Abstract
Adenoviruses for gene or oncolytic therapy are under development. Notable among these strategies is adenoviral delivery of the tumor suppressor p53. Since all therapeutics have limitations in certain settings, we have undertaken retroviral suppressor screens to identify genes conferring resistance to adenovirus-delivered p53. These studies identified the tumor antigen LRRC15, which is frequently overexpressed in multiple tumor types, as a repressor of cell death due to adenoviral p53. LRRC15, however, does not impede p53 function per se but impedes adenoviral infection. Specifically, LRRC15 causes redistribution of the coxsackievirus-adenovirus receptor away from the cell surface. This effect is manifested in less adenoviral binding to the surfaces of LRRC15-expressing cells. This discovery, therefore, not only is important for understanding adenoviral biology but also has potentially important implications for adenovirus-based anticancer therapeutics.
Adenoviral vectors have been developed as agents for either intratumoral or systemic gene or oncolytic therapy (6, 13). One approach has been the development of adenoviral vectors that carry the tumor suppressor p53 (15). p53 is the most frequently mutated gene in human cancer (22). When activated, or delivered virally, p53 can bring about two major phenotypic effects, growth arrest or apoptosis (4). Many tumor cells have a higher predisposition to die in response to p53 than their normal counterparts, making delivery of p53 a selective way to induce tumor cell death. However, as with all therapeutic strategies, not all agents will work equally well in all tumor settings. Differences in genetic backgrounds and gene expression profiles in tumors, differences in the tissues of origin of the tumor, and differences between individuals can render the use of any particular therapeutic strategy ineffective. For example, the use of adenovirus-delivered p53 in many sarcomas that overexpress the p53 negative regulator Mdm2 (12), which causes degradation of p53 (7, 8), appears to be limited. The identification of high levels of Mdm2 in tumors, however, not only points to informed selective use of adenovirus-delivered p53 but also suggests the development of combination therapies involving adenoviral p53 together with agents that perturb the negative regulation of p53 by Mdm2.
Many other tumor types that retain wild-type p53 are considered to contain mutational events, in addition to overexpression of Mdm2, that in some way impede p53 function. We set out, therefore, to search for such factors using genome-wide retroviral insertional mutagenesis screens. We considered that retrovirus-infected cell clones that were resistant to the death-inducing action of adenovirus-delivered p53 would contain retroviral insertions that affected either p53 function per se, cell death pathways downstream of p53, or the effective gene delivery or expression of p53 from the viral vector. Genes identified by this approach, like mdm2, would not only be useful indicators for the selective use of adenovirus-delivered p53 but may also represent potential novel targets for combination therapy.
MATERIALS AND METHODS
Plasmids.
Plasmids involved in the generation of ERM (enhanced retroviral mutagenesis) virus-infected cells were kindly provided by Zhou Songyang (Baylor College of Medicine, Houston, TX) and have been described previously (9). pWZLneo-EcoR and pcDNA3-LRRC15-hemagglutinin (HA) were kind gifts from Gordon Peters (Cancer Research UK—London Research Institute) and Daniel Haber (Harvard Medical School), respectively. pBabePuro-LRRC15-HA was generated by digestion of pcDNA-LRRC15-HA with HindIII and BamHI. The fragment was then blunt-ended and cloned into the SnaBI site of pBabePuro. Expression vectors for p53 (pCB6+p53) and Mdm2 (pCHDM21A) have been described previously (3, 11). The green fluorescent protein (GFP) expression plasmid EGFP-N1 is from BD Biosciences.
Cell culture and generation of cell lines.
Saos2-EcoR, A549-EcoR, and HCT-116-EcoR cells were generated by amphotropic retroviral infection of the respective parental cell lines with pWZLneo-EcoR as previously described (16). The cells were then infected using an ecotropic packaging system with either pBabePuro-LRRC15-HA or an empty pBabePuro vector as a control. Saos2-ERM cells were generated as previously described (9). All lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and incubated at 37°C under an atmosphere of 5% CO2 in air. Transgene expression was induced by addition of 1 μg/ml doxycycline (Dox) (Sigma). For p53 degradation assays, Saos2 cells were transiently transfected by calcium phosphate precipitation as described elsewhere (2). After 16 h, cells were washed and then incubated for a further 24 h before harvesting for Western blotting.
Generation of replication-defective adenoviruses.
Plasmid EGFP-N1-p53 was made by inserting wild-type p53 sequences into the EcoRI and BamHI restriction sites of pEGFP-N1 (Clontech). pShuttleCMV-GFP and pShuttleCMV-p53 were made by inserting the GFP and WTp53-GFP fragments from plasmids EGFP-N1 and EGFP-N1p53 into pShuttleCMV (Stratagene) using BglII and NotI restriction sites. Linearized “Shuttle” plasmids were electroporated into BJ5183-AD-1 electrocompetent cells (Stratagene) containing the Adeasy-1 adenoviral construct (Stratagene). Recombinant plasmids were then amplified in XL10-Gold cells (Stratagene) and transfected into HEK293 cells after restriction digestion with PacI. Following infectious adenovirus amplification in 293 cells, purified virus was isolated by freeze-thaw extraction and titered using the BD Bioscience Adeno-X Rapid Titer kit. Adenoviruses were added to cell cultures at the multiplicities of infection indicated in the figures and legends.
Western blotting.
Cells were lysed in a 2× Western sodium dodecyl sulfate sample buffer and transferred to nitrocellulose membranes as previously described (2). Membranes were probed using standard immunoblotting techniques with antibodies that recognize LRRC15 (horseradish peroxidase-conjugated HA; Roche), p53 (DO-1; Pharmingen), GFP (Covance), Hdm2 (Ab1; Oncogene Science), the coxsackievirus-adenovirus receptor (CAR) (RmcB; ATCC), and actin (clone 1A4; Sigma).
Flow cytometry and cell death assays.
Total populations of cells, including floating and adherent cells, were processed for flow cytometric analysis (FACScan; Becton Dickinson) as described previously (17). Acquired events were analyzed and quantified with CellQuest software (Becton Dickinson). The percentage of cells with a sub-G1 DNA content was taken as a measure of the extent of apoptosis in the cell population at that time.
Identification of ERM clones.
Pools of cells infected with ERM viruses were challenged with three rounds of infection with adenoviral p53. Each round of infection was separated by a period of 7 to 10 days. RNA was isolated from surviving clones and subjected to seminested reverse transcription-PCRs (RT-PCRs) using the following primers and conditions. The RNAs were reverse transcribed using a random primer, RT3 (5′-GCAAATACGACTCACTATAGGGATCCNNNNSTGG-3′), and the GeneAmp RNA PCR core kit (Applied Biosystems). Thirty cycles of PCR amplification were carried out using ProofStart polymerase (Qiagen) and PCR primers HA1 (5′-CACCAAGGCGCGCCAAGCACTATCCGTACGA-3′) and T7 (5′-GGCAAATACGACTCACTATAGGG-3′) according to the manufacturer's manual. PCR conditions (MJ Research DNA Engine PTC-200) were as follows: 95°C for 5 min; 30 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min; and finally 72°C for 10 min. Products from these reactions were sequenced and integration sites identified using the “Ensembl” genome browser (http://www.ensembl.org) and the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST).
qPCR.
DNA and RNA were prepared using TRIzol reagent (Invitrogen). Quantitative PCR (qPCR) analysis was carried using the DyNAmo Sybr green 2-step qRT-PCR kit (Finnzymes). Data were collected using a Chromo4 real-time PCR detector and were analyzed with Opticon Monitor 3. Primers for LRRC15 were as follows: forward, GCCTGTATGTACTGCTTTAACTC; reverse, GGATAATGCCATTTCAGTGGT. Primers for GFP were from Qiagen (QT01171611). Primers for 18S rRNA have been described previously (5). qPCR cycling parameters were 95°C for 15 min; 34 cycles of 94°C for 10 s, 55°C for 30 s, and 72°C for 30 s; and 72°C for 10 min. Expression levels of genes analyzed by qPCR were normalized relative to levels of 18S rRNA.
Analysis of CAR and integrin levels by flow cytometry.
Cells at 80% confluence were trypsinized or removed using phosphate-buffered saline (PBS)-2.5 mM EDTA and pelleted at 1,200 rpm for 5 min. The pellet was washed once with ice-cold fluorescence-activated cell sorter (FACS) buffer (PBS, 2% fetal bovine serum, 0.5 mM EDTA [pH 8.0]). Cells were spun down, resuspended in ice-cold FACS buffer containing 5% fetal bovine serum and a monoclonal antibody (2 μg/ml), and incubated on ice for 60 min. Antibodies against CAR (RmcB; ATCC), integrin αv (ab16821; Abcam), integrin β3 (catalog no. 555752; BD Pharmingen), and integrin αvβ5 (MAB2019Z; Chemicon) were used. Cells were pelleted and washed three times with FACS buffer. Cells were then resuspended in FACS buffer containing goat anti-mouse immunoglobulin G conjugated with fluorescein isothiocyanate (catalog no. 0479; Dako) and incubated at room temperature for 30 min. Cells were pelleted and again washed three times with FACS buffer, and the expression of CAR or integrins on the cell surface was determined using a Becton Dickinson FACSCalibur flow cytometer.
Immunofluorescence.
Cells were seeded on glass coverslips at 50% confluence. The following day, cells were infected with adenovirus (90 min on ice); washed three times, for 5 min each time, with PBS; and fixed in PBS containing 4% paraformaldehyde. For staining, coverslips were rinsed in PBS-0.1% Triton X-100, incubated in blocking solution (PBS, 5% milk, 10% fetal bovine serum, 0.1% Triton X-100) for 30 min at room temperature, washed in blocking solution without milk, and then incubated with an anti-adenovirus type 5 antibody (ab6982; Abcam) for 60 min at 37°C. The coverslips were washed in PBS-0.1% Triton X-100, incubated in a goat anti-rabbit secondary antibody conjugated with Alexa Fluor 488 (A11008; Molecular Probes) for 30 min at 37°C, washed in PBS-0.1% Triton X-100, and refixed in PBS-4% paraformaldehyde for 20 min at room temperature. Cells were then washed again and mounted on slides. Microscopy was carried out using a Zeiss Axioplan 2IE microscope and ISIS software.
RESULTS AND DISCUSSION
A screen for factors causing resistance to adenoviral p53 identifies LRRC15.
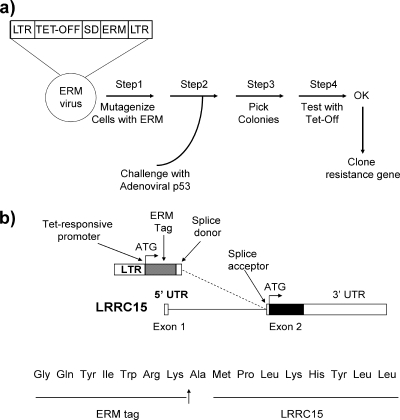
In order to identify genes that cause resistance to adenoviral p53, we utilized the ERM system, which has been described previously (9, 10). In brief, this system comprises tetracycline-responsive exon trap retroviruses (in three reading frames) that integrate randomly throughout the target cell genome when they are used to infect cells. Gene fusions occur between exons of endogenous genes and a sequence tag on the retrovirus (ERM tag) via a splice donor site in the viral vector. This causes up-regulation of expression of the endogenous gene due to the enhancer effect of the viral long terminal repeat (Fig. 1a). The ERM tag can then be used to facilitate identification of the viral integration site by PCR.
FIG. 1.
A screen for suppressors of p53-induced cell death identified LRRC15. (a) Cells containing mutagenizing retroviruses were generated and then challenged with adenoviral p53 as depicted. (b) RT-PCR identified an in-frame viral integration at the beginning of exon 2 of LRRC15. LTR, long terminal repeat; SD, splice donor site; Tet-Off, tetracycline-responsive repressor element; UTR, untranslated region.
Large pools of Saos2 cells, which are null for endogenous p53, were infected with this viral system as previously described (9, 16) in order to attain a high diversity of viral integration. Pools of these cells were challenged with a regimen of infection with adenoviral p53 that is just sufficient to kill an entire population of Saos2 cells without ERM (data not shown). Following adenoviral infection of ERM cells, however, a number of surviving colonies were found to emerge. Sequencing of one of these clones using the viral tag as previously described (9) revealed an in-frame integration at the beginning of exon 2 of the LRRC15 gene (Fig. 1b). Since LRRC15 has a noncoding first exon, this integration involved the entire reading frame of LRRC15.
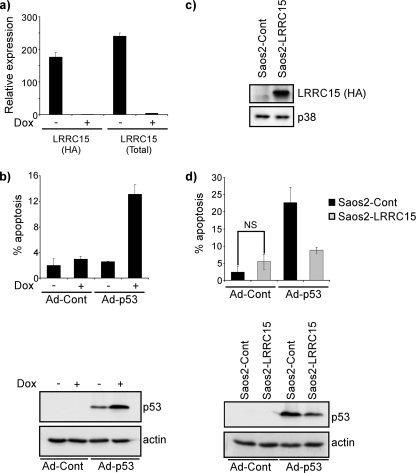
LRRC15 (also known as hLib) belongs to the leucine-rich repeat superfamily, members of which are involved in cell-cell and cell-extracellular matrix interactions. While it was first identified as a protein induced by β-amyloid (19), subsequent studies have indicated that LRRC15 is frequently overexpressed in various tumor types and, in particular, is associated with high-grade, aggressive breast and prostate tumors (14, 18, 20, 23). This association with tumor development caused us to examine further the clones of cells with an integration in LRRC15. First, we retested to determine whether the cells were resistant to death by adenoviral p53 and, since expression from ERM viruses is tetracycline responsive, whether resistance to cell death was dependent on a viral integration. For this purpose, cells were incubated with the tetracycline analog Dox (1 μg/ml for 48 h) to switch off ERM integrations (the ERM constructs are “Tet-Off” regulated). This caused a marked reduction in the levels of both ERM-tagged LRRC15 and total LRRC15 (Fig. 2a). Cells were then challenged with adenoviral p53 and assessed for cell death induction after 48 h by flow cytometry as previously described (17). This revealed clearly that an ERM integration was causing resistance to the effects of adenoviral p53. In the absence of Dox, the amount of cell death from adenoviral p53 was similar to that in control cells. Following Dox treatment, however, cell death from adenoviral p53 was markedly increased (Fig. 2b). Interestingly, this increase in cell death from adenoviral p53 when LRRC15 was switched off occurred concomitantly with an increase in the levels of p53 protein (Fig. 2b).
FIG. 2.
LRRC15 impedes cell death due to adenoviral p53 and decreases p53 levels. (a) ERM-LRRC15 cells were treated with Dox for a period of 48 h prior to infection with adenoviral p53, and the levels of ERM-tagged and total LRRC15 were determined by qPCR. (b) ERM-LRRC15 cells that had been treated with Dox for a period of 48 h were then, where indicated, infected with adenoviral p53 (Ad-p53) or a control “empty” adenovirus (Ad-Cont). Cells were harvested for flow cytometry 48 h postinfection and for Western blotting 24 h postinfection. (c) Saos2 cells were infected with a retrovirus expressing LRRC15 (Saos2-LRRC15) or an empty vector as a control (Saos2-Cont), and the presence of HA-tagged LRRC15 was determined by Western blotting. (d) Saos2-LRRC15 cells and Saos2-Cont cells were infected with Ad-p53 or Ad-Cont and analyzed after 48 h for cell death by flow cytometry and after 24 h for p53 expression by Western blotting. In cell death assays, the percentage of cells with a sub-G1 DNA content was taken as a measure of the extent of apoptosis at the indicated time. NS, not significant; P = 0.12 by a two-tailed t test.
Since the possibility remained that the ERM-LRRC15 cells contained more than one viral integration, such that the effects on cell death and p53 levels were independent of LRRC15, we reestablished a cell line that constitutively expresses LRRC15 from a heterologous promoter. Cells were infected with a retroviral construct that had been generated using a cDNA for LRRC15 that was previously described (14) (Fig. 2c). These cells (Saos2-LRRC15) and empty vector control cells (Saos2-pBabe) were infected with adenoviral p53 and analyzed for cell death induction and p53 protein levels. As with the ERM-LRRC15 cells in the absence of Dox, the presence of LRRC15 caused a marked reduction in the amount of cell death seen following infection with adenoviral p53 (Fig. 2d), confirming that the decreased level of cell death seen in ERM-LRRC15 cells is attributable to the viral integration in the LRRC15 locus. When the levels of p53 protein were analyzed following adenoviral infection, the result was also similar to that observed for ERM-LRRC15 cells: less p53 was detectable in LRRC15-expressing cells than in vector-only controls (Fig. 2d).
LRRC15 impedes adenoviral infection.
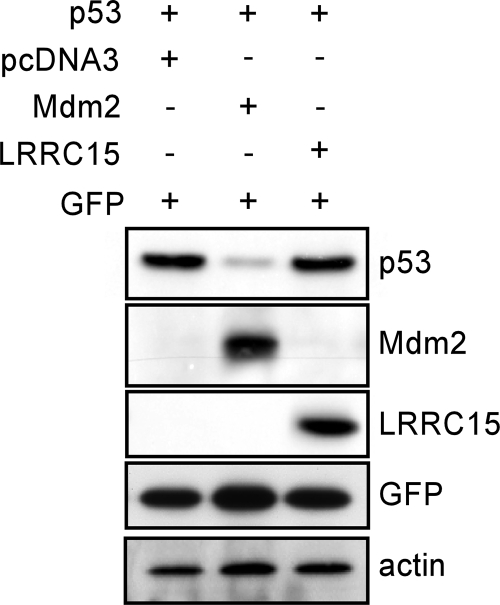
The lower levels of p53 in LRRC15-expressing cells caused us to consider whether LRRC15 was impeding p53 function, in a manner similar to that of Mdm2, by causing p53 degradation. To test this, Saos2 cells were transiently transfected as previously described (2) with p53 in combination with either Mdm2, LRRC15, or an empty vector as a control. In line with previous studies (7, 8), coexpression of Mdm2 caused a marked reduction in p53 levels (Fig. 3). In contrast, coexpression of LRRC15 had no effect on p53 protein levels, indicating that LRRC15 does not promote p53 degradation (Fig. 3).
FIG. 3.
LRRC15 does not affect p53 stability. Saos2 cells were transiently transfected with the combinations of plasmids indicated. After 24 h, cells were harvested and analyzed for protein expression by Western blotting. Equal amounts of total protein were added to each lane. The results shown are representative of five separate experiments. GFP was included in each transfection as a control for transfection efficiency.
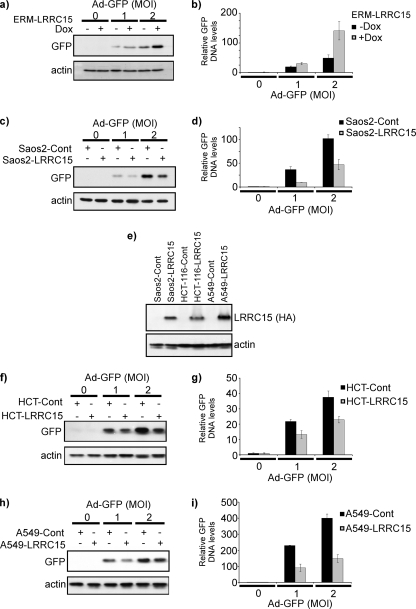
Since we observed that expression of LRRC15 had no effect on the levels or death-inducing activity of p53 following transfection of cells by calcium phosphate precipitation (data not shown), we next considered whether LRRC15 may affect either the efficiency of infection or transgene expression by the adenoviral vector. If either of these were possible, we would expect to see similar effects on the expression of any adenovirally delivered transgene. Saos2-LRRC15 cells, Saos2-pBabe cells, and ERM-LRRC15 cells (with or without Dox) were therefore infected with a previously described adenovirus containing a transgene that expresses GFP (1). As was observed with p53, GFP levels were indeed lower in Saos2-LRRC15 cells and ERM-LRRC15 cells than in Saos2-pBabe cells or Dox-treated ERM-LRRC15 cells, respectively (the ERM-driven integration of LRRC15 in these cells is “Tet-Off” and hence is switched off by treatment with Dox) (Fig. 4a and b). These results therefore indicate that LRRC15 impedes the efficacy of adenoviral gene transfer per se, since the effects we originally saw are not specific to p53. Since LRRC15 is a plasma membrane protein, we considered it most likely that LRRC15 impedes the infectivity of the adenoviral particle as opposed to having an effect on, for example, transgene expression. If the latter were the case, we would expect fewer adenoviral genomes in LRRC15-expressing cells, which would result in lower levels of transgene expression at the protein level. As a reflection of the number of GFP-containing adenoviral genomes in an infected cell population, we analyzed GFP gene levels in adenovirus-infected Saos2-LRRC15 cells, Saos2-pBabe cells, and ERM-LRRC15 cells (with or without Dox) by qPCR as previously described (5). This analysis indeed revealed that LRRC15-expressing cells contain fewer copies of the GFP transgene than control cells (Fig. 4c and d), and since the adenoviral vectors used are replication defective, this finding confirms that LRRC15 impedes adenoviral infection.
FIG. 4.
LRRC15 impedes adenoviral infection. (a and b) After 48 h of pretreatment with Dox (to switch off LRRC15, which is driven by the ERM integration), ERM-LRRC15 cells were infected with an adenovirus containing a GFP transgene (Ad-GFP). Twenty-four hours postinfection, GFP and GFP gene levels were determined by Western blotting (a) and qPCR (b), respectively. MOI, multiplicity of infection (number of virus particles per cell as determined by plaque assay). (c and d) Saos2-LRRC15 cells and control (Saos2-Cont) cells were infected with an adenovirus expressing GFP. Twenty-four hours postinfection, GFP and GFP gene levels were determined by Western blotting (c) and qPCR (d), respectively. (e) HCT-116 and A549 cells were infected either with a retrovirus expressing HA-tagged LRRC15 or with an empty viral vector as a control. The expression of HA-tagged LRRC15 was assessed and compared to that in Saos2-LRRC15 cells by Western blotting. (f and g) HCT-116-LRRC15 and HCT-116-Cont cells were infected with an adenovirus expressing GFP. Twenty-four hours postinfection, GFP and GFP gene levels were determined by Western blotting (f) and qPCR (g), respectively. (h and i) A549-LRRC15 and A549-Cont cells were infected with an adenovirus expressing GFP. Twenty-four hours postinfection, GFP and GFP gene levels were determined by Western blotting (h) and qPCR (i), respectively.
To test the generality of this observation, we extended our study to two other cell lines from different tissues of origin—HCT-116 and A549, which are derived from human colonic and lung tumors, respectively. These cell lines were infected in an identical manner to Saos2 cells with either a retrovirus expressing LRRC15 or an empty viral vector as a control (Fig. 4e). In both cases, as was observed for Saos2 cells (Fig. 4a to d), the presence of LRRC15 reduced both the amount of GFP expressed in cells (Fig. 4f and g) and the number of GFP transgenes upon transduction with adenovirus relative to those observed for the controls (Fig. 4h and i).
LRRC15 affects surface CAR expression and cell surface adenoviral binding.
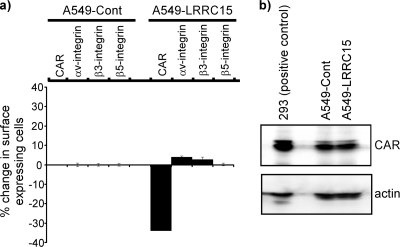
We were interested in understanding how LRRC15 was affecting the delivery of adenovirus to cells. Since changes in cellular morphology and cell cycle stage have been reported to affect adenoviral infectivity (21), we first explored whether LRRC15 was affecting either of these cellular phenotypes, but no significant differences were observed between cells expressing LRRC15 and those containing an empty viral vector as a control (see Fig. S1 in the supplemental material). A number of cell surface proteins have also been shown to affect different stages of adenoviral infectivity. CAR is known to mediate adenoviral attachment to the cell surface, while integrins αvβ3 and α3β5 control internalization (24). Since we observed the most robust effects of LRRC15 expression in A549 cells, we chose to use these cells to explore these cellular parameters of adenoviral infection in the context of LRRC15. Surface expression of integrins and CAR was therefore analyzed by flow cytometry in A549 cells expressing either LRRC15 or an empty viral vector. This revealed that while the expression of LRRC15 had limited effects on the surface expression of integrins αv, β3, and β5, surface expression of CAR was markedly reduced in cells expressing LRRC15 relative to that in controls (Fig. 5a). Interestingly, total levels of CAR were not affected by the expression of LRRC15 (Fig. 5b), indicating that CAR must be redistributed away from the cell surface in LRRC15-expressing cells. The destination(s) of the relocalized CAR, however, has yet to be determined.
FIG. 5.
LRRC15 decreases cell surface, but not total, CAR levels. (a) A549-LRRC15 and A549 control (Cont) cells were analyzed for surface expression of the following parameters, which are known to affect adenoviral transduction: CAR, integrin αv, integrin β3, and integrin β5. Surface expression was quantified by flow cytometry, and the percentage of cells showing a change in surface expression in A549-LRRC15 cells is shown relative to that observed in A549-Cont cells. (b) Total CAR levels in A549-LRRC15 and A549-Cont cells were measured by Western blotting. A lysate from 293 cells is included as a positive-control guideline for detection of CAR expression.
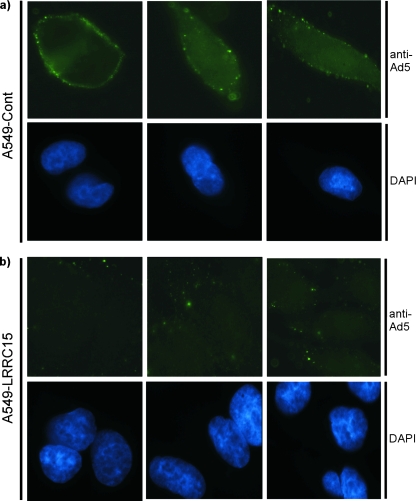
Since CAR is involved in the attachment of adenoviral particles to the cell surface, we postulated that LRRC15 may directly affect this process. To test this, LRRC15-expressing cells and controls were incubated on ice in order to permit adenoviral attachment, but not internalization, such that attachment could be assessed exclusively (24). Cells were then incubated with the adenovirus that contains the GFP transgene. Since the GFP is expressed only when the adenovirus is within the host cells, cells were subsequently fixed and stained with an antibody raised against whole adenovirus (type 5). This clearly revealed, as would be predicted by the decrease in surface CAR expression that we had observed, that LRRC15 markedly reduced the extent of attachment of adenoviral particles to the surfaces of cells (Fig. 6).
FIG. 6.
LRRC15 impedes adenoviral attachment. (a and b) A549-LRRC15 (b) and A549 control (Cont) (a) cells were incubated with Ad-GFP for 90 min on ice to permit adenoviral attachment but not internalization. Cells were subsequently stained with an anti-adenovirus type 5 (Ad5) antibody and 4′,6′-diamidino-2-phenylindole (DAPI) before being visualized by fluorescent microscopy.
We believe that our findings, taken together with the fact that LRRC15 is overexpressed in multiple tumor types, report a new factor that not only is involved in the mediation of adenoviral infectivity but also may have important implications for the successful application of adenoviruses for either gene therapy or oncolytic viral therapy. The further analysis of LRRC15 in other cancers will undoubtedly determine how far-reaching this effect of LRRC15 may be and whether it is a factor for consideration in treating multiple tumor types. Identification of high levels of LRRC15 in a tumor could be used as a criterion in decisions relating to the selective use, or the strategy of administration, of an adenovirus-based therapy in any individual case. Moreover, future studies to further explore the mechanism of action of LRRC15 will also be rewarding and may lead to strategies to enhance adenoviral infectivity in cases where LRRC15 is either at normal levels or overexpressed.
Supplementary Material
Acknowledgments
We thank Zhou Songyang and Daniel Haber for reagents. We are also grateful to Alan Bilsland, Jim Norman, Clodagh O'Shea, Yaohe Wang, and Nicol Keith for advice and for critical reading of the manuscript.
Work in the Tumour Cell Death Laboratory is supported by Cancer Research UK. K.M.R. is a Cancer Research-UK Senior Cancer Research Fellow.
Footnotes
Published ahead of print on 2 April 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bell, H. S., C. Dufes, J. O'Prey, D. Crighton, D. Bergamaschi, X. Lu, A. G. Schatzlein, K. H. Vousden, and K. M. Ryan. 2007. A p53-derived apoptotic peptide derepresses p73 to cause tumor regression in vivo. J. Clin. Investig. 1171008-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, L. A., J. O'Prey, and K. M. Ryan. 2006. DNA-binding independent cell death from a minimal proapoptotic region of E2F-1. Oncogene 255656-5663. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J., X. Wu, J. Lin, and A. J. Levine. 1996. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol. Cell. Biol. 162445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crighton, D., and K. M. Ryan. 2004. Splicing DNA-damage responses to tumour cell death. Biochim. Biophys. Acta 17053-15. [DOI] [PubMed] [Google Scholar]
- 5.Crighton, D., S. Wilkinson, J. O'Prey, N. Syed, P. Smith, P. R. Harrison, M. Gasco, O. Garrone, T. Crook, and K. M. Ryan. 2006. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126121-134. [DOI] [PubMed] [Google Scholar]
- 6.Everts, B., and H. G. van der Poel. 2005. Replication-selective oncolytic viruses in the treatment of cancer. Cancer Gene Ther. 12141-161. [DOI] [PubMed] [Google Scholar]
- 7.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387296-299. [DOI] [PubMed] [Google Scholar]
- 8.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387299-303. [DOI] [PubMed] [Google Scholar]
- 9.Liu, D., X. Yang, and Z. Songyang. 2000. Identification of CISK, a new member of the SGK kinase family that promotes IL-3-dependent survival. Curr. Biol. 101233-1236. [DOI] [PubMed] [Google Scholar]
- 10.Liu, D., X. Yang, D. Yang, and Z. Songyang. 2000. Genetic screens in mammalian cells by enhanced retroviral mutagens. Oncogene 195964-5972. [DOI] [PubMed] [Google Scholar]
- 11.Marston, N. J., T. Crook, and K. H. Vousden. 1994. Interaction of p53 with MDM2 is independent of E6 and does not mediate wild type transformation suppressor function. Oncogene 92707-2716. [PubMed] [Google Scholar]
- 12.Rayburn, E., R. Zhang, J. He, and H. Wang. 2005. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr. Cancer Drug Targets 527-41. [DOI] [PubMed] [Google Scholar]
- 13.Relph, K. L., K. J. Harrington, and H. Pandha. 2005. Adenoviral strategies for the gene therapy of cancer. Semin. Oncol. 32573-582. [DOI] [PubMed] [Google Scholar]
- 14.Reynolds, P. A., G. A. Smolen, R. E. Palmer, D. Sgroi, V. Yajnik, W. L. Gerald, and D. A. Haber. 2003. Identification of a DNA-binding site and transcriptional target for the EWS-WT1(+KTS) oncoprotein. Genes Dev. 172094-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth, J. A. 2006. Adenovirus p53 gene therapy. Expert Opin. Biol. Ther. 655-61. [DOI] [PubMed] [Google Scholar]
- 16.Ryan, K. M., M. K. Ernst, N. R. Rice, and K. H. Vousden. 2000. Role of NF-κB in p53-mediated programmed cell death. Nature 404892-897. [DOI] [PubMed] [Google Scholar]
- 17.Ryan, K. M., and K. H. Vousden. 1998. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol. Cell. Biol. 183692-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satoh, K., M. Hata, and H. Yokota. 2004. High Lib mRNA expression in breast carcinomas. DNA Res. 11199-203. [DOI] [PubMed] [Google Scholar]
- 19.Satoh, K., M. Hata, and H. Yokota. 2002. A novel member of the leucine-rich repeat superfamily induced in rat astrocytes by beta-amyloid. Biochem. Biophys. Res. Commun. 290756-762. [DOI] [PubMed] [Google Scholar]
- 20.Schuetz, C. S., M. Bonin, S. E. Clare, K. Nieselt, K. Sotlar, M. Walter, T. Fehm, E. Solomayer, O. Riess, D. Wallwiener, R. Kurek, and H. J. Neubauer. 2006. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 665278-5286. [DOI] [PubMed] [Google Scholar]
- 21.Seidman, M. A., S. M. Hogan, R. L. Wendland, S. Worgall, R. G. Crystal, and P. L. Leopold. 2001. Variation in adenovirus receptor expression and adenovirus vector-mediated transgene expression at defined stages of the cell cycle. Mol. Ther. 413-21. [DOI] [PubMed] [Google Scholar]
- 22.Soussi, T., and G. Lozano. 2005. p53 mutation heterogeneity in cancer. Biochem. Biophys. Res. Commun. 331834-842. [DOI] [PubMed] [Google Scholar]
- 23.Stanbrough, M., G. J. Bubley, K. Ross, T. R. Golub, M. A. Rubin, T. M. Penning, P. G. Febbo, and S. P. Balk. 2006. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 662815-2825. [DOI] [PubMed] [Google Scholar]
- 24.Wang, K., T. Guan, D. A. Cheresh, and G. R. Nemerow. 2000. Regulation of adenovirus membrane penetration by the cytoplasmic tail of integrin β5. J. Virol. 742731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.