Abstract
A longstanding enigmatic feature of the group 1 coronaviruses is the uncleaved phenotype of their spike protein, an exceptional property among class I fusion proteins. Here, however, we show that some group 1 coronavirus spike proteins carry a furin enzyme recognition motif and can actually be cleaved, as demonstrated for a feline coronavirus. Interestingly, this feature can be lost during cell culture adaptation by a single mutation in the cleavage motif; this, however, preserves a heparan sulfate binding motif and renders infection by the virus heparan sulfate dependent. We identified a similar cell culture adaptation for the human coronavirus OC43.
Enveloped viruses use different types of fusion proteins to realize the membrane fusion by which they initiate their infection. For coronaviruses, it is the spike (S) protein that is responsible for cell entry, and this S protein has been shown to belong to the class I fusion proteins (4). These proteins typically occur in virions as homotrimeric complexes primed for fusion through cleavage by furin-like enzymes. Membrane fusion by these activated proteins can then be triggered upon receptor binding (e.g., human immunodeficiency virus type 1) or by conditions such as low pH after endosomal uptake (e.g., influenza A virus) (for a recent review, see reference 50).
One of the puzzling questions about coronavirus S protein-mediated membrane fusion regards the cleavage requirement of the S protein. Coronaviruses have been assigned to different groups based on antigenic and genetic criteria (41). Interestingly, while the group 1 coronaviruses carry uncleaved S proteins, the S proteins of almost all viruses from groups 2 and 3 are furin activated (10) by processing at a characteristic multibasic motif (often RRXRR) present in these proteins. The importance of cleavage for infectivity was underscored recently by the revelation that the two prominent group 2 viruses lacking such a furin recognition site, and hence carrying uncleaved spikes, appeared to depend on a different, new processing mechanism. Thus, the severe acute respiratory syndrome coronavirus (SARS-CoV) and the murine hepatitis virus strain 2 (MHV-2) were both shown to require proteolytic cleavage in their target cell, which is mediated by cathepsin enzymes (23, 36, 42). The cathepsin cleavage site of the SARS-CoV spike protein was mapped to the same region as that in which, in other viruses, the S protein is activated by furin (B. J. Bosch and P. J. M. Rottier, unpublished observations), hence similarly generating an amino-terminal, receptor binding domain (S1) and a membrane-anchored carboxy-terminal domain (S2) responsible for membrane fusion (for reviews, see references 3 and 9).
When looking closer into the enigmatic lack of cleavage of the group 1 coronavirus spike proteins, we established that the infection of cells by two of those viruses, human coronavirus (HCoV) NL63 (23) and feline infectious peritonitis virus strain 79-1146 (our unpublished observations), is insensitive to cathepsin inhibitors. However, we also noted the presence of a multibasic furin cleavage motif, or an apparent remnant thereof, in reported S protein sequences of some group 1 viruses (Table 1), specifically among the related feline coronaviruses (FCoVs) and canine coronaviruses (CCoVs) of serotype I (20, 35). Thus, a perfect furin cleavage motif (RRXRR) (11, 18) occurs in the FCoV strains UCD (33, 46) and UCD8 (24, 31) and in the CCoV strain Elmö/02 (35). Interestingly, while otherwise almost identical, the S proteins of FCoV strains UCD and UCD1 differ by a conspicuous R-to-G substitution in the furin motif (RRSRG). The combined observations prompted us to investigate the cleavage phenotype of serotype I FCoV spike proteins to further our insight into the role of this characteristic feature of class I fusion proteins in infections by coronaviruses.
TABLE 1.
S1-S2 junction sequences of coronavirus S proteins and their predicted and observed furin cleavage
| Group | Virus straind | Sequencea | Predictionb | Confirmation of protein cleavagec | Reference(s) |
|---|---|---|---|---|---|
| 1 | FCoV UCD | NHTHSRRSRRSTLTSV | + (0.835) | + | This study; 33, 46 |
| 1 | FCoV UCD1 | NHTHSRRSRGSTSTSV | − (0.383) | − | This study; 31, 32 |
| 1 | FCoV UCD8 | NHTQPRRSRRSTPNSV | + (0.819) | ND | 24, 31 |
| 1 | CCoV Elmö | HTHTVRRARRAVQTGT | + (0.787) | ND | 35 |
| 1 | FCoV 1146 | NGALVFINTVTHSDGD | − (0) | − | 6, 45 |
| 1 | HCoV NL63 | DGSLIPVRPRNSSDNG | − (0.105) | ND | 44 |
| 2a | MHV A59 | DYSKSRRAHRSVSTGY | + (0.802) | + | 15, 29 |
| 2a | MHV/BHK | DYSKSRRAHRLGSTGY | − (0.359) | − | 8, 39 |
| 2a | MHV-2 | NYSTSRHARSSVSTGY | − (0.207) | − | 36, 52 |
| 2a | HCoV OC43 BE03 | DYSKNRRSRRAITTGY | + (0.751) | ND | 48 |
| 2a | HCoV OC43 ATCC | DYSKNRRSRGAITTGY | + (0.551) | − | 21, 27 |
| 2a | BCV Mebus | DYSTKRRSRRAITTGY | + (0.774) | + | 1 |
| 2b | SARS-CoV | SYHTVSLLRSTSQKSI | − (0.123) | − | 14, 23, 42, 51 |
| 3 | IBV Bdtt | ITNGTRRFRRSITENV | + (0.833) | + | 2, 5 |
Basic amino acids are in bold; residues immediately downstream of the (putative) cleavage site are italicized; residues mutated, presumably as a result of cell culture adaptation, are underlined.
+ and − indicate whether a furin-specific cleavage site is or is not predicted (11) between the S1 and S2 domains of the spike protein, respectively. The numbers in parentheses indicate the probability scores of furin cleavage; 0.5 is used as the cutoff value.
+ and − indicate whether or not the spike protein cleavage during virus assembly and release has been experimentally confirmed, respectively. ND (not determined) indicates the lack of experimental data.
MHV-2 and SARS-CoV, but not HCoV NL63 or MHV A59, require endosomal proteolysis by cathepsins for their entry. BCV, bovine coronavirus; IBV, infectious bronchitis virus.
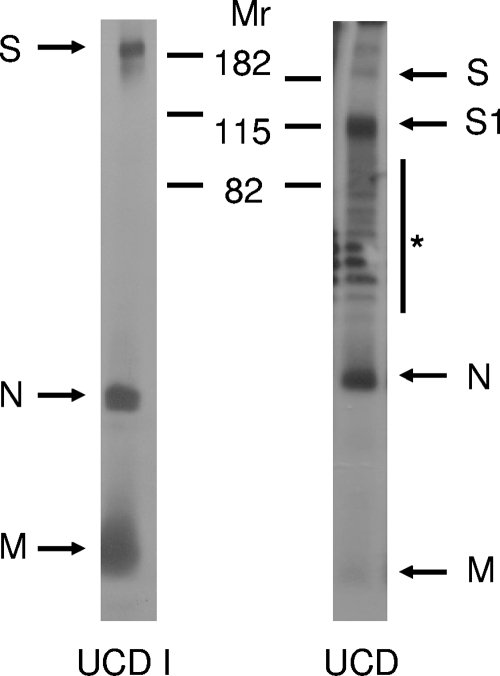
In contrast to the serotype II FCoVs, serotype I FCoVs are notorious for their poor growth in cell culture, which may be related to their use of a different, yet unidentified, cell receptor (12, 22). An exception is FCoV strain UCD1, which has been successfully adapted to cell culture (32) and of which we thus grew (low-titer) stocks in Felis catus whole-fetus (FCWF) cells (ATCC). FCoV UCD was purified from feces collected from experimentally infected cats by using sucrose gradient centrifugation. To examine their S protein cleavage state, cell culture-derived FCoV UCD1 and feces-derived FCoV UCD particles were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12.5% gels followed by Western blotting using serum from a serotype I FCoV (strain RM)-infected cat (34). The results for the analysis of FCoV UCD1, shown in Fig. 1, reveal a full-length S protein, in addition to the nucleocapsid (N) protein and the membrane (M) protein; no S protein cleavage products could be observed. In contrast, in the case of FCoV UCD, hardly any full-length S proteins could be detected; rather, a prominent lower-molecular-weight protein with an electrophoretic mobility corresponding to that of the S1 subunit was observed. No clear S2 subunit could be observed due to its known inefficient recognition by the antiserum used (data not shown). These initial results were consistent with prediction (Table 1).
FIG. 1.
FCoV UCD virions, but not FCoV UCD1 virions, contain cleaved spike proteins. Radiolabeled FCoV UCD1 virions were immunoprecipitated from infected cell culture supernatant using an FCoV RM antiserum and subjected to SDS-PAGE. FCoV UCD virions were subjected to SDS-PAGE, followed by Western blotting using the same antiserum to visualize FCoV-specific proteins. The positions of the molecular weight markers (Mr; in thousands) are indicated. Arrows indicate the FCoV-specific S, S1, N, and M proteins. The asterisk indicates a ladder of nonspecific bands that was consistently observed with feces-derived viral samples.
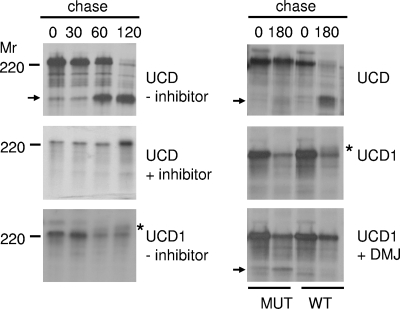
To independently confirm the observations, we next studied the cleavage of these spike proteins when expressed using the vaccinia virus vTF7-3 system (16). The UCD and UCD1 spike genes were obtained from genomic RNA by reverse transcriptase (RT)-PCR and cloned as a 4.5-kb SacI-XhoI fragment into pWSK29 (49) under the control of bacteriophage T7 transcription regulatory elements. The resulting plasmids were used to transfect OST7-1 cells (13) that had been infected 1 h earlier with vaccinia virus vTF7-3. At 5 h postinfection, the cells were pulse-labeled with 35S-amino acids for 1 h and subsequently chased for different time periods. To analyze whether furin-like host cell proteases are responsible for the cleavage of the spike protein, we made use of a decanoyl peptidyl chloromethyl ketone (dec-RVKR-cmk), a chemically modified peptide known to inhibit furin cleavage activity in cultured cells (47). The cells were lysed and processed for immunoprecipitation as described before (7), again using the serotype I FCoV RM antiserum. The immunoprecipitates were analyzed by SDS-12.5% PAGE. As shown in Fig. 2 (left panel), hardly any cleavage of the UCD spike protein was observed after the 60-min labeling period. However, during the chase, an increasing fraction of the S protein became cleaved, as judged by the appearance of the lower-molecular-weight protein. After deglycosylation, the apparent molecular weight of this protein species corresponded with the predicted size of the S1 subunit (data not shown). Cleavage of the spike protein was complete after the 2-h chase. In the presence of the furin peptide inhibitor, cleavage of the UCD spike protein was severely inhibited. This is consistent with furin or a furin-like enzyme being responsible for the cleavage. In contrast, no sign of cleavage could be observed for the UCD1 spike protein, even though the protein was able to traffic through the secretory pathway, as indicated by the characteristic appearance of the mature form of the glycoprotein (Fig. 2). To confirm that the mutations in or close to the multibasic motif of the UCD1 S protein are detrimental for recognition by the furin enzymes, they were introduced into the UCD S protein (RRSRRSTL → RRSRGSTS [mutated residues are indicated in bold]). Cleavage of the UCD mutant protein was indeed severely impaired (Fig. 2, right panel). Conversely, when the multibasic furin cleavage site was restored in the UCD1 spike protein, the characteristic high-molecular-weight mature form of the glycoprotein (Fig. 2) could no longer be detected, while its cleavage could be observed after treatment of the cells with 1-deoxymannojirimycin (DMJ). DMJ inhibits Golgi body-resident mannosidases (17) and, as a consequence, facilitates the detection of otherwise heterogeneously glycosylated proteins. In summary, our results demonstrate that the S protein of FCoV UCD, but not that of FCoV UCD1, is cleaved by a furin-like protease.
FIG. 2.
The spike protein of FCoV UCD, but not that of FCoV UCD1, is cleaved by a furin-like protease. OST7-1 cells infected with vaccinia virus vTF7-3 were transfected with plasmids carrying either the UCD or UCD1 spike gene under the control of bacteriophage T7 transcription regulatory elements. The cells were pulse-labeled with 35S-amino acids for 1 h and subsequently chased for 0, 30, 60, 120, or 180 min. Where indicated by a plus, the furin inhibitor dec-RVKR-cmk (75 μM) (left panel) or the mannosidase I inhibitor DMJ (1 mM) (right panel) was added to the culture media and kept present throughout the experiment. Spike cleavages of the wild-type (WT) and mutant (MUT) UCD and UCD1 proteins were also compared (see text for details). The cells were lysed and processed for immunoprecipitation using FCoV RM antiserum. The immunoprecipitates were analyzed by SDS-PAGE. The position of a molecular weight marker (Mr) is shown on the left side of the panels. The asterisks indicate the positions of the mature UCD1 spike protein. The arrows indicate the positions of the S1 cleavage products. Only the relevant portion of the gel is shown.
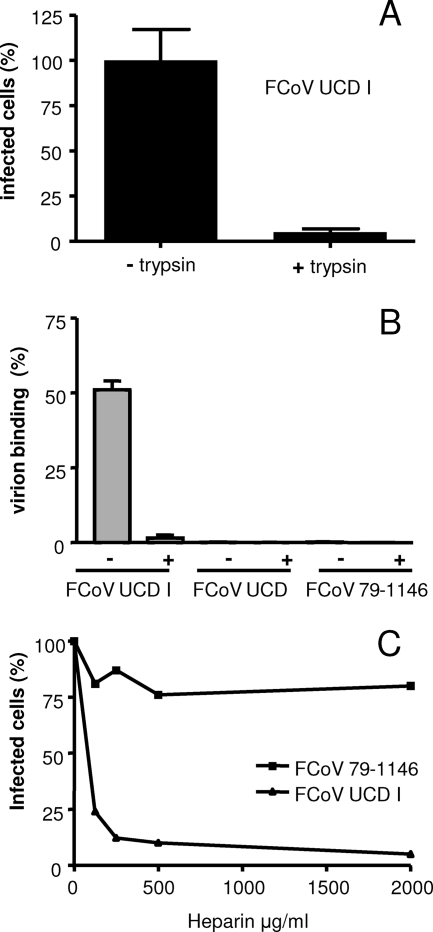
In view of the remarkably different in vitro growth capacities of the UCD and UCD1 strains despite their virtually identical S protein sequences, we hypothesized that the subtle R-to-G mutation in the UCD1 multibasic motif might have been critical for the cell culture adaptation. The situation is reminiscent of a previously described MHV A59 variant obtained from a persistently infected cell culture (38), which appeared to have developed an extended host range (39, 40). This virus, named MHV/BHK, had acquired the ability to use heparan sulfate as an entry receptor (8), due to the acquisition in its spike protein of two putative heparan sulfate binding sites (XBXBBX and XBXXBBBX, with B being a basic amino acid) (28). In addition, the spike protein had lost the ability to be cleaved, although the basic residues within the furin cleavage motif, which also make up a heparan sulfate binding motif, had been preserved (8). Similarly, the basic motif at the S1-S2 boundary of the FCoV UCD1 spike protein (XBBXBX) also corresponds to a heparan sulfate binding motif (28). Therefore, we analyzed whether the lack of cleavage of the UCD1 spike protein is indeed required for cell culture infectivity. While treatment of coronavirus particles with trypsin generally results in increased infectivity (3), treatment of purified FCoV UCD1 particles with trypsin prior to inoculation decreased the number of infected cells (Fig. 3A), which could be visualized by immunoperoxidase staining (37).
FIG. 3.
Heparin binds to FCoV UCD1 and inhibits its infectivity. (A) Sucrose gradient-purified FCoV UCD1 particles were treated with TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (25 μg/ml) for 30 min at room temperature (+trypsin) or mock treated (−trypsin) prior to the inoculation of FCWF cells. The cells were fixed at 18 h postinfection. The number of infected cells relative to the number of cells infected after inoculation with mock-treated viruses was determined by immunoperoxidase staining using the FCoV RM antiserum. (B) Similar quantities of FCoV UCD1, UCD, and 79-1146 virions (as determined by quantitative TaqMan RT-PCR [19]) were incubated for 1 h at 4°C with heparin-agarose beads in the presence (+) or absence (−) of heparin (500 ng/ml; added 1 h before addition of the beads). After three washes, the amounts of virions adsorbed to the beads, expressed again as viral genome equivalents, were determined relative to the amount of input RNA. (C) FCoV UCD1 and 79-1146 were incubated with different concentrations of heparin for 1 h at 4°C prior to the inoculation of FCWF cells. The cells were fixed at 8 h postinfection. The number of infected cells relative to the number of cells infected after inoculation with mock-treated viruses was determined by immunoperoxidase staining using the FCoV RM antiserum.
Next, we analyzed the ability of the virions of FCoV strains UCD1, UCD, and 79-1146 (30), the last being a serotype II FCoV, to bind to heparin-agarose beads. Heparin, a product of mast cells, is commonly used as an analog of cellular heparan sulfate in receptor-ligand interaction assays (25). Similar quantities of the virions, as determined by quantitative TaqMan RT-PCR (19), were incubated for 1 h at 4°C with the heparin-agarose beads in the presence or absence of heparin (500 ng/ml; added 1 h before addition of the beads). After three washes, the amounts of viral genomic RNA adsorbed to the beads were again determined by TaqMan RT-PCR (19). The results are shown in Fig. 3B. As expected, FCoV UCD1 was found attached to the heparin beads, and this binding could efficiently be blocked by the addition of heparin, confirming the specificity of the assay. FCoV UCD, which also contains a heparan sulfate binding motif at its furin cleavage site, was found not to adsorb to the beads. Apparently, the basic motif functions as a heparan sulfate binding site only as long as the motif is not subject to furin cleavage, lending further support to the cleavage phenotype of the UCD spike protein. FCoV 79-1146, which does not contain a multibasic motif, did not bind to the heparin beads.
In order to more directly test the involvement of heparan sulfate in the entry process of FCoV UCD1, the ability of heparin to inhibit viral infectivity was studied. FCoV strains UCD1 and 79-1146 were incubated with different concentrations of heparin for 1 h at 4°C prior to the inoculation of FCWF cells. FCoV UCD was not tested, as this virus does not grow in cell culture. In each case, the number of infected cells relative to the number of cells infected after inoculation with a mock-treated virus was determined. We found that incubation of the FCoVs with heparin prior to infection dramatically decreased the infectivity of FCoV UCD1 (>90%) but not that of FCoV 79-1146. These results indicate that the cell culture-adapted serotype I FCoV strain UCD1, but not the serotype II FCoV strain 79-1146, is critically dependent on binding to heparan sulfate for entry into FCWF cells.
Our observations may resolve some conflicting reports regarding the receptor used by serotype I FCoVs. Based primarily on the ability of FCoV UCD1 to infect cells expressing feline aminopeptidase N (fAPN), a cell surface glycoprotein known to serve as the receptor for serotype II FCoVs, Tresnan and coworkers (43) concluded that fAPN apparently also functions in the entry of serotype I viruses. However, we and others (12, 22) found that serotype I FCoVs do not recognize fAPN as a functional receptor. While the infection of fAPN-expressing cells by serotype II FCoVs could effectively be inhibited with a monoclonal antibody directed against fAPN, serotype I FCoV infections were not affected by this antibody (22). Furthermore, feline cells expressing fAPN could be infected with retroviral pseudovirus particles containing serotype II, but not serotype I, spikes (12). Yet, FCoV UCD1 was able to infect FCWF cells, which express fAPN, as well as Vero and HeLa cells, which do not express fAPN, and these infections were hence insensitive to the fAPN-blocking antibody (B. J. Haijema and P. J. M. Rottier, unpublished results). The results are consistent with the cell culture-adapted FCoV UCD1 infecting cells in a heparan sulfate-dependent manner, with the consequent extension of its host range.
Heparan sulfate is an attractive target for viruses because of its appropriate physiological location on the surface of most animal cells, where the initial interactions with viruses occur. Viruses from widely different families, including alpha-, herpes-, flavi-, picorna-, and retroviruses (for a review, see reference 28), have been shown to interact with glycosaminoglycans, in most cases heparan sulfate, often as a result of cell culture adaptation. Our data suggest that for FCoV-UCD1, the cleavability of its spike protein by a furin-like protease was traded off against heparan sulfate binding. In this way, the cell culture-adapted FCoV UCD1, but not primary isolates like FCoV UCD, is able to infect cells that do not contain the natural receptor, the identity of which is as yet unknown.
One important lesson that the examples of FCoV UCD1 and MHV/BHK taught us is that one has to be cautious when passaging viruses in cell cultures, always being aware of possibly subtle, but often significant, genomic alterations. A possible illustration of such a cell culture adaptation that has gone unnoticed so far involves HCoV OC43, a group 2 HCoV strain. The spike protein of the OC43 strain provided by the ATCC, which has been passaged in cell culture, contains a multibasic motif (RRSRG) that is identical to the one present in the S protein of FCoV UCD1 (Table 1) (27). Cleavage of the OC43 spike protein could not be observed (21), which is consistent with the low theoretical probability thereof (Table 1) and with our observations with FCoV UCD1. Sequence analysis of the genomes of more-recent, primary OC43 isolates, which had not been passaged in cell culture, showed the presence of a perfect furin cleavage motif (RRSRR) in the S protein (48). Based on our observations with the serotype I FCoVs, we hypothesize that in the ATCC strain of HCoV OC43, the S protein has lost its ability to be cleaved by a furin-like protease yet has gained the capacity to bind to heparan sulfate. A similar but inverse correlation between furin cleavage and heparan sulfate binding of a multibasic motif has also been observed with the unrelated Sindbis virus (26).
There are as yet no indications that the furin cleavability of the spike protein somehow contributes to the virulence phenotype of FCoVs. FCoVs of both serotypes occur as two distinct pathotypes. FCoV UCD is an example of the pathotype that causes a mild enteric infection in cats, while FCoV UCD1 causes feline infectious peritonitis, a lethal systemic infection (32, 33). However, the lack of cleavage of the FCoV UCD1 spike protein and its ability to bind to heparan sulfate are not necessarily associated with high virulence, as demonstrated by the occurrence of other serotype I FCoV isolates that have not been passaged in cell culture and are known to cause feline infectious peritonitis but that contain a perfect furin cleavage site in their S proteins (e.g., FCoV UCD8 [Table 1]) (24).
In conclusion, our data show that cleavage of the spike protein is not restricted to group 2 and group 3 coronaviruses but does occur, as well, in group 1 viruses, hence breaking a longstanding dogma. Whether spike protein cleavage is essential for the infectivity of group 1 coronaviruses, as it is for the viruses of groups 2 and 3, is as yet unclear. The case of FCoV UCD1, which apparently lost its furin cleavability while remaining infectious, seems to argue against the idea. However, we have to keep in mind the example of the group 2 coronavirus MHV-2, infection by which is critically dependent on cathepsin cleavage due to the presence of a defective furin cleavage motif in its spike protein. Repair of this motif rendered the virus cathepsin independent, as the S protein was now cleaved by furin (36). Likewise, the introduction of a furin cleavage site at the S1-S2 boundary into the SARS-CoV spike protein enhanced its cell-cell fusion activity (14), while protease treatment of SARS-CoV S protein-pseudotyped retroviruses rendered their cell entry process insensitive to cathepsin inhibitors (42). Similarly, FCoV UCD1 might use an alternative mechanism to ensure cleavage of its spike protein, and this might generally hold for group 1 coronaviruses with S proteins not cleaved by furin. Thus, the great challenge will be to demonstrate whether this hypothesis is correct and, if so, to elucidate the cleavage mechanism(s) and identify the proteases involved, as this would confirm that cleavage activation is indeed a universal feature of class I fusion proteins.
Acknowledgments
We thank Berend Jan Bosch for stimulating discussions.
This work was supported by a grant from The Netherlands Organization for Scientific Research (NWO-VIDI-700.54.421) to C.A.M.D.H. and by a grant from The Netherlands Organization for Scientific Research and The Netherlands Foundation for Applied Sciences to B.J.H., P.S., and P.J.M.R.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Abraham, S., T. E. Kienzle, W. Lapps, and D. A. Brian. 1990. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology 176296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binns, M. M., M. E. Boursnell, D. Cavanagh, D. J. Pappin, and T. D. Brown. 1985. Cloning and sequencing of the gene encoding the spike protein of the coronavirus IBV. J. Gen. Virol. 66719-726. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, B. J., and P. J. Rottier. 2008. Nidovirus entry into cells, p. 157-177. In S. Perlman, T. Gallagher, and E. J. Snijder (ed.), Nidoviruses. ASM Press, Washington, DC.
- 4.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 778801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanagh, D., P. J. Davis, D. J. Pappin, M. M. Binns, M. E. Boursnell, and T. D. Brown. 1986. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 4133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Groot, R. J., J. Maduro, J. A. Lenstra, M. C. Horzinek, B. A. van der Zeijst, and W. J. Spaan. 1987. cDNA cloning and sequence analysis of the gene encoding the peplomer protein of feline infectious peritonitis virus. J. Gen. Virol. 682639-2646. [DOI] [PubMed] [Google Scholar]
- 7.de Haan, C. A., L. Kuo, P. S. Masters, H. Vennema, and P. J. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 726838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, C. A., Z. Li, E. te Lintelo, B. J. Bosch, B. J. Haijema, and P. J. Rottier. 2005. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 7914451-14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Haan, C. A., and P. J. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A., K. Stadler, G. J. Godeke, B. J. Bosch, and P. J. Rottier. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 786048-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duckert, P., S. Brunak, and N. Blom. 2004. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 17107-112. [DOI] [PubMed] [Google Scholar]
- 12.Dye, C., N. Temperton, and S. G. Siddell. 2007. Type I feline coronavirus spike glycoprotein fails to recognize aminopeptidase N as a functional receptor on feline cell lines. J. Gen. Virol. 881753-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elroy-Stein, O., and B. Moss. 1990. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. USA 876743-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Follis, K. E., J. York, and J. H. Nunberg. 2006. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology 350358-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frana, M. F., J. N. Behnke, L. S. Sturman, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 56912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuhrmann, U., E. Bause, G. Legler, and H. Ploegh. 1984. Novel mannosidase inhibitor blocking conversion of high mannose to complex oligosaccharides. Nature 307755-758. [DOI] [PubMed] [Google Scholar]
- 18.Garten, W., S. Hallenberger, D. Ortmann, W. Schafer, M. Vey, H. Angliker, E. Shaw, and H. D. Klenk. 1994. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76217-225. [DOI] [PubMed] [Google Scholar]
- 19.Gut, M., C. M. Leutenegger, J. B. Huder, N. C. Pedersen, and H. Lutz. 1999. One-tube fluorogenic reverse transcription-polymerase chain reaction for the quantitation of feline coronaviruses. J. Virol. Methods 7737-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haijema, B. J., P. J. Rottier, and R. J. de Groot. 2007. Feline coronaviruses: a tale of two-faced types, p. 183-203. In V. Thiel (ed.), Coronaviruses: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom.
- 21.Hogue, B. G., and D. A. Brian. 1986. Structural proteins of human respiratory coronavirus OC43. Virus Res. 5131-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hohdatsu, T., Y. Izumiya, Y. Yokoyama, K. Kida, and H. Koyama. 1998. Differences in virus receptor for type I and type II feline infectious peritonitis virus. Arch. Virol. 143839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, I. C., B. J. Bosch, F. Li, W. Li, K. H. Lee, S. Ghiran, N. Vasilieva, T. S. Dermody, S. C. Harrison, P. R. Dormitzer, M. Farzan, P. J. Rottier, and H. Choe. 2006. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2813198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiss, I., A. M. Poland, and N. C. Pedersen. 2004. Disease outcome and cytokine responses in cats immunized with an avirulent feline infectious peritonitis virus (FIPV)-UCD1 and challenge-exposed with virulent FIPV-UCD8. J. Feline Med. Surg. 689-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjellén, L., and U. Lindahl. 1991. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 60443-475. [DOI] [PubMed] [Google Scholar]
- 26.Klimstra, W. B., H. W. Heidner, and R. E. Johnston. 1999. The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate. J. Virol. 736299-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Künkel, F., and G. Herrler. 1993. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology 195195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, J., and S. C. Thorp. 2002. Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 221-25. [DOI] [PubMed] [Google Scholar]
- 29.Luytjes, W., L. S. Sturman, P. J. Bredenbeek, J. Charite, B. A. van der Zeijst, M. C. Horzinek, and W. J. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKeirnan, A. J. E., A. Hargis, L. M. Miller, and R. L. Ott. 1981. Isolation of feline coronaviruses from two cats with diverse disease manifestations. Feline Pract. 1116-20. [Google Scholar]
- 31.Motokawa, K., T. Hohdatsu, H. Hashimoto, and H. Koyama. 1996. Comparison of the amino acid sequence and phylogenetic analysis of the peplomer, integral membrane and nucleocapsid proteins of feline, canine and porcine coronaviruses. Microbiol. Immunol. 40425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen, N. C., J. F. Boyle, and K. Floyd. 1981. Infection studies in kittens, using feline infectious peritonitis virus propagated in cell culture. Am. J. Vet. Res. 42363-367. [PubMed] [Google Scholar]
- 33.Pedersen, N. C., J. F. Boyle, K. Floyd, A. Fudge, and J. Barker. 1981. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am. J. Vet. Res. 42368-377. [PubMed] [Google Scholar]
- 34.Poland, A. M., H. Vennema, J. E. Foley, and N. C. Pedersen. 1996. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J. Clin. Microbiol. 343180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratelli, A., V. Martella, N. Decaro, A. Tinelli, M. Camero, F. Cirone, G. Elia, A. Cavalli, M. Corrente, G. Greco, D. Buonavoglia, M. Gentile, M. Tempesta, and C. Buonavoglia. 2003. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Methods 1109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu, Z., S. T. Hingley, G. Simmons, C. Yu, J. Das Sarma, P. Bates, and S. R. Weiss. 2006. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 805768-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottier, P. J., K. Nakamura, P. Schellen, H. Volders, and B. J. Haijema. 2005. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 7914122-14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawicki, S. G., J. H. Lu, and K. V. Holmes. 1995. Persistent infection of cultured cells with mouse hepatitis virus (MHV) results from the epigenetic expression of the MHV receptor. J. Virol. 695535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schickli, J. H., L. B. Thackray, S. G. Sawicki, and K. V. Holmes. 2004. The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells. J. Virol. 789073-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schickli, J. H., B. D. Zelus, D. E. Wentworth, S. G. Sawicki, and K. V. Holmes. 1997. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 719499-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siddell, S. G. 1995. The Coronaviridae: an introduction, p. 1-10. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, NY.
- 42.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 10211876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tresnan, D. B., R. Levis, and K. V. Holmes. 1996. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 708669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vennema, H., L. Heijnen, A. Zijderveld, M. C. Horzinek, and W. J. Spaan. 1990. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 64339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vennema, H., A. Poland, J. Foley, and N. C. Pedersen. 1998. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology 243150-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vey, M., W. Schafer, B. Reis, R. Ohuchi, W. Britt, W. Garten, H. D. Klenk, and K. Radsak. 1995. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediated by the human endoprotease furin. Virology 206746-749. [DOI] [PubMed] [Google Scholar]
- 48.Vijgen, L., E. Keyaerts, P. Lemey, E. Moes, S. Li, A. M. Vandamme, and M. Van Ranst. 2005. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology 33785-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 50.Weissenhorn, W., A. Hinz, and Y. Gaudin. 2007. Virus membrane fusion. FEBS Lett. 5812150-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, X., S. Chakraborti, A. S. Dimitrov, K. Gramatikoff, and D. S. Dimitrov. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 3121159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada, Y. K., K. Takimoto, M. Yabe, and F. Taguchi. 1997. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology 227215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]