Abstract
We describe the first “Mycobacterium paraffinicum” (unofficial taxon) pseudo-outbreak in a tertiary-care medical center. Fifteen clinical nontuberculous mycobacterium isolates from 10 patients were initially identified by biochemical tests and high-performance liquid chromatography as Mycobacterium scrofulaceum. However, further testing by molecular analysis revealed “M. paraffinicum.” Epidemiological and environmental investigation determined that the ice machine was the source of the pseudo-outbreak.
Nontuberculous mycobacteria (NTM) are common in the environment and are typically found in water and soil. Pseudo-outbreaks of NTM species in healthcare institutions have been reported (11). Molecular techniques have been useful for identification and strain typing and have assisted in confirming outbreaks and pseudo-outbreaks but have also identified new NTM species. We describe the first reported pseudo-outbreak of “Mycobacterium paraffinicum” (unofficial taxon) at a tertiary-care medical center.
During a 1-year period, a total of 15 specimens (14 respiratory and 1 stool) from 10 patients admitted to the same patient care unit were initially identified as Mycobacterium scrofulaceum. The identification of M. scrofulaceum was based on the scotochromogenic nature of the organisms; high-performance liquid chromatography (HPLC); negative gene probe results for the Mycobacterium avium/Mycobacterium intracellulare complex, Mycobacterium kansasii, Mycobacterium gordonae, and Mycobacterium tuberculosis; and a biochemical profile that included negative nitrate reduction, Tween hydrolysis, urease activity, and sodium chloride tolerance. Fifty-seven percent of the isolates had positive semiquantitative catalase test results, and 67% had positive tellurite reduction reactions. The cultures took approximately 6 weeks to grow.
The 10 patients submitted a total of 62 specimens for mycobacterial cultures: 44 respiratory, 2 stool, 5 pleural fluid, 2 cerebral spinal fluid, 6 surgical pathology tissue, and 3 blood specimens. All specimens were acid-fast-bacillus-smear negative. All of the patients were discharged with alternative diagnoses prior to the identification or growth of M. scrofulaceum, and none of the patients were subsequently treated for this NTM. Environmental investigation of the hospital water sources identified an ice machine located on the patient care unit as also positive for M. scrofulaceum.
We pursued 16S rRNA gene sequencing and PCR restriction analysis (PRA) of a 440-bp region of hsp65 (7) to identify the specific NTM species because some M. scrofulaceum isolates in patients with pulmonary disease have recently been reidentified as M. parascrofulaceum by using newer molecular methods (9, 10). Although a pseudo-outbreak was suspected, since all of the patients had pulmonary symptoms, it was important to rule out M. parascrofulaceum as a potential cause of true infection. Amplification of the 16S rRNA genes of isolates from selected patients and the ice machine identified the mycobacterium as “M. paraffinicum.” The hsp65 PRA patterns included 240-bp, 125-bp, and 100-bp BstEII fragments and 145-bp and 125-bp HaeIII fragments. This pattern also matched the American Type Culture Collection (ATCC) type strain of “M. paraffinicum” (ATCC 12670) (data not shown).
Strain typing of isolates from patients and the ice machine was performed by pulsed-field gel electrophoresis (PFGE) with a CHEF-DR III (Bio-Rad, Hercules, CA) and by repetitive-sequence-based PCR (rep-PCR) with the DiversiLab system (BioMerieux, Durham, NC), using methods previously described (2, 6). For PFGE, large DNA restriction fragment patterns were generated using restriction enzyme XbaI and pulse times of 3 to 12 seconds for 20 h at 6 V/cm. As defined by Tenover et al., a unique PFGE strain consists of four or more band differences from at least one other isolate in the outbreak group (8). For rep-PCR, the modified Kullback-Leibler (KL) distance method was used to create a pair-wise percent similarity matrix, and a dendrogram was generated using the unweighted-pair group method of arithmetic averages. The DiversiLab system allows two options for the calculation of percent similarities: the Pearson correlation and the modified KL methods. Both are “curve-based” methods that calculate similarity based on the relative intensity at each data point; however, the Pearson correlation method is more intensity based, whereas the KL method weighs the presence/absence of bands more heavily. With the complex “M. paraffinicum” fingerprints generated here, the KL method was chosen for analysis due to the large number of weaker-intensity bands observed. For the interpretation of similarities between the isolates, sample relationships were designated following the manufacturer's suggestion for heterogeneous species: “indistinguishable” (no band differences), “similar” (one or two band differences), and “different” (three or more band differences on the electropherogram). Isolates designated rep-PCR clusters were similar to each other, with one or two band differences, and shared >95% similarity.
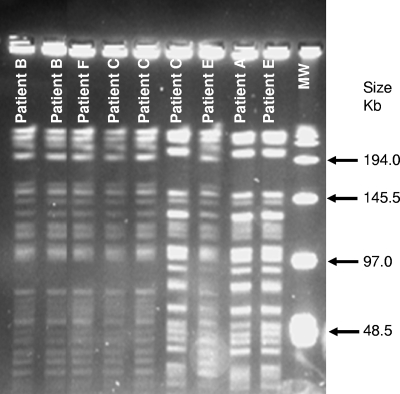
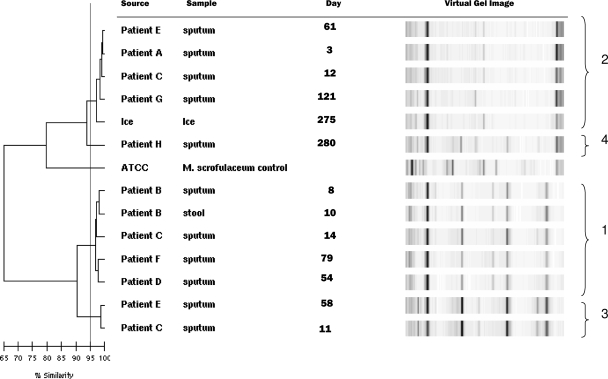
The results for PFGE and rep-PCR molecular typing conducted on the ice machine isolate and the clinical isolates from eight patients correlated well (Table 1 and Fig. 1). In the tables and figures, each date is the number of days from the first patient's admission date (day 0). Two primary strains (designated “types” for PFGE and “clusters” for rep-PCR) were identified. Strain type 1/cluster 1 included patients B, C, D, and F, and strain type 2/cluster 2 included patients A, C, E, and G and the ice machine water sample. A third pattern, representing a mixture of the two strains, which highlighted differences between PFGE and rep-PCR typing, was noted (Table 1). Patients C and E both had multiple patterns recovered at different times by both methods. Interestingly, the sputum from patient C on day 11, which was PFGE type 1, was subdivided by rep-PCR into a third cluster (rep-PCR cluster 3). This patient had all three rep-PCR clusters on different days (Table 1 and Fig. 2). Patient E likewise had rep-PCR cluster 2 and cluster 3 and PFGE type 1 and a mixture of type 1 and type 2 on two different dates. Patient H, in rep-PCR cluster 4, had >90% similarity to rep-PCR cluster 2, but this result was considered “different” due to the presence of more than three band differences.
TABLE 1.
Comparison of genotyping of selected clinical isolates and an ice machine isolate for “M. paraffinicum” by PFGE and rep-PCRa
| Patient or source | Specimen type | Initial culture day | Final culture day | PFGE type(s) | Rep- PCR cluster |
|---|---|---|---|---|---|
| A | Respiratory | 3 | 92 | 2 | 2 |
| B | Respiratory | 8 | 91 | 1 | 1 |
| Stool | 10 | 92 | 1 | 1 | |
| C | Respiratory | 11 | 113 | 1 | 3 |
| Respiratory | 12 | 113 | 2 | 2 | |
| Respiratory | 14 | 113 | 1 | 1 | |
| D | Respiratory | 54 | 123 | 1 | 1 |
| E | Respiratory | 58 | 113 | 1, 2 | 3 |
| Respiratory | 61 | 113 | 2 | 2 | |
| F | Respiratory | 79 | 137 | 1 | 1 |
| G | Respiratory | 121 | 182 | 2 | 2 |
| H | Respiratory | 280 | 339 | 2 | 4 |
| Patient care unit ice machine | Ice | 275 | 354 | 2 | 2 |
All respiratory specimens were expectorated except for that of patient D, which was induced.
FIG. 1.
PFGE patterns of clinical and environmental isolates of the “M. paraffinicum” pseudo-outbreak. PFGE type 1: lanes 1 and 2, patient B (day 8, and stool day 10); lane 3, patient F (day 79); lanes 4 and 5, patient C (days 11 and 14). PFGE type 2: lane 6, patient C (day 12); lane 8, patient E (day 61); lane 9, patient A (day 3). Mixed PFGE type 1 and type 2: lane 7, patient E (day 58). Lane MW, molecular weight standard.
FIG. 2.
Rep-PCR-based dendrogram illustrating the relationship among clinical and environmental isolates of a “M. paraffinicum” pseudo-outbreak. The ATCC M. scrofulaceum strain was included as an unrelated control. The KL distance method was used to create a pair-wise percent similarity matrix, and the tree was generated using the unweighted-pair group method of arithmetic averages. The horizontal scale bar indicates the percents similarity among strains. Clusters 1 to 4, shown at the right, were defined by a 95% similarity threshold (vertical line).
Besides confirming the pseudo-outbreak, the results of the PFGE and rep-PCR conducted on 13 isolates from patients and the ice machine also confirmed that the two primary strains, i.e., PFGE type 1 and type 2 and rep-PCR cluster 1 and cluster 2, were likely two clonally different strains of “M. paraffinicum” that were either coexistent in the water system at the same time or transiently dominant at different times. PFGE type 1 and type 2 had a >4-band difference, while a third PFGE type appeared to be a mixture of PFGE types 1 and 2. The rep-PCR phylogenetic analysis on the mixed PFGE type, identified as rep-PCR cluster 3, was closely related to rep-PCR cluster 1, with nearly 90% DNA similarity to cluster 1 strains as opposed to 75% DNA similarity with cluster 2 strains.
There is a paucity of clinical and epidemiologic information on “M. paraffinicum,” which may be due to the difficulty in identifying this species by routine biochemical procedures and the requirement of molecular methods for species identification. Three clinical isolates of “M. paraffinicum” have been previously identified at the CDC (R. Cooksey, unpublished data). The existence of these isolates, in addition to our clinical and environmental “M. paraffinicum” isolates from this pseudo-outbreak, substantiates the need to formalize the naming of this species.
“M. paraffinicum” isolates were first recovered from the soil in 1956 using an enriched ethane culture medium (3). The colonies were described as yellow, waxy, and wrinkled on mineral salt agar and stained acid fast. Interestingly, this species does not utilize bacteriological organic media, such as peptone, yeast extract, or glucose, but rather uses paraffinic hydrocarbons other than methane. “M. paraffinicum” is very similar to M. scrofulaceum. They share approximately 99% 16S rRNA sequence and have similar HPLC patterns (1). While M. scrofulaceum and the M. avium/M. intracellulare complex also have similar HPLC patterns, they are differentiated by a gene probe specific for M. avium/M. intracellulare. Biochemically, M. paraffinicum has a negative urease test reaction whereas M. scrofulaceum may or may not be urease negative (4, 5, 12). All of our isolates were urease negative. Combining molecular amplification of the 16S-23S rRNA internal transcribed spacer region with confirmation by 16S rRNA gene sequencing may be the most precise method for identifying “M. paraffinicum” (5). Our patient and ice machine isolates were conclusively identified as “M. paraffinicum” by 16S rRNA gene sequence analysis and hsp65 PRA.
Our study is limited by the assumption that the patient-specific isolates were most likely due to oral consumption of contaminated ice or water and the inability to consistently isolate “M. paraffinicum” from the environmental water sources. Because the same water system distributes water to the ice machine and the patient rooms, we cannot state for certain that only the ice machine and not the general water system contained this organism. The transient nature of the contaminant in the ice machine and/or the delay in environmental sampling may be reasons why we were not able to recover this species consistently.
In summary, “M. paraffinicum” was shown by molecular analysis to be associated with this pseudo-outbreak. The clinical significance of “M. paraffinicum” is unknown, as this species has not been previously associated with clinical disease or pseudo-outbreak. An epidemiological investigation with molecular analysis was needed to confirm the suspicion of a pseudo-outbreak. Molecular methods are necessary to efficiently and accurately identify “M. paraffinicum.”
Acknowledgments
Special thanks to Tom Pulchaski for isolation and processing of the specimens, David Taylor for assistance with epidemiology data, and Kevin Sohner and Lena Fischer (Ohio Department of Health Tuberculosis Laboratory) for performing HPLC analysis.
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Butler, W. R., L. Thibert, and J. O. Kilburn. 1992. Identification of Mycobacterium avium complex strains and some similar species by high-performance liquid chromatography. J. Clin. Microbiol. 302698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cangelosi, G. A., R. J. Freeman, K. N. Lewis, D. Livingston-Rosanoff, K. S. Shah, S. J. Milan, and S. V. Goldberg. 2004. Evaluation of high-throughput repetitive-sequence-based PCR system for DNA fingerprinting Mycobacterium tuberculosis and Mycobcterium avium complex strains. J. Clin. Microbiol. 422685-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, J. B., H. H. Chase, and R. L. Raymond. 1956. Mycobacterium paraffinicum n. sp., a bacterium isolated from soil. Appl. Microbiol. 4310-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Häfner, B., H. Haag, H. K. Geis, and O. Nolte. 2004. Different molecular methods for the identification of rarely isolated non-tuberculous mycobacteria and description of new hsp65 restriction fragment length polymorphism patterns. Mol. Cell Probes 1859-65. [DOI] [PubMed] [Google Scholar]
- 5.Lebrun, L., F. X. Weill, L. Lafendi, F. Houriez, F. Casanova, M. C. Gutierrez, D. Ingrand, P. Lagrange, V. Vincent, and J. L. Herrmann. 2005. Use of the INNO-LiPA-MYCOBACTERIA assay (version 2) for identification of Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex isolates. J. Clin. Microbiol. 432567-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazurek, G. H., S. Hartman, Y. S. Zhang, B. A. Brown, J. S. Hector, D. Murphy, and R. J. Wallace. 1993. Large DNA restriction fragment polymorphism in the Mycobacterium avium-M. intracellulare complex: a potential epidemiologic tool. J. Clin. Microbiol. 31390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulse-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tortoli, E., L. Chianura, L. Fabbro, A. Mariottini, N. Martin-Casabona, G. Mazzarelli, C. Russo, and M. Spinelli. 2005. Infections due to the newly described species Mycobacterium parascrofulaceum. J. Clin. Microbiol. 434286-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turenne, C. Y., V. J. Cook, T. V. Burdz, R. J. Pauls, L. Thibert, J. N. Wolfe, and A. Kabani. 2004. Mycobacterium parascrofulaceum sp. nov., novel slowly growing, scotochromogenic clinical isolates related to Mycobacterium simiae. Int J. Syst. Evol. Microbiol. 541543-1551. [DOI] [PubMed] [Google Scholar]
- 11.Wallace, R. J., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52453-490. [DOI] [PubMed] [Google Scholar]
- 12.Wayne, L. G., and H. A. Sramek. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin. Microbiol. Rev. 5 1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]