Abstract
Two attenuated vaccines, Ingelvac PRRS MLV and Ingelvac PRRS ATP, derived from VR2332 and JA142, respectively, have been used to control porcine reproductive and respiratory syndrome (PRRS) virus. However, there have been several field reports concerning the reversion of the vaccine virus to virulence. Furthermore, viruses genetically indistinguishable from the vaccines and wild-type parental viruses have been detected in clinical PRRS cases, raising the need for a better differential tool. As the vaccine viruses replicated better and produced bigger plaques in MARC-145 cells than did the wild-type parental strains, the following study was conducted to determine if the growth difference in MARC-145 cells can be utilized to differentiate a vaccine-like virus (VLV) from a wild-type virus and to identify genetic markers corresponding to such phenotype of the vaccine viruses. The relatedness of 83 field isolates collected between 1996 and 2005 to VR2332 and JA142 was classified genetically and antigenically. Thirteen of 25 VR2332-related viruses and 9 of 10 JA142-related viruses were determined as VLVs, since those viruses produced plaques similar to those by the vaccine viruses. Four unique amino acids each were identified throughout structural genes for MLV and ATP. Among those, F10 in open reading frame 2 (ORF2) of MLV and E85 and Y165 in ORF3 of ATP were stable during pig passages. When the sequences unique for MLV were incorporated into an infectious clone constructed based on VR2332, the virus growth and resultant plaque size in MARC-145 cells were increased, suggesting that these sequences can be used as genetic markers for VLVs.
Porcine reproductive and respiratory syndrome (PRRS) is characterized by reproductive failure in breeding animals and respiratory distress in all ages of pigs (1, 3). The syndrome was first reported in the United States in 1987 (11). The causative agent of PRRS was, however, isolated first in The Netherlands in 1991 and named the Lelystad virus, the European prototype PRRS virus (PRRSV). Isolation of VR2332, the North American prototype PRRSV, was subsequently reported in 1992 (3). North American PRRSV (type II) and European PRRSV (type I) share less than 70% sequence homology and comprise two distinct genotypes and serotypes of PRRSV (6).
PRRSV is a small, enveloped virus and belongs to the family Arteriviridae in the order Nidovirales. PRRS virion contains a positive, single-strand RNA genome of approximately 15 kb in length. The genome encodes at least nine open reading frames (ORFs) (2, 7). ORF1a and -1b encode nonstructural proteins required for virus replication (2). ORF2a, -3, and -4 encode three N-glycosylated minor envelope proteins designated GP2, -3, and -4, respectively, while ORF2b, which is completely embedded in ORF2a, encodes a nonglycosylated minor envelope protein named 2b, which is equivalent to E protein of equine arteritis virus (24). ORF5, -6, and -7 encode the major envelope (GP5 or E), membrane (M), and nucleocapsid (N) proteins, respectively (7).
PRRS has been identified worldwide and caused a significant economic loss to the swine industry. The economic impact of PRRS to U.S. swine producers has been estimated at an approximate $560 million loss per year (19). In response to the economic significance of PRRS, two attenuated live-virus vaccines (Ingelvac PRRS MLV and Ingelvac PRRS ATP) have been used most to control the disease (12). Ingelvac PRRS MLV (hereafter “MLV”) vaccine, which was derived from VR2332 after sequential passage of the virus in a monkey kidney cell line (CL2621), was first introduced to the U.S. market in 1996 (16). This vaccine has been demonstrated to reduce the rate of PRRSV-associated reproductive failure in sows and lessen clinical symptoms in younger pigs (8, 18). Nonetheless, a severe form of PRRS (a.k.a. acute or atypical PRRS), which was described as “abortion storm” or “sow mortality syndrome,” has been reported for MLV-vaccinated farms. The incidence of this acute form of disease in vaccinated pigs resulted in the introduction of the Ingelvac PRRS ATP (hereafter “ATP”) vaccine, which is made of an attenuated derivative of JA142, to the market in 2000 (12). The JA142 strain was initially isolated from a severe abortion storm case in Iowa in 1997 (17).
Since the vaccines use live viruses even though attenuated, the reversion of vaccine virus to virulence has been a concern and was documented by Danish pig producers after the MLV vaccine was introduced to Denmark in 1995 as part of the national PRRS control program. After the introduction, viruses closely related to the MLV vaccine virus were isolated from clinically ill pigs in 1996 (16). Since no North American PRRSV was reported in Denmark or other European countries until the spring of 1996, it was concluded that these field isolates originated from the vaccine virus (16, 25). Unlike the Danish experience, the differentiation of some field isolates of PRRSV from wild-type parental and vaccine viruses has been challenging in the U.S (21) due to the antigenic and genetic similarity between the vaccine and wild-type parental viruses (25). A few molecular assays, such as sequencing or restriction fragment polymorphism analysis, have been extensively employed for the differential purpose (28). However, a great number of field isolates which are genetically indistinguishable from both a vaccine virus and its parental strain have been continuously identified from clinical PRRS cases (29), raising the need for a better way to differentiate wild-type viruses from the vaccine viruses.
Due to the fact that the vaccine virus was attenuated by sequential passages in cell culture, the vaccine viruses tend to grow much better in MARC-145 cells, a highly permissive clone of African monkey kidney cell line MA104 (13), than in their wild-type parental viruses (14). Recently it was also observed in our laboratory that the vaccine virus produced a significantly bigger-sized plaque in MARC-145 cells than what was seen for their parental strains. The following study was to evaluate if the difference in growth characteristics (i.e., phenotype) between the vaccine strains and their wild-type parental viruses in MARC-145 can be utilized to aid strain differentiation by compensating for the limitation of genetic differentiation. In addition, attempts were made to identify sequence elements associated with the phenotype of the vaccine or related viruses (i.e., the attenuated form of PRRSV). Furthermore, the utility of such sequence elements as surrogate markers for the rapid detection of vaccine-like viruses (VLVs) was assessed.
MATERIALS AND METHODS
Study design.
Field isolates (n = 83) of PRRSV were classified into the VR2332 group, the JA142 group, and an unrelated wild-type virus group based on their genetic (i.e., sequencing) and/or antigenic (i.e., one-way cross virus neutralization [VN]) similarities. Then, a plaque assay was applied to the isolates in order to determine how well the virus grows (i.e., the highest progeny virus titer and time to peak titer) and the size of plaque produced in MARC-145 cells compared to those of the vaccine and their wild-type parental viruses. The stability of the phenotypic characteristics of the wild-type and vaccine viruses were assessed by sequentially passing the viruses in cell culture and animals, respectively. In addition, structural genes (ORF2 to -7) of vaccine viruses or VLVs and wild-type viruses were compared to identify sequence elements for the vaccine viruses or VLVs. A reverse genetics system constructed based on the sequence of VR2332 (20) was then employed to determine the biological role of the identified genetic markers unique for MLV.
Viruses and cells.
Eighty-three field isolates of PRRSV which were collected from submissions to Iowa State University Veterinary Diagnostic Laboratory during the period from 1999 to 2005 were employed for the study. Besides the field isolates, two vaccine viruses (MLV and ATP) and their parental strains (VR2332 and JA142, respectively) were used as reference viruses. The vaccine viruses were kindly provided by Boehringer-Ingelheim Vetmedica, Inc., in Ames, IA (14). All field and reference viruses were propagated in MARC-145. All viruses represented five passages in the cell.
Characterization of growth of reference viruses in MARC-145 cells.
To determine the one-step growth of VR2332 and MLV, confluent monolayers of MARC-145 prepared in 25-cm2 flasks and 48-well plates were inoculated with VR2332 or MLV at a multiplicity of infection of 0.01. After a 1-hour incubation at 37°C, the inoculums were discarded and the cells were replenished with RPMI-1640 medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (Sigma), 20 mM l-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml gentamicin, and 0.25 μg/ml amphotericin B (hereafter “RPMI growth medium”). Every 3 h postinoculation (p.i.), cell culture fluid and cells were collected separately from each flask for 24 h. The virus titer (50% tissue culture infective dose [TCID50]/0.1 ml) in each of the cell culture fluids or cell lysate supernatants was determined by a microtitration infectivity assay (9, 23) in MARC-145 cells. At each collection time, cells in the corresponding 48-well plate were simultaneously fixed with cold 80% acetone aqueous solution for a fluorescent antibody test to assess the production of viral proteins.
Growths of the VR2332, MLV, JA142, and ATP strains were also assessed for a multistep growth curve. Each of the viruses was inoculated onto confluent monolayers of MARC-145 prepared in 25-cm2 flasks at a multiplicity of infection of 0.01. After a 1-hour incubation at 37°C, the inoculum was discarded and the cells were replenished with RPMI growth medium. One flask of MARC-145 cells inoculated with each strain was frozen at 6, 12, and 24 h and thereafter every 24 h until 4 days p.i. After three rounds of freeze-thawing at −80°C and 37°C, respectively, the cell culture fluid was harvested for virus titration.
Plaque assay.
The assay was performed as previously described (22) with some modifications. One hundred or 10 fluorescence focus-forming units (FFUs) of each virus in 0.1 ml of RPMI growth medium were inoculated on confluent monolayers of MARC-145 prepared in 24-well plates. The cells were incubated for 1 h at 37°C and the inoculum was replaced with overlay medium (RPMI growth medium containing 0.6% of agar). The cells were further incubated for up to 4 days at 37°C until plaques were clearly observed. The overlay medium was removed from each well of the plates and cell monolayers were stained for 30 min with 0.25% Coomassie brilliant blue R250 staining solution (Bio-Rad, Hercules, CA), which was prepared in the mixed solution of absolute acetic acid and 50% methanol at a ratio of 1:9 (vol/vol). The diameter (mm) of plaque produced by each virus was determined by averaging the sizes of the three well-defined plaques observed. A plaque bigger than 3 mm in diameter was considered as a big-sized plaque, and one smaller than 2 mm in diameter was considered as a small-sized plaque. A plaque between 2 and 3 mm in diameter was regarded as a medium-sized plaque. The assay was repeated at least three times for all viruses examined.
VN assay.
A florescent focus neutralization assay was conducted as previously described (30) with some modifications to assess the susceptibility of field PRRSV isolates to the neutralizing activity of antisera produced in pigs against the VR2332 and JA142 strains. The antisera had VN titers of 1:64 and 1:32 against the homologous virus, respectively. Sera collected from PRRSV-naïve age-matched pigs served as a negative control. A pair of anti-PRRSV antibody-positive and -negative sera was serially diluted twofold in 100 μl of RPMI growth medium and mixed with an equal volume of each virus at a rate of 100 FFU/μl. Mixtures of virus and serum were then incubated for 1 h at 37°C. Each mixture was inoculated onto MARC-145 cell monolayers prepared in 96-well plates and incubated for another hour at 37°C. The plates were then replenished with 100 μl of fresh RPMI growth medium per well and further incubated for 20 h at 37°C in a humid 5% CO2 atmosphere. After the incubation, the cells were fixed with cold 80% acetone aqueous solution for 2 min at ambient temperature and incubated with PRRSV-specific monoclonal antibody SDOW-17 conjugated with fluorescent isothiocyanate (Rural Technologies, Brookings, SD). After a 1-h incubation, the cells were washed three times with phosphate-buffered saline, and the number of virus-specific fluorescent foci in each well was counted. VN titer was expressed as the reciprocal of the highest dilution in which a 90% or higher reduction in the number of FFU was observed.
Sequencing and sequence analysis.
All 83 field isolates were sequenced for ORF2, -3, -5, and -6, and selected viruses (n = 50) were sequenced for ORF2 to -7. PCR and sequencing primers were designed as follows: ORF2 to 6 Forward (PSF), 5′-CCACTGCCACCAGCTTGAAGTT-3′; ORF2 to 6 Reverse (PSR), 5′-CAGACACAATTGCCGCTCACTAGG-3′; ORF2 Forward (P2F), 5′-AAACGGTGAGGACTGGGAGGATTA-3′; ORF2 Reverse (P2R), 5′-TCGAAAGAAAAATTGCCCCTAACC-3′; ORF3 Forward (P3F), 5′-CCGGTTGGCTGGTGGTCT-3′; ORF3 Reverse (P3R), 5′-CAAAACAGAACGGCACGATACACC-3′; ORF5 Forward (P5F), 5′-CCTGAGACCATGAGGTGGG-3′; ORF5 Reverse (P5R), 5′-TTTAGGGCATATATCATCACTGG-3′; ORF6 Forward (P6F), 5′-GCGGTCGCCTGTCATCATAG-3′; ORF6 Reverse (P6R), 5′-GGCTGGCCATCCCCCTTCTTTCT-3′; ORF7 Forward (P7F), 5′-TCGTGTTGGGTGGCAGAAAAGC-3′; and ORF7 Reverse (P7R), 5′-GCCATTCACCACACATTCTTCC-3′.
Viral RNA was extracted using the QIAamp viral RNA mini kit (Qiagen, Valencia, CA). For sequencing the full length of ORF2 to -6, reverse transcription (RT) was conducted using Superscript III (Invitrogen, Grand Island, NY) according to the manufacturer's protocol, and cDNA was amplified with the GeneAmp XL long PCR kit (Applied Biosystems, Foster City, CA). The cycling conditions were 94°C for 1 min, 16 cycles of 94°C for 15 s, and 68°C for 4 min followed by 14 cycles of 94°C for 15 s and 68°C for 4 min, with time increased by 15 s per cycle, and then final extension at 68°C for 10 min. To sequence individual ORFs, the target ORF was directly amplified with the Qiagen one-step RT-PCR kit (Qiagen). The reaction conditions were 50°C for 30 min for RT and 95°C for 15 min followed by 30 cycles of 95°C for 30 s, 50°C for 45 s, and 72°C for 1 min; final extension was conducted at 72°C for 10 min.
All PCR products were purified using the QIAquick PCR purification kit (Qiagen) and submitted to the Iowa State University Nucleic Acid Facility for sequencing. Once sequence data were available, multiple alignments and sequence comparisons were conducted using Lasergene software (DNASTAR Inc., Madison, WI). Unrooted dendrograms were generated by the distance-based neighbor-joining method using Molecular Evolutionary Genetic Analysis (MEGA) software (15). Bootstrap values were calculated on 1,000 replicates of the alignment to determine the reliability of the phylogram.
Assessment of the stability of phenotypic characteristics.
The stability of cell growth phenotypes (i.e., attenuated versus wild) of PRRSVs was determined in vitro and in vivo. For an in vitro assessment, the VR2332 and JA142 strains were sequentially passed in MARC-145 cells for 20 passages. Sequential passages were repeated three different times for each virus, resulting in a total of 120 viruses derived from the two strains for plaque assay and sequence analysis.
For an in vivo assessment, the CC-01 strain, a plaque-cloned virus derived from VR2332 through cell passages, was sequentially passed in pigs housed in HEPA-filtered isolation units for 13 times. The CC-01 strain shared high sequence homology with MLV (99.6% for entire structural genes) and showed phenotypic characteristics similar to those for MLV virus in cells and animals. It produced large plaques (>4 mm) similar to those seen for the vaccine viruses and induced a low level of viremia with minimum virulence in the inoculated pigs. Details of the pig-to-pig passage study were reported elsewhere (6). In brief, at each passage, a total of three pigs were used for inoculation of the virus material, and the fourth pig served as a sham-inoculated control. At the completion of the 60-day observation period in each passage, all pigs were euthanized and a variety of tissues was collected. Homogenates of mixed tissues were used to inoculate pigs in the subsequent passage in a manner to keep three independent lines of pig passage (A, B, and C). After seven passages, line A was discontinued. Two of 15 plaque-cloned viruses isolated from each pig's serum collected at day 7 p.i. in each passage were used for plaque assay and sequence analysis. A total of 66 plaque-cloned isolates (three pigs/passage times 2 isolates/pig times seven passages plus two pigs/passage times 2 isolates/pig times six passages) were examined.
Generation of mutant virus.
The cDNA infectious clone constructed based on the sequence of VR2332 was used for this study (20). Mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Stratagene, West Cedar Creek, TX) as per the manufacturer's manual. A shuttle vector (TOPO XL PCR cloning vector; Invitrogen, Carlsbad, CA) containing the whole structural genes (ORF2 to -7) of VR2332 was constructed and used as a template for mutagenesis to prevent any unexpected mutations during the process. The targeted mutation was confirmed by sequencing the whole structural genes in the shuttle vector, and then the whole fragment of the structural genes in the shuttle vector was substituted for those genes in the plasmid of the infectious clone by use of BsrGI and HpaI sites. The infectious clone containing only a targeted mutation in the right position was selected by sequencing the entire structural genes of the plasmid purified from competent cells (XL10-Gold ultracompetent cells; Stratagene). The plasmid of mutant infectious clone was then linearized by AclI and purified using DNAclear kit (Ambion, Austin, TX). The linearized plasmid was transcribed using T7 promoter by the mMESSAGE mMACHINE T7 kit (Ambion). The transcribed RNA was purified by the MEGAclear kit (Ambion) and 1 to 10 μg of purified RNA was transfected into MARC-145 cells (5 × 106 cells/ml) prepared in chilled Dulbecco's modified Eagle's medium (Sigma) containing 1.25% dimethyl sulfoxide (Sigma) by electroporation at 250 V and 950 μF (Gene Pulser Xcell electroporation system; Bio-Rad) (27). The transfected cells were plated on a six-well plate and the medium was changed with RPMI growth medium 16 to 18 h after plating. At 48 h, 200 μl of the supernatant was inoculated on MARC-145 cells prepared in a 24-well plate. After incubation for 1 h at 37°C, the inoculum was replaced with RPMI growth medium and the cells were further incubated at 37°C until a cytopathic effect became evident. Rescued mutants were grown once in 25-cm2 flasks and stored at −80°C until used. An aliquot of each mutant was sequenced to confirm the introduction of mutations and the integrity of other sequences.
Data analysis and sequence comparison.
The repeated measurements of virus growth were analyzed with multivariate analysis of variance to define the growth difference among the mutants by use of JMP (SAS Institute Inc., Cary, NC). Sequences were aligned and compared using Lasergene MegAlign software (DNASTAR Inc., Madison, WI).
RESULTS
Phenotypic characteristic of wild-type and vaccine viruses in MARC-145.
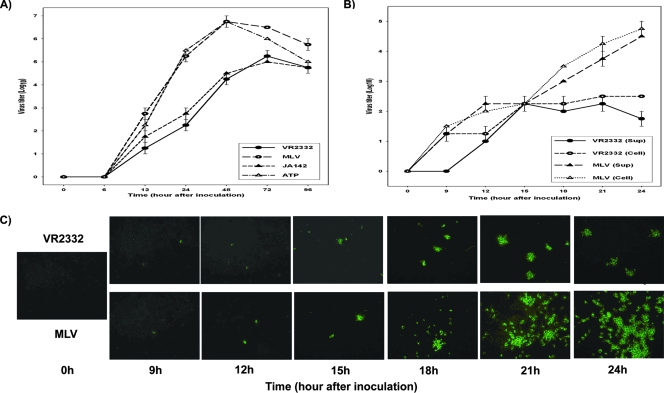
The vaccine strains (MLV and ATP) grew more efficiently than the wild-type parental viruses (VR2332 and JA142) in MARC-145 cells (Fig. 1A). The progeny virus titers of all four viruses were similar up to 12 h after inoculation. After 12 h p.i., MLV and ATP started to replicate much faster than their wild-type parental strains as the vaccine viruses produced a higher titer of progeny viruses. At 48 h p.i., the progeny virus titer from cells inoculated with the vaccine viruses reached 106.5 TCID50/0.1 ml, whereas the progeny virus titer from cells inoculated with the wild-type parental strains was approximately 104.5 TCID50/0.1 ml (P < 0.001).
FIG. 1.
Different growth characteristics of wild-type PRRSVs (VR2332, JA142) and their cell-attenuated vaccine viruses (MLV, ATP) in MARC-145 cells as measured by multistep growth (A) and one-step (B) curves and immunofluorescence microscopy (C). Sup, supernatant.
Similar results were observed in the one-step growth curve and immunofluorescence assessment of virus replication. The infectious progeny virus titers of VR2332 and MLV in both the culture fluid fraction (i.e., cell-free form) and the cell fraction (cell-associated form) were similar for each virus during the observation period. However, the kinetics of progeny virus production were significantly different between the two viruses. The MLV virus grew rapidly and steadily to 105 TCID50/0.1 ml during 24 h p.i., forming a linear growth curve overall, whereas the VR2332 virus grew slowly and reached a plateau (5 × 102 TCID50/0.1 ml) after 15 h p.i. (Fig. 1B). In immunofluorescence microscopy, the sizes and numbers of foci produced by VR2332 and MLV were similar until 15 h, but those by MLV increased drastically by 24 h compared to what was seen for VR2332 (Fig. 1C).
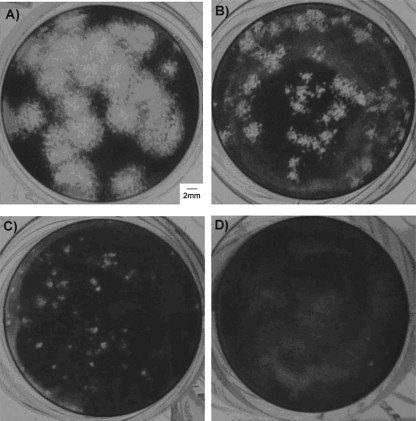
The different growth characteristics of the vaccine viruses and their wild-type parental strains yielded the production of differently sized plaques in MARC-145 cells (Fig. 2). The vaccine strains, i.e., MLV and ATP (Fig. 2A), produced larger plaques (diameter, >4 mm on average), while some field strains (Fig. 2B) generated medium-sized plaques (diameter, between 2 and 3 mm on average). In contrast, the parental viruses, i.e., JA142 (Fig. 2C) and VR2332 (Fig. 2D), produced much smaller plaques (diameter, <1 ml on average) within the same incubation time (4 days) and under the same propagation conditions.
FIG. 2.
Photomicroscopy of representative plaques produced by wild-type and attenuated PRRSVs in MARC-145 cells. The vaccine strains or VLVs produce bigger plaques (≥2 mm in diameter) (MLV [A] and 64955-01 [B]), whereas wild-type viruses including the parental strains of the vaccine viruses produce small-sized plaques (<1 mm in diameter) (JA142 [C] and VR2332 [D]).
Stability of the cell growth phenotypic characteristic of virus.
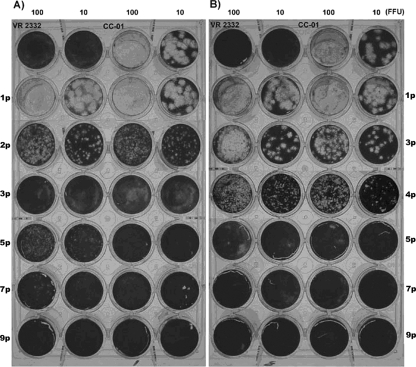
When the CC-01 strain was sequentially passed in three independent lines of pigs 13 times (a total of 726 days of in vivo replication), the faster growth and higher level of viremia began to be observed at the second passage of the virus (4). In contrast to the CC-01 virus that produced large-sized plaques (diameter, >4 mm) in MARC-145, its descendants collected from subsequent pig passages started to produce plaques smaller than those by the CC-01 after two or three pig passages (Fig. 3). After the third (Fig. 3, lines A and B) or fourth (Fig. 3, line C) passage, all recovered progeny viruses produced plaques with diameters of less than 2 mm. Once the plaque size became small, the viruses recovered from subsequent pig passages produced the same small-sized plaques until the termination of pig-to-pig passages.
FIG. 3.
The change in the size of plaques produced by a MLV vaccine-like PRRSVs (CC-01) during sequential pig-to-pig passages which were maintained in three independent lines (A, B, and C). Two plaque-cloned viruses isolated from each serum sample collected 7 days p.i. at each passage were used for the plaque assay. (A and B) Viruses collected from pig line B (results for line A were identical) and line C, respectively, during the passages. Numbers on top indicate the amounts of virus inoculated to MARC-145 cells (duplicate wells/virus) as expressed in FFU per 0.1 ml. Numbers with suffix “p” represent the number of pig passages of the CC-01 strain. The viruses after 9 passages (9p) consistently produced small-sized plaques.
During sequential cell culture passages of the VR2332 and JA142 viruses, the size of plaques produced by the viruses began to get bigger after 17 passages. However, overall plaque size still remained smaller than 2 mm in diameter, indicating that the cell growth phenotype (i.e., smaller-sized plaque) of wild-type PRRSVs is relatively stable during cell passages.
Genetic, antigenic, and phenotypic characteristics of PRRSV field isolates.
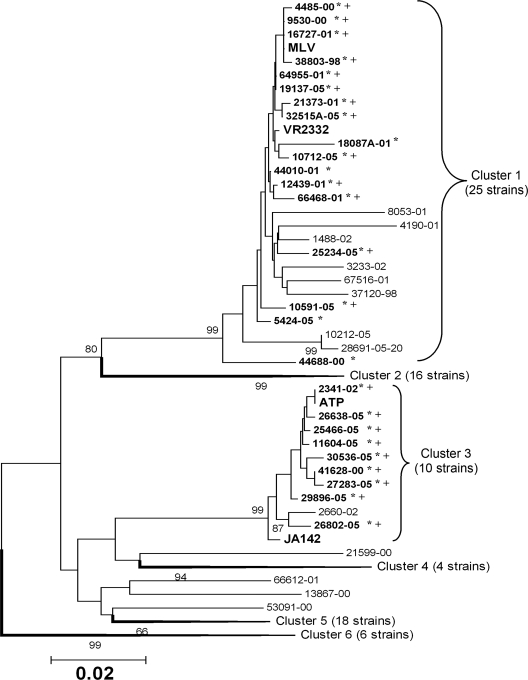
Eighty-three field isolates were assayed for their susceptibilities to antisera raised against VR2332 or JA142. Then, the genetic relatedness of the field isolates to VR2332 and JA142 was assessed based on the ORF5 sequence. Six genotypic clusters were identified among the isolates examined. Twenty-five viruses (≥96.5% homology to VR2332/MLV) and 10 viruses (≥98.5% homology to JA142/ATP) were classified into clusters related to VR2332/MLV and JA142/ATP, respectively (Fig. 4). In the VN test, the infection of MARC-145 cells by 17 of the 25 VR2332/MLV-like virus and 9 of the 10 JA142/ATP-like viruses was significantly affected (P < 0.05) by VR2332 or JA142 antiserum, respectively (i.e., a less than fourfold decrease in the susceptibility of tested virus to antisera compared to that of the control viruses, VR2332 or JA142). In contrast, the infectivities of the remaining 48 viruses to MARC-145 were not significantly affected by the VR2332 or JA142 antiserum.
FIG. 4.
Phylogenetic relationship of 83 PRRSV field isolates with the vaccine viruses (MLV, ATP) and their parental viruses (VR2332, JA142) based on ORF5 nucleotide sequence. Boldface and asterisks indicate isolates whose infections were significantly affected by VR2332 or JA142 antiserum. The symbol + indicates VLVs, which produced medium- to big-sized plaques. The reliability of analysis was determined by 1,000-times-repeated bootstraps.
Thirteen of the 17 VR2332/MLV-like viruses and all 9 JA142/ATP-like viruses produced medium-sized (2- to 3-mm) to big-sized (>3-mm) plaques, which were similar to those produced by the MLV and ATP strains. Therefore, these 22 viruses were defined as VLVs, since they were antigenically and genetically close to VR2332/MLV or JA142/ATP and still maintained the phenotype of the vaccine virus (i.e., bigger-sized plaques). All 61 field isolates except one produced small-sized plaques (<2 mm), similar to those produced by VR2332 and JA142. Interestingly, the one virus designated 2M11715 was not closely related to either VR2332 or JA142 (90.9% or 90% ORF5 nucleotide homology to each virus) and produced medium-sized plaques (2.5 mm).
Relationship between phenotypic and genetic similarities of field isolates.
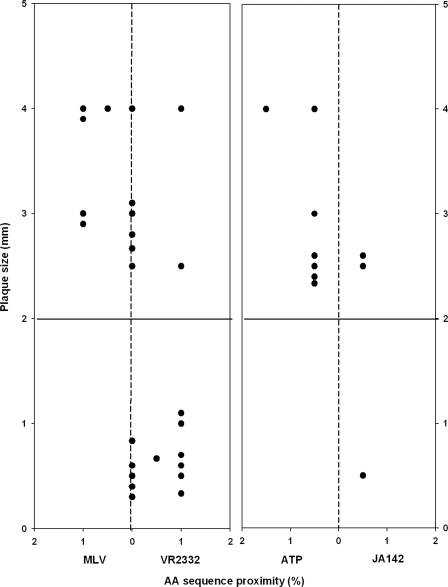
As shown in Fig. 4 and 5, the genetic proximity of the ORF5 amino acid sequence of the viruses to VR2332 or MLV was not always well correlated with phenotypic characteristics. Five of the 12 viruses that were genetically close to MLV (99.5% homology) produced medium-sized (2- to 3-mm) or big-sized (>3-mm) plaques and the other seven viruses that were closely related to VR2332 (95 to 99.5% homology) produced small-sized plaques (<2 mm). However, the remaining 11 VR2332/MLV-like viruses had the same genetic distance from both VR2332 and MLV. Among those, six viruses (98.5 to 99.5% homology to both viruses) produced medium- or big-sized plaques, whereas the remaining five viruses (95% to 99% homology to both viruses) produced small-sized plaques. In addition, two viruses, which were genetically closer to VR2332 (98 and 99% homology, respectively), produced medium- or big-sized plaques.
FIG. 5.
The relationship in plaque size and sequence proximity of between 35 field isolates defined as VLVs and the vaccine strains (MLV and ATP) or their wild-type parental strains (VR2332 and JA142). The horizontal solid line at 2 mm of plaque size indicates the minimum size for medium-sized plaques. The vertical dotted lines at 0% indicate that the virus on the line is located at the identical genetic distance from both viruses. AA, amino acid.
In contrast, the genetic proximity of the ORF5 amino acid sequence of the viruses to JA142 or ATP appeared to have a better correlation with phenotypic characteristics. Seven viruses closely related to ATP (98.5 to 99.5% homology) produced a medium- or big-sized plaque, and one virus closely related to JA142 (97% homology) produced a small-sized plaque. Nonetheless, two viruses genetically closer to JA142 based on ORF5 amino acid sequence (98% homology) produced a big-sized plaque, indicating that ORF5 sequence homology may not be able to differentiate VLVs from wild-type viruses.
Genetic markers for PRRSVs with vaccine virus-like phenotype.
By comparing sequences of ORF2 to -7 among vaccine viruses, VLVs, and wild-type viruses, four unique amino acids were identified for each of the MLV and ATP vaccine strains (Tables 1 and 2). Phenylalanine at position 10 (F10) in ORF2a and tyrosine at 9 (Y9) in ORF2b, which arose from the same nucleic acid change, glycine at 151 (G151) in ORF5, and glutamate at 16 (E16) in ORF6 were found only in the MLV. Glutamate at 85 (E85) and tyrosine at 165 (Y165) in ORF3, serine at 80 (S80) in ORF5, and cysteine at 62 (C62) in ORF6 were found only in the ATP. Among these unique sequences, F10 in ORF2a and E85 and Y165 in ORF3 were the most stable and consistent sequence elements for MLV-like and ATP-like PRRSVs, respectively, since those amino acids were identified only in the vaccine viruses and VLVs that produced medium- to big-sized plaques among the 83 field isolates examined (Table 2).
TABLE 1.
Unique amino acid sequences for MLV-like viruses
| ORF (aaa no.) | Plaque sizeb (classification) | No. of viruses | No. of viruses with indicated amino acid at ORF position
|
||
|---|---|---|---|---|---|
| VR2332 | MLV | Other sequences | |||
| ORF2a (10) | L | Fc | S | ||
| Big to medium (MLV-like) | 13 | 2 | 11 | ||
| Small (VR2332-like) | 12 | 11 | 1 | ||
| Small (others)d | 58 | 25 | 33 | ||
| ORF2b (9) | D | Y | N or H | ||
| Big to medium (MLV-like) | 13 | 2 | 7 | 4 (H) | |
| Small (VR2332-like) | 12 | 11 | 1 (N) | ||
| Small (others)d | 58 | 50 | 8 (N) | ||
| ORF5 (151) | R | G | K, I, or N | ||
| Big to medium (MLV-like) | 13 | ≈10 | 3 | ||
| Small (VR2332-like) | 12 | 11 | 1 (I) | ||
| Small (others)d | 58 | 52 | 6 | ||
| ORF6 (16) | Q | E | |||
| Big to medium (MLV-like) | 13 | 6 | 7 | ||
| Small (VR2332-like) | 12 | 12 | |||
| Small (others)d | 58 | 58 | |||
aa, amino acid.
Big to medium is bigger than or equal to 2 mm in diameter and small is smaller than 2 mm in diameter (plaque size).
The unique sequences found only in MLV are in boldface.
One isolate (2M11715) not closely related to either VR2332 or JA142 but producing a medium sized-plaque was included in this group.
TABLE 2.
Unique amino acid sequences for ATP-like viruses
| ORF (aaa no.) | Plaque sizeb (classification) | No. of viruses | No. of viruses with indicated amino acid at ORF position
|
||
|---|---|---|---|---|---|
| JA142 | ATP | Other sequences | |||
| ORF3 (85) | D | Ec | |||
| Big to medium (ATP-like) | 9 | 9 | |||
| Small (JA142-like) | 1 | 1 | |||
| Small (others)d | 74 | 74 | |||
| ORF3 (165) | F | Y | L | ||
| Big to medium (ATP-like) | 9 | 9 | |||
| Small (JA142-like) | 1 | 1 | |||
| Small (others)d | 74 | 71 | 3 | ||
| ORF5 (80) | G | S | |||
| Big to medium (ATP-like) | 9 | 2 | 7 | ||
| Small (JA142-like) | 1 | 1 | |||
| Small (others)d | 74 | 74 | |||
| ORF6 (62) | F | C | |||
| Big to medium (ATP-like) | 9 | 2 | 7 | ||
| Small (JA142-like) | 1 | 1 | |||
| Small (others)d | 74 | 74 | |||
aa, amino acid.
Big to medium is bigger than or equal to 2 mm in diameter and small is smaller than 2 mm in diameter (plaque size).
The unique sequences found only in ATP are in boldface.
One isolate (2M11715) not closely related to either VR2332 or JA142 but producing a medium sized-plaque was included in this group.
Stability of unique amino acid sequences of vaccine viruses or VLVs during passages in animals and MARC-145.
Amino acid sequences F10 in ORF2a and Y9 in ORF2b of the CC-01 strain (MLV-like virus) reverted to the sequences of VR2332, i.e., leucine (L) and aspartate (D), respectively, after three passages in the pigs (lines A and C). In line B, however, F10 was still observed in ORF2a without reversion to L even after 13 sequential pig-to-pig passages of the virus, suggesting that F10 in ORF2a may be stable during in vivo passages of the MLV strain. The Y9 in ORF2b was, on the other hand, changed into histamine (H) instead of D after three passages, which remained unchanged until the end of 13 pig passages. The H9 in ORF2b was found in field isolates which produced a medium- to big-sized plaque (Table 1) and thus was considered to be a marker amino acid alternative to Y9.
Similarly, the amino acid sequence E16 in ORF6 was substituted with glutamine (Q) after three passages in the pigs. In line C, however, the E16 was substituted with G instead of Q, which remained without any further alteration until the completion of 13 pig-to-pig passages. Therefore, G16 in ORF6 would be an intermediate form between E and Q, similar to H9 in ORF2b, even though such an amino acid was not found in ORF6 of any of the field isolates examined in this study. In contrast, the amino acid sequence G151 in ORF5 quickly reverted to that of the parental virus, i.e., arginine (R), after the first passage in the pigs.
None of the amino acid sequences unique to the vaccine viruses were found in progeny viruses produced during 20 sequential passages of VR2332 or JA142 in MARC-145. All descendant viruses had the same amino acid sequences as VR2332 or JA142 at the determined sites during the passages, suggesting that over 20 passages might be required to have vaccine-specific sequences at the identified positions.
Biological role of the identified genetic markers.
The identified MLV genetic markers were incorporated into the VR2332 infectious clone (Table 3). To characterize the mutants, the multistep growth curve (Fig. 6A) and the size of plaques they produced (Fig. 6B) were determined with MARC-145 cells. All of the mutants appeared to grow better than VR2332E which was rescued from the original infectious clone. The effect of those identified sequences on the replication of the virus in MARC-145 cells was, however, accumulative. The growth of mutants with single-site changes (i.e., P2L10F, P5R151G, and P6G16E) was not significantly enhanced compared to that of VR2332E (P = 0.07). In contrast, mutants with changes in two sites (i.e., P25, P26, and P56) grew significantly better than did single-site-changed mutants (P = 0.01) and VR2332E (P = 0.002). Likewise, P256, i.e., a mutant with changes in three sites, grew significantly better than the mutants with changes in two sites (P = 0.008), although its growth was still less than that of MLV.
TABLE 3.
List of mutants and their mutation profiles
| Category | Name of mutant | Target ORF(s) | Mutation |
|---|---|---|---|
| Single-site mutation | P2L10F | ORF2a and -2b | L-to-F mutation at position 10 of ORF2a and D-to-Y mutation at position 9 of ORF2b |
| P5R151G | ORF5 | R-to-G mutation at position 151 of ORF5 | |
| P6Q16E | ORF6 | Q-to-E mutation at position 16 of ORF6 | |
| Multiple-site mutation | P25 | ORF2 and -5 | Combination of P2L10F and P5R151G |
| P26 | ORF2 and -6 | Combination of P2L10F and P6Q16E | |
| P56 | ORF5 and -6 | Combination of P5R151G and P6Q16E | |
| P256 | ORF2, -5, and -6 | Combination of P2L10F, P5R151G, and P6Q16E |
FIG. 6.
Growth characteristics of a modified live PRRSV vaccine (MLV), the parental strain of the vaccine (VR2332E), and mutants constructed from a VR2332-based infectious cDNA clone with amino acid substitution(s) in one or more structural proteins of VR2332 toward MLV, as determined by multistep growth curve (A) and the size of plaque they produced (B) with MARC-145 cells. Asterisks in panel A indicate that the viruses grew significantly better than VR2332E, as determined by the multiple analysis of variation test. Numbers at the top of panel B indicate the amounts of virus inoculated to MARC-145 cells (duplicate wells/virus), as expressed in FFU per 0.1 ml.
A similar observation was also made for the effect of the identified sequences on the size of plaques. The size of the plaques produced by the mutants became bigger and bigger when more of the identified genetic markers for MLV were incorporated into the VR2332 infectious clone (Fig. 6B).
DISCUSSION
A high level of genetic and antigenic variability that exists among PRRSVs is well documented. Therefore, genetic analysis tools such as sequencing and restriction fragment length polymorphism have been utilized to aid in better decision making on the introduction of a new strain into the farm, the use of pig flow as a preventive measure, and intervention (i.e., commercial vaccine versus autogenous vaccine). Unfortunately, it has been common to detect field isolates which are genetically indistinguishable from both the vaccine virus and its wild-type parental strain because the vaccine strains are genetically very close to their parental strains. There is only a 0.3 or 1% difference in the sequence of ORF5, which is one of the most variable genes of PRRSV, between MLV and VR2332 or between ATP and JA142, respectively. MLV differs in full-length sequence from VR2332 by only 44 nucleotides (31). Considering that all commercial PRRS vaccines are modified live virus products without any marker, it is reasonable to expect that many of those indistinguishable field isolates may have been derived from the vaccine strains, since the vaccine virus is known to spread to unvaccinated animals and PRRSV is known to be continuously mutated during in vivo replication (26). However, the possibility that the parental strain of the vaccine virus, such as VR2332 or JA142, has been circulating in the pig population still exists. At present, no convenient way to reliably discriminate the vaccine viruses from their wild-type parental viruses is available.
In the present study, the vaccine viruses demonstrated highly efficient growth in the cell line MARC-145 (monkey kidney cell line), resulting in plaques of sizes significantly bigger than those seen for their wild-type parental strains. As the vaccine viruses are known to be attenuated through sequential passages in the cell line, the observed biotypic characteristic (i.e., better growth in the cells) was an expected outcome. It was interesting that such a difference of viral growth in MARC-145 was also observed for some of the field isolates, which provided the basis for identifying viruses derived from the vaccine strains in conjunction with sequence analysis. The plaque assay in combination with sequencing of ORF5 can be a very powerful tool for detecting VLVs, since it utilizes both genotypic and phenotypic characteristics of the virus. Several difficulties would be expected if the plaque assay alone is used for the purpose of virus differentiation. First, the plaque assay requires virus isolates, which is not always easy to get from clinical cases. Second, field isolates which are genetically as different from MLV as ATP (approximately 10% nucleotide difference) can produce a medium-sized plaque, as demonstrated in this study. Third, the phenotype of VLV can revert to that of a wild-type when the virus passed through animals, as evident in the present study (Fig. 3). These foreseeable difficulties necessitate the combined use of plaque assay and sequence comparison, so that the plaque assay can be performed on viruses which are indistinguishable from both vaccine and parental strains by gene sequencing (e.g., ORF5).
It was somewhat unexpected that a vaccine-like plaque phenotype of the virus (CC-01) completely reverted to the wild-type plaque phenotype (i.e., small-sized plaque) after three or four sequential passages in the pigs. Such faster reversion is believed to be attributed to the fact that the virus was not highly attenuated in cell culture like MLV vaccine viruses and was passed in pigs via an unnatural process (i.e., intramuscular injection of tissue homogenate containing the virus) (6). Vaccine viruses neither replicate well in pigs nor produce high levels of viremia after vaccination (10). Also, low levels of virus are detected from semen and respiratory tracts after vaccination compared to what is seen for wild-type viruses (5, 26). Therefore, the reversion of the vaccine phenotype to the wild-type phenotype during natural processes is expected to be slower than what was observed in the present study, since the vaccine virus likely needs considerable time to be adapted to the host and evolves slowly by maintaining a low level of replication rather than spreading through many pigs in a short time.
A noteworthy finding in the present study was the identification of amino acid sequences in ORF2, -3, -5, and -6, which were unique for the vaccine viruses or VLVs among the field isolates examined. Reverse genetics demonstrated their significant role in the plaque phenotype of the virus, suggesting that these sequence elements can be utilized as surrogate genetic markers for the vaccine viruses or VLVs. The identification of these surrogate markers for the vaccine viruses or VLVs should make it possible to determine genetic and phenotypic traits of a virus simply by gene sequencing without performing the laborious plaque assay. Among the sequence elements, G151 in ORF5 and E16 in ORF6 have been suggested as cell attenuation markers (21), implying that some or all of the identified markers could be related to PRRSV virulence. This postulate could be substantiated by the presence of VR2332 sequences at all of these identified sites in MLV-like viruses isolated from Danish pigs, which caused severe PRRS outbreaks after vaccination (16). Hence, the identified surrogate genetic markers also would provide a quick way to monitor the reversion of the vaccine virus to a wild type. Nonetheless, the role of nonstructural genes and their products in the virulence of PRRSV remains to be further studied.
Acknowledgments
We thank Christine Meraz for excellent assistance in manuscript preparation. Kay Faaberg at the University of Minnesota (St. Paul, MN) is acknowledged for providing the VR2332 infectious clone, which was very valuable to this study. The vaccine viruses were kindly provided by Boehringer-Ingelheim Vetmedica, Inc., in Ames, IA.
The study was supported in part by funding from the USDA CSREES National Research Initiative (2002-35204-12772), a USDA formula grant, the Iowa Livestock Health Advisory Council, the National Pork Board, and the Iowa Pork Producers Association.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Albina, E. 1997. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet. Microbiol. 55309-316. [DOI] [PubMed] [Google Scholar]
- 2.Allende, R., T. L. Lewis, Z. Lu, D. L. Rock, G. F. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80307-315. [DOI] [PubMed] [Google Scholar]
- 3.Benfield, D. A., E. Nelson, J. E. Collins, L. Harris, S. M. Goyal, D. Robison, W. T. Christianson, R. B. Morrison, D. Gorcyca, and D. Chladek. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 4127-133. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C. C., K. J. Yoon, J. J. Zimmerman, K. M. Harmon, P. M. Dixon, C. M. Dvorak, and M. P. Murtaugh. 2002. Evolution of porcine reproductive and respiratory syndrome virus during sequential passages in pigs. J. Virol. 764750-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christopher-Hennings, J., E. A. Nelson, J. K. Nelson, and D. A. Benfield. 1997. Effects of a modified-live virus vaccine against porcine reproductive and respiratory syndrome in boars. Am. J. Vet. Res. 5840-45. [PubMed] [Google Scholar]
- 6.Dea, S., C. A. Gagnon, H. Mardassi, and G. Milane. 1996. Antigenic variability among North American and European strains of porcine reproductive and respiratory syndrome virus as defined by monoclonal antibodies to the matrix protein. J. Clin. Microbiol. 341488-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dea, S., C. A. Gagnon, H. Mardassi, B. Pirzadeh, and D. Rogan. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145659-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewey, C. E., S. Wilson, P. Buck, and J. K. Leyenaar. 2004. Effects of porcine reproductive and respiratory syndrome vaccination in breeding-age animals. Prev. Vet. Med. 62299-307. [DOI] [PubMed] [Google Scholar]
- 9.Greig, A. 1975. The use of a microtitration technique for the routine assay of African swine fever virus. Arch. Virol. 47287-289. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, W., M. Roof, E. Vaughn, J. Christopher-Hennings, C. R. Johnson, and M. P. Murtaugh. 2004. Pathogenic and humoral immune responses to porcine reproductive and respiratory syndrome virus (PRRSV) are related to viral load in acute infection. Vet. Immunol. Immunopathol. 102233-247. [DOI] [PubMed] [Google Scholar]
- 11.Keffaber, K. 1989. Reproductive failure of unknown aetiology. Am. Assoc. Swine Pract. Newsl. 11-10. [Google Scholar]
- 12.Key, K. F., D. K. Guenette, K. J. Yoon, P. G. Halbur, T. E. Toth, and X. J. Meng. 2003. Development of a heteroduplex mobility assay to identify field isolates of porcine reproductive and respiratory syndrome virus with nucleotide sequences closely related to those of modified live-attenuated vaccines. J. Clin. Microbiol. 412433-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133477-483. [DOI] [PubMed] [Google Scholar]
- 14.Kim, W. I., D. S. Lee, W. Johnson, M. Roof, S.-H. Cha, and K.-J. Yoon. 2007. Effects of genotypic and biotypic difference among PRRS viruses on the serologic assessment of pigs for virus infection. Vet. Microbiol. 1231-14. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 171244-1245. [DOI] [PubMed] [Google Scholar]
- 16.Madsen, K. G., C. M. Hansen, E. S. Madsen, B. Strandbygaard, A. Botner, and K. J. Sorensen. 1998. Sequence analysis of porcine reproductive and respiratory syndrome virus of the American type collected from Danish swine herds. Arch. Virol. 1431683-1700. [DOI] [PubMed] [Google Scholar]
- 17.Mengeling, W. L., K. M. Lager, and A. C. Vorwald. 1998. Clinical consequences of exposing pregnant gilts to strains of porcine reproductive and respiratory syndrome (PRRS) virus isolated from field cases of “atypical” PRRS. Am. J. Vet. Res. 591540-1544. [PubMed] [Google Scholar]
- 18.Mengeling, W. L., K. M. Lager, A. C. Vorwald, and D. F. Clouser. 2003. Comparative safety and efficacy of attenuated single-strain and multi-strain vaccines for porcine reproductive and respiratory syndrome. Vet. Microbiol. 9325-38. [DOI] [PubMed] [Google Scholar]
- 19.Neumann, E. J., J. B. Kliebenstein, C. D. Johnson, J. W. Mabry, E. J. Bush, A. H. Seitzinger, A. L. Green, and J. J. Zimmerman. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227385-392. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen, H. S., G. Liu, J. Nielsen, M. B. Oleksiewicz, A. Botner, T. Storgaard, and K. S. Faaberg. 2003. Generation of an infectious clone of VR-2332, a highly virulent North American-type isolate of porcine reproductive and respiratory syndrome virus. J. Virol. 773702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opriessnig, T., P. G. Halbur, K. J. Yoon, R. M. Pogranichniy, K. M. Harmon, R. Evans, K. F. Key, F. J. Pallares, P. Thomas, and X. J. Meng. 2002. Comparison of molecular and biological characteristics of a modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine (Ingelvac PRRS MLV), the parent strain of the vaccine (ATCC VR2332), ATCC VR2385, and two recent field isolates of PRRSV. J. Virol. 7611837-11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, B.-K., T. W. Molitor, and H.-S. Joo. 1999. Viral characteristics of plaque variants of porcine reproductive and respiratory syndrome virus. Korean J. Vet. Res. 39751-759. [Google Scholar]
- 23.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoint. Am. J. Hyg. 27493-497. [Google Scholar]
- 24.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. Raamsman, and A. A. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 736335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storgaard, T., M. Oleksiewicz, and A. Botner. 1999. Examination of the selective pressures on a live PRRS vaccine virus. Arch. Virol. 1442389-2401. [DOI] [PubMed] [Google Scholar]
- 26.Thanawongnuwech, R., P. G. Halbur, M. R. Ackermann, E. L. Thacker, and R. L. Royer. 1998. Effects of low (modified-live virus vaccine) and high (VR-2385)-virulence strains of porcine reproductive and respiratory syndrome virus on pulmonary clearance of copper particles in pigs. Vet. Pathol. 35398-406. [DOI] [PubMed] [Google Scholar]
- 27.Truong, H. M., Z. Lu, G. F. Kutish, J. Galeota, F. A. Osorio, and A. K. Pattnaik. 2004. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology 325308-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wesley, R. D., W. L. Mengeling, K. M. Lager, D. F. Clouser, J. G. Landgraf, and M. L. Frey. 1998. Differentiation of a porcine reproductive and respiratory syndrome virus vaccine strain from North American field strains by restriction fragment length polymorphism analysis of ORF 5. J. Vet. Diagn. Investig. 10140-144. [DOI] [PubMed] [Google Scholar]
- 29.Wesley, R. D., W. L. Mengeling, K. M. Lager, A. C. Vorwald, and M. B. Roof. 1999. Evidence for divergence of restriction fragment length polymorphism patterns following in vivo replication of porcine reproductive and respiratory syndrome virus. Am. J. Vet. Res. 60463-467. [PubMed] [Google Scholar]
- 30.Wu, W. H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287183-191. [DOI] [PubMed] [Google Scholar]
- 31.Yuan, S., D. Mickelson, M. P. Murtaugh, and K. S. Faaberg. 2001. Complete genome comparison of porcine reproductive and respiratory syndrome virus parental and attenuated strains. Virus Res. 79189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]