Abstract
Repetitive-sequence-based PCR (rep-PCR) using the DiversiLab system was investigated for identification of Aspergillus. Ninety-five clinical isolates, identified by conventional methods, and five ATCC strains were tested. Sequencing of the internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2) was performed on 2 isolates with discrepant rep-PCR and conventional results and 15 randomly selected outlier isolates. One isolate not identified by sequencing was excluded from analysis. After resolving discrepancies, all but one A. glaucus strain had ≥85% similarity to one or more strains of the same Aspergillus species in the mold database, thereby providing accurate identification. No isolate was misidentified.
Invasive aspergillosis is a serious opportunistic infection in the immunocompromised host. Although Aspergillus fumigatus is the most common pathogen, other species are being isolated with increased frequency (1, 4, 5, 8). Some species (e.g., Aspergillus niger and Aspergillus versicolor) are more often colonizers than a cause of invasive disease, and for a few species, the susceptibility pattern has therapeutic implications (e.g., Aspergillus terreus is innately resistant to amphotericin B). For these reasons, identification of Aspergillus to the species level is important.
Once growth is apparent, an isolate of Aspergillus can be identified to the species level in several days to a few weeks, based on growth rate, colony morphology, and microscopic features. In most clinical laboratories, A. fumigatus, A. niger, and perhaps A. flavus and A. terreus are easily recognized, but identification of the less commonly encountered species is more difficult, requiring considerable training in clinical mycology. Healy et al. (2) showed that the automated DiversiLab system (Bacterial Barcodes, Inc., Athens, Ga.), which uses repetitive-sequence-based PCR (rep-PCR) to discriminate among bacterial and fungal isolates, accurately identifies A. fumigatus, A. flavus, and A. terreus in less than 5 h from the appearance of colonies on a solid medium. A limitation of that study, however, was the inclusion of only three species, all from a single geographical location. The objective of this study was to more rigorously challenge the DiversiLab system by testing more Aspergillus species and including isolates from diverse geographic locations.
Isolates.
Ninety-five clinical isolates of Aspergillus (one isolate per patient: 17 of A. fumigatus; 17 of A. terreus; 13 of A. niger; 9 of A. flavus; 8 of A. nidulans; 5 each of A. sydowii, A. ochraceous, and A. versicolor; 4 of A. ustus; 4 of A. glaucus; 2 of A. candidus; 2 of A. versicolor group; and 1 each of A. carbonarius, A. parasiticus, A. japonicus, and A. clavatus), previously identified to the species level based on conventional testing (colony morphology and microscopic characteristics), and 5 ATCC strains (A. niger 16888, A. niger 10535, A. nidulans 10074, A. fumigatus 36607, and A. terreus 1012) were included in the study. All patient information associated with the clinical isolates was removed. Isolates were selected to include both commonly and infrequently encountered species, and for A. niger and A. terreus, additional isolates were chosen for further comparison because geographical data were available. Cultures, which had been stored at room temperature for up to 2 years, were transferred to Sabouraud dextrose agar slants, which were labeled with the species name and a code number, and incubated at 30°C.
rep-PCR.
Fungal samples were subcultured to Sabouraud dextrose agar plates and incubated at 30°C for 2 to 5 days. DNA was extracted from a 10-μl loop of each fungal mass using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Solona Beach, CA), extending the bead beat step to 30 min as recommended by Bacterial Barcodes for use with the DiversiLab mold kit when testing fungi. Using agarose gel electrophoresis and a molecular weight standard, DNA solutions were standardized to approximately 25 ng/μl, and genomic integrity was visualized. The fungal DNA was amplified using the DiversiLab mold kit (Bacterial Barcodes, Inc.) for DNA fingerprinting following the manufacturer's instructions. Briefly, 50 ng of genomic DNA was added to the rep-PCR master mix, along with 2.5 U AmpliTaq and 10× PCR buffer (Applied Bio System, Foster City, CA) for a 25-μl total reaction. Thermal cycling parameters were as follows: initial denaturation at 94°C for 2 min; 35 cycles of denaturation at 92°C for 30 s, annealing at 50°C for 30 s, and extension at 70°C for 90 s; and a final extension at 70°C for 3 min. rep-PCR products were detected and analyzed using the DiversiLab system (Bacterial Barcodes, Inc.). Further analysis was performed with the web-based DiversiLab software, version 3.3, which uses the Pearson correlation coefficient and unweighted-pair group method with arithmetic mean (UPGMA), to automatically compare the rep-PCR-based DNA fingerprints of unknown isolates. An isolate was given a species identification by rep-PCR if its fingerprint pattern had at least 85% similarity to one or more isolates of the same species in the mold library, which includes several species of Aspergillus, Fusarium, and Candida; some more commonly encountered dermatophytes; Blastomyces dermatitidis; Coccidioides species; and Histoplasma capsulatum.
Sequence analysis.
Sequencing of the internal transcribed spacer 1 (ITS1)-5.8S-ITS2 region was performed, as described by White et al. (9), to confirm the identification based on rep-PCR clustering in the following situations: (i) the identification differed from conventional testing, (ii) the fingerprint pattern of randomly selected isolates did not match that of any other isolate in the study with at least 85% similarity, and (iii) the fingerprint pattern did not match any species in the mold library with ≥85% similarity. Briefly, the fragments with an average size of 580 bp containing ITS regions were amplified. The amplicons were purified using the High Pure PCR product purification kit (Roche Diagnostics Co., Indianapolis, IN), and bidirectional sequencing was carried out using ITS1 and ITS4 primers and BigDye terminator V3.1 cycle sequencing kit. The products were purified by PERFORMA DTR gel filtration cartridges (EdgeBioSystems, Gaithersburg, MD) and sequenced on an ABI-377 sequencer (Applied Biosystems, Foster City, CA). Results were analyzed using Sequencing Analysis v3.3. The contiguous sequences were constructed using SeqMan (DNAStar, Inc., Madison, WI) and identified using BLAST on the NCBI website (http://www.ncbi.nlm.nih.gov). An UPGMA-based tree was created based on total ITS sequence identity. Average sequence similarity within ITS1 and ITS2 regions of each sample was tabulated using a reference sequence with the closest match. The identification provided by sequencing was considered correct.
Findings.
After rep-PCR testing, 2 of the 100 isolates clustered at >85% similarity with different species, as identified by conventional testing. For both of these isolates, sequencing confirmed the rep-PCR result: one isolate identified as A. ustus by conventional testing was A. flavus, and one identified as A. versicolor was A. niger. Sequencing also was performed to confirm the identification of 15 outlier isolates that did not cluster with any other isolate(s) in the study. One of these 15 isolates (initially called A. clavatus) was not identified by sequencing and, therefore, was excluded from the study. The original identification of two outlier isolates was in error: one A. nidulans isolate was A. unguis, and the A. japonicus isolate was A. aculeatus. Both isolates initially called members of the A. versicolor group were identified to the species level by sequencing (one was A. sydowii and the other was A. versicolor); rep-PCR correctly identified both to the species level. The identification of the remaining 10 outlier isolates was confirmed.
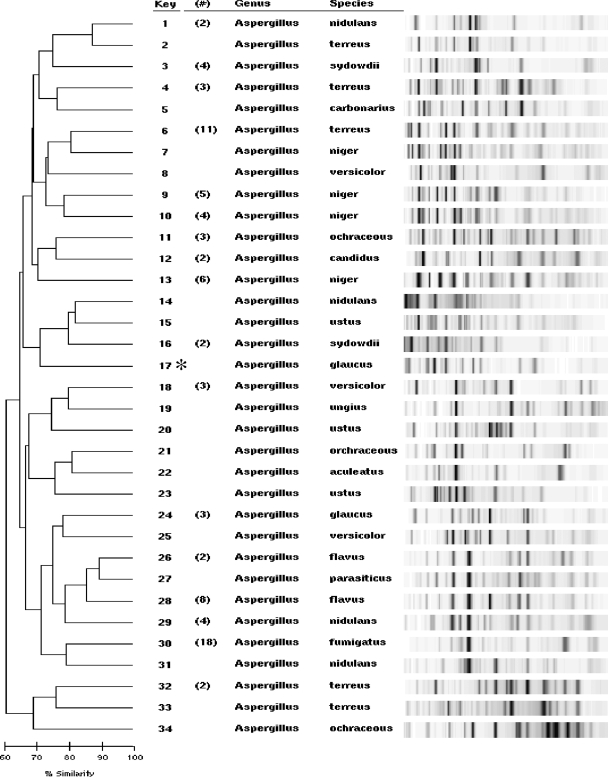
Figure 1 shows rep-PCR profiles of the Aspergillus isolates; the identification shown is based on conventional testing, sequencing, or both, as discussed above. The dendrograms were generated automatically using the DiversiLab software and pairwise groupings with the Aspergillus DNA fingerprint patterns. Eighty-two of the 99 (83%) isolates had ≥85% similarity to at least one additional isolate of the same species (Fig. 1), providing an accurate identification. The fingerprint patterns of the study isolates were then compared to all of those in the mold database. All except one A. glaucus isolate (99%) clustered with one or more isolates in the database with at least 85% similarity, and no isolate was identified incorrectly. Sequencing confirmed the identification of the A. glaucus isolate that rep-PCR did not identify.
FIG. 1.
Collapsed dendrogram and gel-like images representing all of the Aspergillus isolates tested. Gel-like images of samples with ≥85% similarity were collapsed so that one representative fingerprint is shown. The numbers in parentheses represent the numbers of samples in those collapsed clusters. The isolate marked with an asterisk (key no. 17, A. glaucus) was the only isolate in the study with a fingerprint pattern that had <85% similarity to at least one isolate in the mold library.
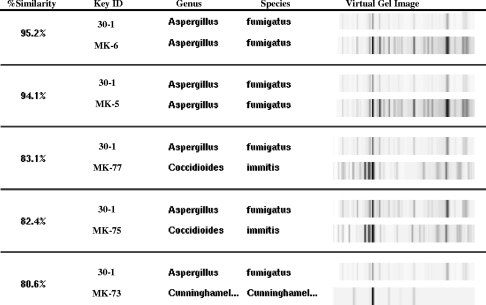
In addition to generating dendrograms, the DiversiLab software allows the user to build custom databases and reports. One example is the “top-five-match” report, which can be used for rapid matching in which the only results shown are the “unknown” sample with five library samples that have the highest percentage match. Figure 2 shows the query “unknown” sample (identified as A. fumigatus by conventional methods) with the five-closest samples in the library. Dendrograms of the first two, both A. fumigatus, have >90% similarity to the “unknown” isolate, suggesting that this is likely the correct choice.
FIG. 2.
The top-five-match feature of the software showing a query or unknown sample, 30-1 (A. fumigatus from the study isolates), compared to the five most-similar samples within the mold library (MK no.). The top two matches with over 90% similarity are also A. fumigatus (MK-6 and MK-5).
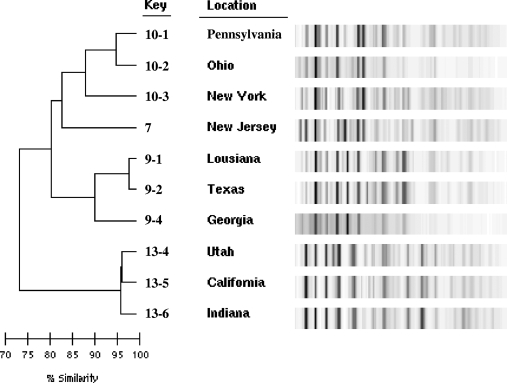
As illustrated in Fig. 1, there were multiple clusters for some species (such as A. niger), whereas all of the A. fumigatus isolates clustered together, perhaps suggesting that the former species has a more heterogeneous genome. To investigate whether subspecies discrimination could be determined by geographical location, dendrograms were generated for the 10 A. niger (Fig. 3) and 7 A. terreus (data not shown) clinical isolates for which the state of residence of the patient at the time of specimen collection was known. Subspecies discrimination is evident as shown by the three clusters of A. niger in Fig. 3. Although no pattern clustering could be discerned from geography, differences in patterns clearly indicate variations in the isolates of the species.
FIG. 3.
Expanded dendrogram of the A. niger samples as shown by similar key identification no. 7, 9, 10, and 13 from the collapsed dendrogram illustrated in Fig. 1. The sample collection location is designated by state.
Conclusions.
Identification of molds based on traditional culture and morphological characteristics requires considerable expertise, gained only after years of experience. Because of the relative shortage of technologists with adequate mycology training, it is becoming increasingly difficult for many laboratories to identify molds. Additionally, culture-based identification can be time-consuming, especially when an isolate does not sporulate after incubation for several weeks.
One alternative to identification based on colony and microscopic morphology is determination of genetic sequence variation. Ribosomal genes, for example, have regions of variability that are useful for species identification. The ITS regions are located between the 18S and 28S rRNA genes. The rRNA gene for 5.8S RNA separates the two ITS regions, and the sequence variation of ITS regions has led to their use in phylogenetic studies of many organisms (9). However, capital costs related to sequencing can be considerable, and highly skilled testing personnel are required to perform the analysis. Additionally, depending on the organism, regions other than ITS (gene specific; i.e., EF-1α for Fusarium) may be required to confirm the identification.
The DiversiLab system is a supplemental tool with the potential to help microbiology laboratories that are struggling with fungal identification due to the absence of experienced mycologists. It may provide a rapid, easy molecular method by which to identify many fungi, including Blastomyces dermatitidis, Coccidioides species, Candida species, Fusarium species, commonly encountered dermatophytes, and perhaps Histoplasma capsulatum (3, 6, 7). The system, however, should not be used in a vacuum. A rep-PCR result should not be reported unless it is consistent with the colony morphology and the microscopic features of the specific mold. Therefore, laboratories should have good mycology reference books. The morphological confirmation of molecular results is usually not possible when isolates remain sterile. In microbiology laboratories where the DiversiLab system is already used as a bacterial strain typing tool, the ability of the system to identify fungal isolates would make it a multiple-use platform for the laboratory. Expanding the test menu of the system also makes it economically more attractive. To be most cost efficient, 12 samples plus a control (13 samples if no control is run) should be tested at one time because the chip contains 13 wells and cannot be reused.
In this study, we confirmed the reliability of rep-PCR for identification of commonly encountered Aspergillus species (i.e., A. fumigatus, A. flavus, and A. terreus, as shown previously by Healy et al. [2]), as well as A. niger, and demonstrated that it can also identify species seen less frequently in a clinical laboratory, such as A. ochraceus, A. candidus, A. sydowii, A. versicolor, and A. nidulans. Although the DiversiLab system did not identify all isolates of Aspergillus, no isolate was incorrectly identified. We also demonstrated that rep-PCR can provide subspecies discrimination. Although no pattern clustering was discerned based on geography, it is interesting that similar fingerprint patterns were seen in different regions.
The major weakness of our study was the small number of isolates of some of the less commonly encountered species tested. Further validation with a larger sample set that includes multiple well-characterized isolates of each species is needed. Continued expansion of the DiversiLab mold library with many subspecies and strains of each species should enhance its identification capabilities.
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Baddley, J. W., T. P. Stroud, D. Salzman, and P. G. Pappas. 2001. Invasive mold infections in allogeneic bone marrow transplant recipients. Clin. Infect. Dis. 321319-1324. [DOI] [PubMed] [Google Scholar]
- 2.Healy, M., K. Reece, D. Walton, J. Huong, K. Shah, and D. P. Kontoyiannis. 2004. Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 424016-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Healy, M., K. Reece, D. Walton, J. Huong, S. Frye, I. I. Raad, and D. P. Kontoyiannis. 2005. Use of the DiversiLab System for species and strain differentiation of Fusarium species isolates. J. Clin. Microbiol. 435278-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubak, B. M. 2002. Fungal infection in lung transplantation. Transpl. Infect. Dis. 424-31. [DOI] [PubMed] [Google Scholar]
- 5.Marr, K. A., R. A. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34909-917. [DOI] [PubMed] [Google Scholar]
- 6.Pounder, J. I., S. Williams, D. Hansen, M. Healy, K. Reece, and G. L. Woods. 2005. Repetitive-sequence-PCR-based DNA fingerprinting using the DiversiLab system for identification of commonly encountered dermatophytes. J. Clin. Microbiol. 432141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pounder, J. I., D. Hansen, and G. L. Woods. 2006. Identification of Histoplasma capsulatum, Blastomyces dermatitidis, and Coccidioides species by repetitive-sequence-based PCR. J. Clin. Microbiol. 442977-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh, N., and D. L. Paterson. 2005. Aspergillus infections in transplant recipients. Clin. Microbiol. Rev. 1844-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White, T., T. Burns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.