 | 0.857 | 0.467 | 0.651 | 0.573 | 0.798 | 0.862 | 0.673 | 0.851 | 0.857 | 1.011 | 0.570 | 0.387 | 0.122 | 0.490 | 0.018 | 1.033 | 1.095 | 0.013 | ||
| Elaphostrongylus alces | 0.683 | 0.923 | 1.266 | 0.865 | 0.554 | 0.757 | 0.619 | 0.843 | 0.924 | 0.663 | 1.021 | 0.980 | 1.020 | 0.560 | 0.425 | 0.055 | 0.545 | 0.143 | 0.991 | 1.105 | 0.129 | 0.135 | |
| Elaphostrongylus cervi | 0.769 | 0.936 | 1.344 | 0.828 | 0.671 | 0.726 | 0.604 | 0.775 | 0.784 | 0.946 | 1.013 | 0.995 | 1.044 | 0.718 | 0.488 | 0.000 | 0.644 | 0.156 | 0.931 | 0.997 | 0.136 | 0.137 | 0.063 |
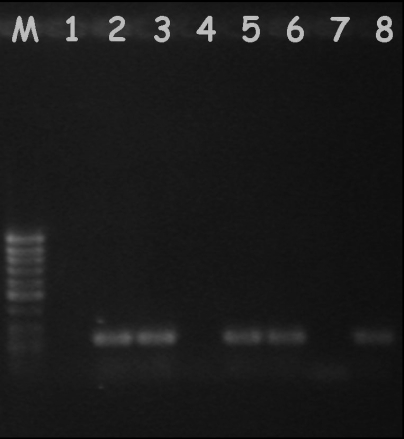
Among each fecal (i.e., feces, flotation supernatant, and Baermann sediment) and pharyngeal mucus sample collected from the 30 infected cats and subjected to the nested PCR, the second round with the primer set AabFor-AabRev yielded amplicons for 24 feces (80%) samples, 28 supernatant (93.3%) samples, and 29 Baermann sediment samples and pharyngeal swabs (96.6%) (Fig. 1). No amplicons were produced from any of the control samples included in the evaluation of the nested-PCR assay (Table 1). PCR amplification conducted on samples spiked with DNA from A. abstrusus demonstrated no evidence of inhibition in the PCR, since an amplicon of the expected size (∼200 bp) was detected.
FIG. 1.
Example of an agarose gel showing results from the nested PCR of fecal and pharyngeal samples. Lane M, size marker; lane 1, negative fecal sample; lanes 2 and 3, positive samples from flotation supernatants and Baermann sediment, respectively; lane 4, negative flotation sample; lanes 5 and 6, positive pharyngeal samples; lane 7, negative control (no-DNA sample); lane 8, positive control (A. abstrusus DNA).
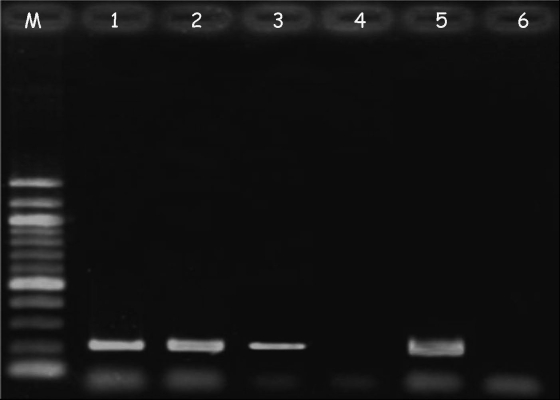
Table 3 reports the results of the fecal examination of the 50 cats with clinical respiratory signs enrolled into the study, together with the results of the molecular diagnosis. In particular, six animals tested positive for L1s of A. abstrusus for at least one of the coprological tests performed (Table 3). The pharyngeal swabs from the same six cats and from five more animals that were negative by the fecal examination were positive by the nested-PCR assay (Fig. 2). The remaining 39 cats were negative by coprological and molecular examinations for A. abstrusus (Table 3). Sequencing of all amplicons obtained by the second round with the primer set AabFor-AabRev confirmed that they represented the appropriate species A. abstrusus.
TABLE 3.
Cats with clinical respiratory signs determined to be infected or not infected with Aelurostrongylus abstrusus, coccidia, ascarids, tapeworms, and hookworms at examination by direct smear, flotation, Baermann method, and nested PCR on pharyngeal swabsa
| Cat nos. | Direct smear result
|
Flotation result
|
Baermann method result
|
Pharyngeal swabs | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. abstrusus | Coccidia | Ascarids | Tapeworms | Hookworms | A. abstrusus | Coccidia | Ascarids | Tapeworms | Hookworms | A. abstrusus | Coccidia | Ascarids | Tapeworms | Hookworms | ||
| 1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 5 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 6 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 7 | + | − | − | − | − | + | − | − | − | − | + | − | − | − | − | + |
| 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 9 | − | − | + | − | − | − | − | + | − | − | − | − | + | − | − | − |
| 10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 11 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 12 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| 13 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| 14 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 15 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 16 | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| 17 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 18 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 19 | + | − | + | − | − | − | − | + | − | − | + | − | + | − | − | + |
| 20 | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + |
| 21 | + | − | + | − | − | − | − | + | − | − | + | − | + | − | − | + |
| 22 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 23 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| 24 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 25 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 26 | + | − | − | − | − | − | − | + | − | − | + | − | − | − | − | + |
| 27 | − | − | + | − | + | − | − | + | − | + | + | − | − | − | − | + |
| 28 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 29 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 30 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 31 | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| 32 | − | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − |
| 33 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + |
| 34 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 35 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 36 | − | + | + | − | − | − | + | + | − | − | − | − | − | − | − | − |
| 37 | − | − | + | − | + | − | − | + | − | + | − | − | − | − | − | − |
| 38 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 39 | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − | + |
| 40 | − | + | + | − | − | − | + | + | − | − | − | − | − | − | − | + |
| 41 | − | − | + | − | + | − | − | + | − | + | − | − | − | − | − | − |
| 42 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 43 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 44 | − | + | + | − | − | − | + | + | − | − | − | − | − | − | − | − |
| 45 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 46 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 47 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 48 | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − | + |
| 49 | − | − | + | + | − | − | − | + | + | − | − | − | − | − | − | − |
| 50 | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − |
+, infected; −, not infected.
FIG. 2.
Example of an agarose gel showing results from the nested PCR of pharyngeal samples collected from symptomatic animals negative at the microscopic fecal examination. Lane M, size marker; lanes 1 to 3, positive samples from animals 13, 33, and 39 (Table 2); lane 4, negative sample from animal 50; lane 5, positive control (A. abstrusus DNA); lane 6, negative control (no-DNA sample).
DISCUSSION
The PCR approach herein validated with the ribosomal ITS2 of A. abstrusus proved to be useful for the molecular detection and identification of the lungworm from a range of biological samples. Indeed, the two-step nested PCR was demonstrated to be a powerful method to identify A. abstrusus from feces and pharyngeal swabs, showing a specificity of 100% and a sensitivity ranging from 80 to 96.6%. All of the PCRs yielded amplicons of the expected size (i.e., ∼200 bp), regardless of coinfections with other endoparasites (i.e., coccidia, tapeworms, ascarids, and hookworms). The 100% specificity of the assay was also confirmed by the sequencing of all amplicons produced by the second round of the nested PCR and by the absence of control sample amplification (Table 1). Conversely, failure to detect A. abstrusus DNA in one Baermann and one pharyngeal sample, coming from 1 of the 30 animals with a history of aelurostrongylosis, might be due to the concentration or the quality of parasitic DNA. The value of diagnostic sensitivity (i.e., ∼97%) is in accordance with those previously recorded for other parasites by using two-step PCR approaches on fecal or pharyngeal samples (27, 28, 30), and it is higher than that recorded by employing classical diagnostic methodologies. The higher percentage of sensitivity shown by the two-step PCR over the conventional diagnostic methods is likely due to the specific amplification of the parasites' water-soluble DNA molecules present in feces and in pharyngeal swabs.
Accordingly, when the molecular approach was employed for the pharyngeal swab samples from 50 symptomatic animals from the field, 5 animals that were negative by classical coprological examination were positive by PCR (Table 3; Fig. 2). Six animals were positive by both coprology and nested PCR, while 39 were negative. Even though no confirmation at necropsy was possible, it is arguable that those 39 cats testing negative were actually not infected by feline lungworms. Indeed, the same animals were later diagnosed ex juvantibus with upper respiratory tract disease complex (i.e., 35 animals received this diagnosis) or with nasopharyngeal polyps (i.e., 4 animals received this diagnosis).
The validation of the nested PCR on the set of samples collected from the 30 cats with known histories of aelurostrongylosis demonstrated that the pharyngeal swab has an overall sensitivity similar to that of Baermann sediment for detecting A. abstrusus DNA. Nonetheless, the present experiment demonstrates that the pharyngeal swab is the most suitable sample instrumental to the molecular diagnosis of feline lungworm infection for a range of practical considerations (i.e., difficulties in collecting adequate stool samples in the field and long parasite prepatent and migration periods), which may lead to a lack of DNA readily available for PCR amplification. Moreover, DNA extraction from feces is more laborious than that from pharyngeal mucus and may imply the presence of PCR inhibitors.
From a clinical standpoint, the assay presented herein can be used as a reliable tool for detecting A. abstrusus DNA in pharyngeal swabs from clinically infected cats, thus having implications for diagnosis. The difficulties inherited with a reliable clinical diagnosis of cat aelurostrongylosis is worthy of note, as this disease may share symptoms with other respiratory feline diseases (e.g., nematode infections by Dirofilaria immitis and Eucoleus aerophilus), presenting with similar respiratory distress and signs and abnormal radiographic and hematologic findings (5, 10, 11, 19, 31). Also, the feline respiratory tract may be infected by several viruses and bacteria causing infections with aelurostrongylosis-like symptoms (10, 11). Hence, as a consequence, several cases of aelurostrongylosis may not be clinically included in differential diagnosis and, thus, cats are often not subjected to coprology.
It is also known that the fecal examinations used routinely (i.e., direct fecal smear and fecal flotation) may be inaccurate, since L1s may not be present and/or the feces amount used is inadequate (Table 3). The coprological procedure of flotation with sugar solution was confirmed to be less sensitive than that with salt solution (Table 3) (15). Nonetheless, flotation with concentrated salt solutions implies limits inherent to L1 sink and dehydration, leading to morphological alteration (3, 23). An unequivocal diagnosis of A. abstrusus infection is of crucial importance to the timeliness of treatments, thus resulting in a positive prognosis for the infected animals.
Based on the present results, the nested PCR validated herein is the most reliable method for the diagnosis of feline aelurostrongylosis in a clinical setting and it represents a new tool for the evaluation of the efficacies of anthelmintic compounds against feline lungworm under laboratory and field conditions. The ethical implications of using such a method to monitor the decline/absence of parasite-specific DNA in feces or pharyngeal mucus in treated animals is of relevance, since it allows us to circumvent the need to sacrifice infected cats.
Other implications of the molecular assay for epidemiological studies of cat aelurostrongylosis are related to better understanding of the ecology, epidemiology, seasonal occurrence, and transmission patterns of A. abstrusus. For instance, this nested assay could be employed in studies to identify mollusks acting as intermediate hosts of A. abstrusus in those geographical regions where their identities are still unknown.
Acknowledgments
We are particularly grateful to Fredric Frye (Cloverdale, CA) for his valuable advice and suggestions for the manuscript. We also thank Sharon Coleman (Louisiana State University, Baton Rouge, LA) for providing some specimens of cat endoparasites.
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. C. 2000. The superfamily Metastrongyloidea, p. 163-164. In Nematode parasites of vertebrates. Their development and trasmission. C.A.B. International, Wallingford, Oxfordshire, United Kingdom.
- 3.Bonsembiante, P., and B. Di Sacco. 1991. Protocollo diagnostico delle parassitosi polmonari e gastrointestinali dei piccoli animali. Suppl. Vet. 425-27. [Google Scholar]
- 4.Bowman, D. D., C. M. Hendrix, D. S. Lindsay, and S. C. Barr. 2002. Feline clinical parasitology, p. 267-271. Iowa State University Press (a Blackwell Science company), Ames, IA.
- 5.Calvert, C. A., and C. P. Mandell. 1982. Diagnosis and management of feline heartworm disease. J. Am. Vet. Med. Assoc. 180550-552. [PubMed] [Google Scholar]
- 6.Chilton, N. B. 2004. The use of nuclear ribosomal DNA markers for the identification of bursate nematodes (order Strongylida) and for the diagnosis of infections. Anim. Health Res. Rev. 5173-187. [DOI] [PubMed] [Google Scholar]
- 7.Epe, C., N. Coati, and T. Schnieder. 2004. Results of parasitological examinations of faecal samples from horses, ruminants, pigs, dogs, cats, hedgehogs and rabbits between 1998 and 2002. Dtsch. Tierarztl. Wochenschr. 6243-247. [PubMed] [Google Scholar]
- 8.Euzeby, J. 1981. Helminthes parasites de l'appareil respiratoire, p. 200-202. In Diagnostic expérimental des helminthoses animales, vol. 2. Informations Techniques des Services Vétérinaires, Paris, France. [Google Scholar]
- 9.Fischer, P., D. W. Büttner, J. Bamuhiiga, and S. A. Williams. 1998. Detection of the filarial parasite Mansonella streptocerca in skin biopsies by a nested polymerase chain reaction-based assay. Am. J. Trop. Med. Hyg. 58816-820. [DOI] [PubMed] [Google Scholar]
- 10.Foster, S. F., P. Martin, G. S. Allan, V. R. Barrs, and R. Malik. 2004. Lower respiratory tract infections in cats: 21 cases (1995-2000). J. Feline Med. Surg. 6167-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, S. F., P. Martin, J. A. Braddock, and R. Malik. 2004. A retrospective analysis of feline bronchoalveolar lavage cytology and microbiology (1995-2000). J. Feline Med. Surg. 6189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandi, G., L. E. Calvi, L. Venco, C. Paratici, C. Genchi, D. Memmi, and L. H. Kramer. 2005. Aelurostrongylus abstrusus (cat lungworm) infection in five cats from Italy. Vet. Parasitol. 134177-182. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton, J. M. 1963. Aelurostrongylus abstrusus infestation of the cat. Vet. Rec. 75417-422. [Google Scholar]
- 14.Hamilton, J. M., and A. W. McCaw. 1968. The output of first stage larvae by cats infested with Aelurostrongylus abstrusus. J. Helminthol. 42295-298. [DOI] [PubMed] [Google Scholar]
- 15.Iorio, R., and D. Traversa. New epidemiological and molecular insights into feline lungworm infection. Ann. N. Y. Acad. Sci., in press. [DOI] [PubMed]
- 16.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 17.López, C., R. Panadero, A. Paz, R. Sanchez-Andrade, P. Diaz, P. Diez-Banos, and P. Morrondo. 2005. Larval development of Aelurostrongylus abstrusus (Nematoda, Angiostrongylidae) in experimentally infected Cernuella (Cernuella) virgata (Mollusca, Helicidae). Parasitol. Res. 9513-16. [DOI] [PubMed] [Google Scholar]
- 18.Miro, G., A. Montoya, S. Jimenez, C. Frisuelos, M. Mateo, and I. Fuentes. 2004. Prevalence of antibodies to Toxoplasma gondii and intestinal parasites in stray, farm and household cats in Spain. Vet. Parasitol. 3249-255. [DOI] [PubMed] [Google Scholar]
- 19.Pechman, R. D., Jr. 1984. Newer knowledge of feline bronchopulmonary disease. Vet. Clin. N. Am. Small Anim. Pract. 141007-1019. [DOI] [PubMed] [Google Scholar]
- 20.Ribeiro, V. M., and W. S. Lima. 2001. Larval production of cats infected and re-infected with Aelurostrongylus abstrusus (Nematoda: Protostrongylidae). Rev. Med. Vet. 152815-820. [Google Scholar]
- 21.Robben, S. R. M., W. E. Nobel, D. Dopfer, W. M. L. Hendrikx, J. H. Boersema, F. Fransen, and M. E. Eysker. 2004. Infections with helminths and/or protozoa in cats in animal shelters in the Netherlands. Tijdschr. Diergeneeskd. 12-6. [PubMed] [Google Scholar]
- 22.Romstad, A., R. B. Gasser, J. R. Monti, A. M. Polderman, P. Nansen, D. S. Pit, and N. B. Chilton. 1997. Differentiation of Oesophagostomum bifurcum from Necator americanus by PCR using genetic markers in spacer ribosomal DNA. Mol. Cell. Probes 11169-176. [DOI] [PubMed] [Google Scholar]
- 23.Scott, D. W. 1973. Current knowledge of Aelurostrongylus abstrusus in the cat. Cornell Vet. 63483-500. [PubMed] [Google Scholar]
- 24.Sharrocks, A. D. 1994. The design of primers for PCR, p. 5-11. In PCR technology: current innovations. CRC Press, Inc., Boca Raton, FL.
- 25.Sloss, M. W., R. L. Kemp, and A. M. Zajac. 1994. Fecal examination: dogs and cats, p. 17-44. In Veterinary clinical parasitology, 6th ed. Iowa State University Press, Ames, IA.
- 26.Stockdale, P. H. G. 1970. The pathogenesis of the lesions elicited by Aelurostrongylus abstrusus during its prepatent period. Pathol. Vet. 7102-115. [DOI] [PubMed] [Google Scholar]
- 27.Traversa, D., A. Giangaspero, R. Iorio, D. Otranto, B. Paoletti, and R. B. Gasser. 2004. Semi-nested PCR for the specific detection of Habronema microstoma or Habronema muscae DNA in horse faeces. Parasitology 129733-739. [DOI] [PubMed] [Google Scholar]
- 28.Traversa, D., and D. Otranto. 2006. A new approach for the diagnosis of myiasis of animals: the example of horse nasal myiasis. Vet. Parasitol. 141186-190. [DOI] [PubMed] [Google Scholar]
- 29.Verweij, J. J., A. M. Polderman, M. C. Wimmenhove, and R. B. Gasser. 2000. PCR assay for the specific amplification of Oesophagostomum bifurcum DNA from human faeces. Int. J. Parasitol. 30137-142. [DOI] [PubMed] [Google Scholar]
- 30.Verweij, J. J., D. S. Pit, L. van Lieshout, S. M. Baeta, G. D. Dery, R. B. Gasser, and A. M. Polderman. 2001. Determining the prevalence of Oesophagostomum bifurcum and Necator americanus infections using specific PCR amplification of DNA from faecal samples. Trop. Med. Int. Health 6726-731. [DOI] [PubMed] [Google Scholar]
- 31.Willard, M. D., R. E. Roberts, N. Allison, R. B. Grieve, and K. Escher. 1988. Diagnosis of Aelurostrongylus abstrusus and Dirofilaria immitis infections in cats from a human shelter. Am. J. Vet. Res. 192913-916. [PubMed] [Google Scholar]
- 32.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92371-373. [DOI] [PubMed] [Google Scholar]