Abstract
Homologs of the UL25 gene product of herpes simplex virus 1 (HSV-1) are highly conserved among the Herpesviridae. However, their exact function during viral replication is unknown. Current evidence suggests that in the alphaherpesvirus pseudorabies virus (PrV) the capsid-associated pUL25 plays a role in primary envelopment of DNA-containing mature capsids at the inner nuclear membrane. In the absence of pUL25, capsids were found in close association with the inner nuclear membrane, but nuclear egress was not observed (B. G. Klupp, H. Granzow, G. M. Keil, and T. C. Mettenleiter, J. Virol. 80:6235-6246, 2006). In contrast, HSV-1 pUL25 has been assigned a role in stable packaging of viral genomes (N. Stow, J. Virol. 75:10755-10765, 2001). Despite these apparently divergent functions, we wanted to assess whether the high sequence homology translates into functional homology. Therefore, we first analyzed a newly constructed HSV-1 UL25 deletion mutant in our assay system and observed a similar phenotype as in PrV. In the nuclei of infected cells, numerous electron-dense C capsids were detected, whereas primary envelopment of these capsids did not ensue. In agreement with results from PrV, vesicles were observed in the perinuclear space. Since these data indicated functional homology, we analyzed the ability of pUL25 of HSV-1 to complement a PrV UL25 deletion mutant and vice versa. Whereas a HSV-1 pUL25-expressing cell line partially complemented the pUL25 defect in PrV, reciprocal complementation of a HSV-1 UL25 deletion mutant by PrV pUL25 was not observed. Thus, our data demonstrate overlapping, although not identical functions of these two conserved herpesvirus proteins, and point to a conserved functional role in herpes virion formation.
Capsid formation is morphologically similar in all herpesviruses and resembles that of tailed bacteriophages (10). After synthesis in the cytosol, capsid proteins are transported into the nucleus, where they assemble in an autocatalytic process. The newly replicated concatemeric DNA is cleaved to unit length during packaging into the preformed capsids with a concomitant loss of the scaffold protein. After encapsidation, capsids contact and bud at the inner nuclear membrane for primary envelopment, thus initiating nucleocapsid transit to the cytosol for final maturation (24, 26, 38). Viral proteins homologous to the products of the HSV-1 genes UL31 and UL34 are required for nuclear egress (8, 9, 15, 35, 39) and are sufficient for the formation of membranous vesicles resembling primary envelopes in PrV (17). pUL34 is a nuclear-membrane-associated phosphoprotein with a predicted type II integral topology that interacts with pUL31 (8, 9, 19, 34, 35, 41). This complex formation is important for proper positioning of both proteins at the inner nuclear membrane (28, 36, 45). Moreover, coexpression of pUL31 and pUL34 alters lamin architecture (3, 27, 28, 33, 37), a prerequisite for intranuclear capsids to access the inner nuclear membrane. However, how intranuclear capsids are directed to the budding site is unknown.
Current evidence suggests that the conserved pUL25 capsid-associated protein is also required for primary envelopment (16, 44). Homologs of the UL25 gene product of HSV-1 have been identified in all three subfamilies of the Herpesviridae (2, 4, 21). The UL25 gene of pseudorabies virus (PrV) encodes a 534-amino-acid protein with a molecular mass of 57 kDa (7, 11, 14). The identity at the amino acid level is high, e.g., 54% between PrV and HSV-1 pUL25. The high degree of sequence conservation might indicate common functions in the herpesvirus life cycle and it has indeed been shown that pUL25 homologs play an essential role in replication of all of the herpesviruses studied thus far. However, their exact function is still unclear. Ogasawara et al. (32) demonstrated that pUL25 is directly associated with the major capsid protein UL19 (VP5) and triplex protein pUL38 (VP19C), while Trus et al. identified an interaction between pUL25 and the essential minor capsid protein pUL17 (44). HSV-1 pUL25 has been proposed to be essential for the packaging of newly replicated viral genomic DNA (22). In cells infected with an UL25-negative HSV-1 mutant, KUL25NS, no DNA-containing capsids were found (22). In contrast, in another study in cells infected with the same mutant, a low level of stably packaged unit-length viral genomes was detectable (42). Interestingly, although DNA-containing capsids were observed, these were not translocated to the cytoplasm, indicating that pUL25 may be essential for a step beyond packaging (42). To shed more light on pUL25 function, we analyzed pUL25 of PrV. In the absence of pUL25, newly replicated genomic PrV DNA was cleaved, and DNA-containing C capsids were detected in infected cell nuclei, albeit at reduced levels. This demonstrated that PrV pUL25 may be involved in but is clearly not essential for the cleavage-packaging process (16). Although numerous DNA-containing C-type capsids were found in close association with the inner nuclear membrane, primary envelopment was not observed. By immunolabeling pUL25 was only found on mature C capsids but not on precursor or defective capsids (16). The HSV-1 pUL25 was also found to be located on the outside of the mature capsid as part of the penton and hexon structures (29, 32), suggesting that it may play a role in capsid egress from the nucleus. Moreover, DNA-containing HSV-1 capsids were found to have higher pUL25 content than capsids lacking DNA (32, 40), supporting the hypothesis that the association of pUL25 with DNA-containing capsids is a signal for their interaction with the nuclear envelopment complex and inclusion into primary envelopes. In line with this proposal, cryoelectron microscopic studies visualized an additional structural component detected on C capsids, designated C-capsid-specific component, which supposedly consists of a pUL25/pUL17 heterodimer. This structure was not observed on empty A capsids (44).
To examine whether pUL25 of PrV and HSV-1 execute similar functions, we generated a HSV-1 pUL25-expressing rabbit kidney cell line and a HSV-1 UL25 deletion mutant paralleling our constructs in PrV (16). We characterized its phenotype ultrastructurally and analyzed matching PrV and HSV-1 UL25 mutants for transcomplementation by the heterologous pUL25.
MATERIALS AND METHODS
Cells and viruses.
All virus mutants analyzed were derived from PrV strain Kaplan (PrV-Ka [12]) or HSV-1 strain KOS (HSV1-KOS [kindly provided by P. Spear, Northwestern University, Chicago, IL]). Viruses were grown on rabbit kidney (RK13) or African green monkey kidney (Vero) cells in minimum essential medium supplemented with 10 or 5% fetal calf serum, respectively. The generation of PrV-ΔUL25 and the corresponding RK13-UL25 cells has been described (16).
Generation of a HSV-1 UL25-expressing cell line and HSV-1 UL25 deletion mutant.
For the generation of a HSV-1 UL25-expressing cell line, the complete UL25 open reading frame was amplified by PCR using Pfx polymerase (Invitrogen, Karlsruhe, Germany), the primers HUL25For (5′-CACAGAATTCCTCTCGCAGATGGACCCG-3′) (nucleotides [nt] 48803 to 48821; GenBank accession number X14112; the start codon is shown in boldface) and HUL25Rev (5′-CACACTCGAGACCACCCACTAAACGCGC-3′) (reverse to nt 50546 to 50563; GenBank accession number X14112; the stop codon is shown in boldface), and a cloned 2.3-kb genomic BamHI fragment containing the complete HSV-1 UL25 open reading frame (Fig. 1) as a template. The primer sequences contained added restriction enzyme recognition sites for EcoRI and XhoI (underlined), which were used for subsequent cloning of the 1.8-kb PCR product into appropriately cleaved eukaryotic expression vector pcDNA3 (Invitrogen). The resulting plasmid pcDNA-UL25(HSV-1) was transfected into RK13 cells by using Superfect transfection reagent (Qiagen, Hilden, Germany). Transfected cell clones were selected in medium containing 500 μg of Geneticin (Invitrogen) per ml and tested for pUL25(HSV-1) expression.
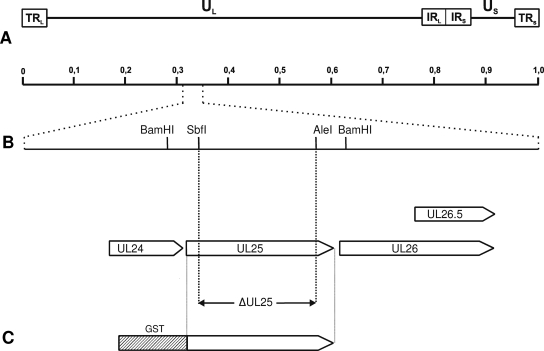
FIG. 1.
Construction of HSV1-ΔUL25. (A) Map of the HSV-1 genome with the unique long (UL), unique short (US), and inverted repeat (TRL, TRS, IRL, and IRS) sequences. Arbitrary map units are indicated. (B) Enlargement of the HSV-1 UL25 gene region. The open reading frames for UL24, UL25, UL26, and UL26.5 genes are shown as pointed rectangles. Relevant restriction sites for construction of HSV1-ΔUL25 are indicated, as is the extent of the introduced deletion. (C) The complete UL25 open reading frame was used for prokaryotic expression as GST-UL25(HSV-1).
For construction of a HSV-1 UL25 deletion mutant, pcDNA-UL25(HSV-1) was digested with SbfI and AleI, thereby removing codons 55 to 516 of the 580-codon open reading frame (Fig. 1A and B). The deleted sequence was replaced with a blunt-ended 1.3-kb BstBI fragment of pKD13 (5), which contains the kanamycin resistance gene flanked by an flp recognition target (FRT) sites. The complete insert of the generated pcDNA-HSV1-ΔUL25KF was amplified by PCR using the vector-specific primers T7 and SP6 (Invitrogen) and Pfx DNA polymerase (Invitrogen). The PCR product was used for Red recombinase-mediated mutagenesis of the HSV-1 bacterial artificial chromosome (BAC) clone pHSV1-ΔgJ. This BAC clone of HSV-1 strain KOS contains the miniF sequences from pMBO131 (31) and a green fluorescent protein expression cassette at the gJ locus. After the isolation of kanamycin-resistant clones, the kanamycin cassette was removed by using flp recombinase provided by plasmid pCP20 (5). The gJ gene was finally restored by cotransfection of pHSV1-ΔUL25 and a plasmid that contains a 6,475-bp BamHI-fragment of the HSV-1 genome, including the authentic gJ gene, thereby removing the bacterial sequences. Several single plaque isolates of the transfection progeny were characterized, and one was randomly chosen and designated HSV1-ΔUL25. Correct deletion of UL25 sequences was verified by Southern blot analysis and sequencing (data not shown), which yielded the expected results.
Generation of an HSV-1 UL25-specific antiserum.
To generate a monospecific antiserum, the complete HSV-1 UL25 ORF was cloned in frame with glutathione S-transferase (GST) into the prokaryotic expression vector pGEX-4T-1 (Amersham Biosciences, Freiburg, Germany). To this end, pcDNA-UL25(HSV-1) was digested with EcoRI and XhoI, and the insert was cloned into appropriately cleaved pGEX-4T-1 (Fig. 1C). Correct in-frame cloning was verified by sequencing. The ∼85-kDa GST-UL25(HSV-1) fusion protein was eluted from sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and used for immunization of a rabbit as described previously (15). Serum obtained after the second immunization was used throughout the study.
Virus purification and immunoblotting.
For virus purification, RK13-UL25(HSV-1) cells were infected with HSV1-KOS or HSV1-ΔUL25 at a multiplicity of infection (MOI) of 1, RK13-UL25(HSV-1) cells were infected with PrV-ΔUL25 at an MOI of 2, and, for control, RK13 cells were infected with HSV1-ΔUL25 at an MOI of 5. Cells were incubated until a complete cytopathic effect developed. Remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), cellular debris was removed by low-speed centrifugation, and virions were sedimented by centrifugation for 1 h at 20,000 rpm (SW-32 rotor; Beckman) through a 40% sucrose cushion in TBSal (200 mM NaCl, 10 mM Tris-HCl [pH 7.4], 20 mM MgCl2, 1.8 mM CaCl2 [pH 7.5]). Cell lysates were harvested 24 h after infection by using a cell scraper. After centrifugation at 16,000 × g for 1 min, the pellet was resuspended in 100 μl of phosphate-buffered saline. Lysates of purified virions or infected cells were separated by SDS-10% polyacrylamide gel electrophoresis, electrotransferred onto nitrocellulose membranes, and reacted for 1 h at room temperature with monospecific antisera against PrV pUL25 (1:50,000 [16]) or HSV-1 pUL25 (1:200,000 [the present study]). Binding of peroxidase-conjugated secondary antibodies (Dianova, Hamburg, Germany) was detected by chemiluminescence (Pierce, Bonn, Germany) and recorded on X-ray films (GE Healthcare, Freiburg, Germany).
One-step growth analysis and plaque assays.
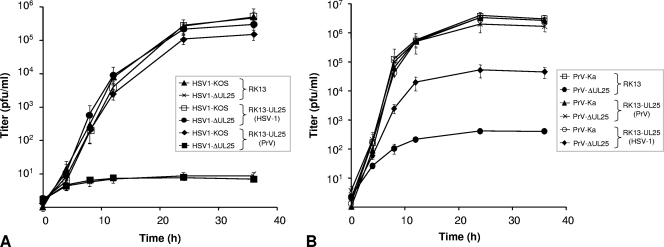
For analysis of one-step growth kinetics, RK13, RK13-UL25(PrV) or RK13-UL25(HSV-1) cells were infected with PrV-Ka, pUL25(PrV) complemented PrV-ΔUL25F, HSV1-KOS, or pUL25(HSV-1) complemented HSV1-ΔUL25 at an MOI of 5, followed by incubation on ice for 1 h. Prewarmed medium was added, and the cells were further incubated for 1 h at 37°C. Thereafter, remaining extracellular virus was inactivated by low pH treatment (23). Immediately (0 h) and after 4, 8, 12, 24, or 36 h, the cells were scraped into the medium and lysed by freezing (−70°C) and thawing (37°C). All samples were centrifuged for 2 min at 16,000 × g, and titers of progeny virus in the supernatant were determined on PrV or HSV-1 pUL25-expressing cells. Mean values of three independent experiments were calculated and plotted with the corresponding standard deviations.
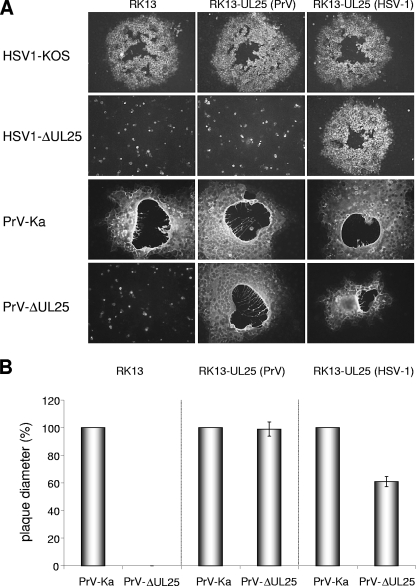
For plaque assays, cells in six-well tissue dishes were infected with 100 PFU per well of PrV-Ka, or PrV-ΔUL25 for RK13-UL25(PrV) and RK13-UL25(HSV-1), or with 1,000 PFU of PrV-ΔUL25 for RK13 and HSV1-ΔUL25 for RK13 and RK13-UL25(PrV). Two (PrV) or four (HSV-1) days after infection, cells were fixed with ethanol and stained with a monoclonal antibody to PrV gC (B16-c8) (30). Due to the Fc-binding activity of the HSV-1 gE/gI complex (20), HSV-1-infected cells could also be labeled with this monoclonal antibody. Fluorescent plaques were documented with a digital camera (C3040 Zoom; Olympus, Hamburg, Germany). Twenty plaques each were measured microscopically, and the average plaque diameters were determined. Values were calculated in comparison to those of PrV-Ka or HSV1-KOS, which were set at 100%. Average percentages and standard deviations were calculated from three independent experiments.
Electron microscopy.
For analysis of ultrathin sections, RK13 cells were infected at an MOI of 1 with PrV-ΔUL25 or HSV1-ΔUL25 which had been propagated on the cell line expressing homologous pUL25. After 14 h (PrV) or 20 h (HSV-1) cells were fixed and embedded as described previously (16). Ultrathin sections were counterstained with uranyl acetate and lead salts and examined with a model 400T microscope (Philips, Eindhoven, The Netherlands).
RESULTS
Isolation and characterization of HSV1-ΔUL25 and transcomplementing cells.
PrV and HSV-1 pUL25 homologs share 54% identical amino acids. To test for a putative functional homology, we generated a HSV-1 pUL25 expressing rabbit kidney (RK13) cell line, as well as a HSV-1 UL25 deletion mutant from strain KOS. To this end, RK13 cells were transfected with pcDNA-UL25(HSV-1) and selected in medium containing Geneticin. pUL25(HSV-1)-expressing cell lines were identified by indirect immunofluorescence and Western blot analysis with the α-UL25(HSV-1) serum (data not shown). They were further characterized by transcomplementation of HSV1-ΔUL25 (see below).
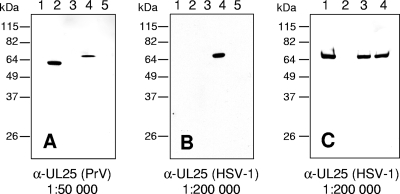
HSV1-ΔUL25 lacking codons 55 to 516 was constructed in a BAC clone of HSV-1 strain KOS. As expected, infectious progeny could only be isolated on RK13-UL25(HSV-1) cells. Southern blot analysis of genomic DNA and sequencing verified correct deletion of the UL25 specific sequences (data not shown). Whereas in cells infected by wild-type HSV-1 (Fig. 2B, lane 4) the monospecific anti-UL25(HSV-1) antiserum detected the 65-kDa pUL25, it was absent from cell lysates infected by HSV1-ΔUL25 (Fig. 2B, lane 5). HSV1-ΔUL25 replicated on RK13-UL25(HSV-1) cells with kinetics similar to those of HSV-1 KOS on either RK13 or RK13-UL25(HSV-1) cells, whereas on RK13 cells only a very few infectious particles were produced (Fig. 3A). Plaque sizes of HSV1-ΔUL25 could also be rescued to wild-type diameters on RK13-UL25(HSV-1) cells. In contrast, only single infected RK13 cells were observed (Fig. 4A). Thus, HSV1-ΔUL25 demonstrated the phenotype expected for a pUL25 deletion mutant, and the results indicated that no other unwanted mutation was introduced during mutagenesis.
FIG. 2.
Western blot analyses. (A and B) Lysates of cells infected for 24 h with PrV-Ka (lanes 2), PrV-ΔUL25 (lanes 3), HSV-1 KOS (lanes 4), or HSV1-ΔUL25 (lanes 5) or mock-infected RK13 cells (lanes 1) were separated on SDS-10% polyacrylamide gels. After electrotransfer onto nitrocellulose membranes, parallel blots were incubated with monospecific antisera against pUL25(PrV) (A) or pUL25(HSV-1) (B). (C) Purified virions of HSV-1 KOS (lane 1), HSV1-ΔUL25 propagated on RK13-UL25(HSV-1) cells (lane 3) and PrV-ΔUL25 propagated on RK13-UL25(HSV-1) cells (lane 4) were analyzed by immunoblotting with antiserum to HSV-1 pUL25. As a control, supernatant of HSV1-ΔUL25-infected RK13 cells was processed identically (lane 2). Molecular masses of marker proteins are indicated on the left.
FIG. 3.
One-step growth analysis. RK13, RK13-UL25(PRV), and RK13-UL25(HSV-1) cells were infected at an MOI of 5 with HSV1-KOS or HSV1-ΔUL25 (A) or with PrV-Ka or PrV-ΔUL25 (B), harvested at the indicated times after infection, and titrated on either RK13-UL25(HSV-1) (A) or RK13-UL25(PrV) (B) cells. Average titers (PFU/ml) and standard deviations from three independent experiments are shown.
FIG. 4.
Plaque formation of PrV and HSV UL25 deletion mutants. RK13, RK13-UL25(PrV), and RK13-UL25(HSV-1) cells were infected with PrV-Ka, PrV-ΔUL25, HSV1-KOS, and HSV1-ΔUL25 under plaque assay conditions and fixed 2 days (for PrV) or 4 days (for HSV-1) after infection. Infected cells were visualized by indirect immunofluorescence with a PrV gC-specific monoclonal antibody. (B) Calculation of plaque diameters. Plaques on RK13, RK13-UL25(PrV), or RK13-UL25(HSV-1) cells infected with PrV-Ka or PrV-ΔUL25 were microscopically measured at 2 days postinfection. Relative plaque sizes were calculated and compared to those of PrV-Ka, which were set as 100%. Average values and standard deviations from three independent experiments are shown.
Ultrastructural phenotype of HSV1-ΔUL25.
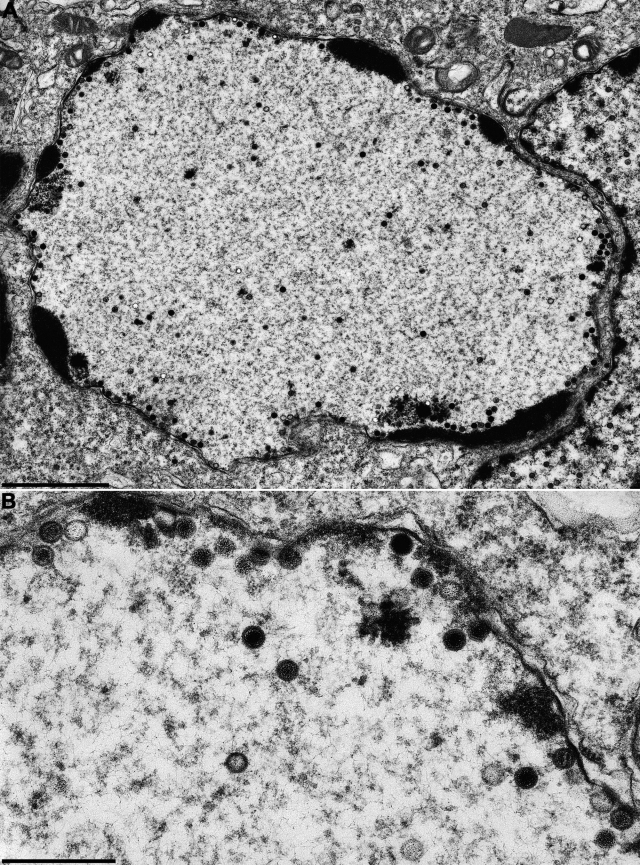
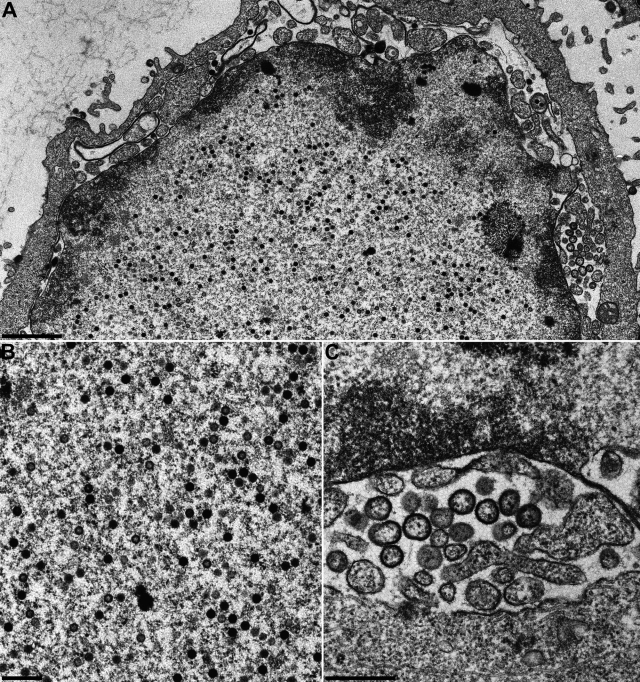
In PrV, in the absence of pUL25 C capsids are produced, but they fail to leave the nucleus, despite close contact with the inner nuclear membrane (16). However, vesicles are formed that accumulate in the perinuclear cleft, indicating that the defect is not associated with an inhibition of the primary envelopment machinery (17). In contrast, the absence of pUL25 in HSV-1 has been reported to inhibit (22) or impair (42) encapsidation of viral genomes into preformed capsids. To analyze whether these differences can also be observed in our assay system, we infected RK13 cells at the same MOI with PrV-ΔUL25 or HSV1-ΔUL25 and examined infected cells 14 h (for PrV) or 20 h (for HSV-1) after infection. The different times of incubation are due to the differences in the length of the replication cycle between PrV and HSV-1. As shown in Fig. 5A, in the absence of pUL25 in PrV, C capsids are observed in abundance in the nucleus, some of which are in intimate contact with the inner nuclear membrane but do not bud (Fig. 5B). Consequently, neither nonenveloped capsids nor enveloped viral particles are visible in the cytosol. C capsids are also present in high numbers in HSV1-ΔUL25-infected cell nuclei (Fig. 6A and B) but not in the cytosol (Fig. 6A), indicating a similar defect in both viruses. In contrast, vesicles were observed within the perinuclear cleft which resemble primary envelopes (Fig. 6C), as has been observed for PrV-ΔUL25 (17). Interestingly, C capsids in contact with the inner nuclear membrane were only very rarely observed in HSV1-ΔUL25-infected cells (Fig. 6A), pointing to a subtle but significant difference in the phenotypes of both mutants.
FIG. 5.
Ultrastructural analysis of PrV-ΔUL25. RK13 cells were infected at an MOI of 1 with transcomplemented PrV-ΔUL25 and analyzed 14 h after infection. (A) Overview of an infected cell; (B) higher-magnification view of intranuclear capsids. Scale bars: A, 2 μm; B, 500 nm.
FIG. 6.
Ultrastructural analysis of HSV1-ΔUL25. RK13 cells were infected at an MOI of 1 with transcomplemented HSV1-ΔUL25 and analyzed 20 h after infection. (A) Overview of an infected cell; (B) higher-magnification view of intranuclear capsids; (C) vesicles within the perinuclear cleft. Scale bars: A, 1.5 μm; B, 500 nm; C, 500 nm.
pUL25(PrV)-specific antiserum recognizes pUL25(HSV-1).
To test whether the high sequence homology correlates with a similar antigenicity of the PrV and HSV-1 pUL25 proteins, we assayed whether the pUL25-specific antisera cross-reacted with the heterologous protein. To this end, lysates of RK13 cells infected with PrV-Ka, PrV-ΔUL25, HSV-1 KOS, or HSV1-ΔUL25 were analyzed by immunoblotting with the α-UL25(PrV) or α-UL25(HSV-1) monospecific sera. As shown in Fig. 2A, the α-UL25(PrV) serum reacted not only with the ∼57-kDa PrV pUL25 but also with an ∼65-kDa protein in lysates of HSV-1 KOS-infected cells. No signal was detectable in lysates of cells infected with the UL25 deletion mutants or mock-infected cells (Fig. 2A). Unlike the UL25(PrV) specific serum, the α-UL25(HSV-1) serum only detected the 65-kDa pUL25 in lysates of HSV-1 KOS-infected cells but not the corresponding PrV protein (Fig. 2B). These data demonstrated an antigenic relationship between the two proteins that was detected by the UL25(PrV) specific serum but not by the UL25(HSV-1) serum.
HSV-1 pUL25 partially complements replication of a PrV UL25 deletion mutant but not vice versa.
To test for heterologous complementation, RK13, RK13-UL25(PrV) and RK13-UL25(HSV-1) cells were infected either with HSV-1 KOS and HSV1-ΔUL25 (Fig. 3A) or with PrV-Ka and PrV-ΔUL25 (Fig. 3B). As described previously (17), RK13-UL25(PrV) cells supported the replication of the PrV deletion mutant to wild-type-like titers, whereas after infection of RK13 cells only very few infectious particles not exceeding 200 PFU/ml were produced. Interestingly, on RK13-UL25(HSV-1) cells, PrV-ΔUL25 was able to replicate to a significant extent, yielding ∼25-fold-reduced final titers compared to cells expressing the homologous pUL25 (Fig. 3B). In contrast, infection of RK13-UL25(PrV) cells with the HSV-1 UL25 deletion mutant did not result in an increase of infectious progeny compared to RK13 cells, while the mutant replicated to wild-type-like titers on RK13-UL25(HSV-1) cells (Fig. 3A).
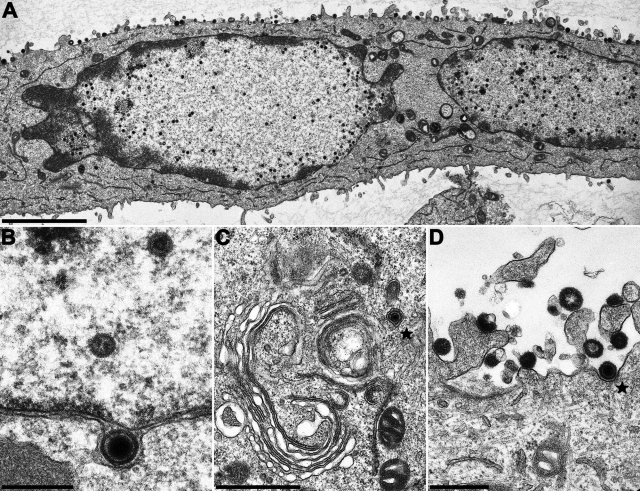
Similar results were obtained in plaque assays. Whereas PrV-ΔUL25 was unable to produce plaques on noncomplementing RK13 cells, plaques were formed on RK13-UL25(HSV-1) cells (Fig. 4A) that reached ca. 60% the size of those produced by PrV-Ka on the same cell line (Fig. 4B). In contrast, HSV1-ΔUL25 produced plaques only on cells expressing the homologous pUL25, and only single infected RK13-UL25(PrV) or nontransfected RK13 cells (Fig. 4A) were detectable. In ultrastructural analyses after infection of RK13-UL25(HSV-1) cells with PrV-ΔUL25 all stages of virion formation were observed (Fig. 7A), including primary (Fig. 7B) and secondary envelopment (Fig. 7C), as well as extracellular virions (besides L-particles; Fig. 7D). However, correlating with the lower titer of infectious progeny on these cells, these stages were observed significantly less frequently than after wild-type PrV infection or infection of RK13-UL25(PrV) cells.
FIG. 7.
Ultrastructural analysis of PrV-ΔUL25 deleted on HSV-1 pUL25-expressing cells. RK13-UL25(HSV-1) cells were infected at an MOI of 1 with transcomplemented PrV-ΔUL25 and analyzed 14 h after infection. (A) Overview of an infected cell; (B) primary enveloped virion; (C) secondary envelopment in the cytosol; (D) extracellular L-particles and an enveloped virion. Scale bars: A, 3 μm; B, 300 nm; C, 1 μm; D, 500 nm. Virions in panels C and D are marked by asterisks.
Together, these data demonstrate that HSV-1 pUL25 can complement to a significant extent the replication defect of PrV-ΔUL25, indicating a functional overlap between both proteins. However, the reverse assays did not show complementation, demonstrating that both proteins are functionally not identical.
pUL25(HSV-1) is incorporated into PrV-ΔUL25 virions.
To directly assay whether the transcomplementing pUL25(HSV-1) is incorporated into PrV virions, PrV-ΔUL25 virions were purified after infection of RK13-UL25(HSV-1) cells and compared to purified virions of HSV-1 KOS or HSV1-ΔUL25 propagated on cells expressing HSV-1 pUL25. Supernatant of HSV1-ΔUL25-infected RK13 cells was processed identically. As shown in Fig. 2C, the ∼65-kDa HSV-1 pUL25 was present in purified HSV-1 KOS virions, as well as in virions of either pUL25 deletion mutant propagated on HSV-1 pUL25-expressing cells. To verify the purity of the virus preparation, PrV-ΔUL25 virions were also probed with antiserum to the pUL34 protein, which is a component of primary virions but absent from mature virus particles. This antiserum yielded the expected negative result (data not shown). Thus, the partial functional complementation was paralleled by incorporation of the heterologous pUL25 into PrV-ΔUL25 virions.
DISCUSSION
The salient findings of this study are as follows: (i) the ultrastructural phenotypes of deletion mutants of pUL25 in PrV and HSV-1 are similar; (ii) pUL25(HSV-1) is incorporated into PrV-ΔUL25 virions; (iii) pUL25(HSV-1) is able to partially complement the replication defect of PrV-ΔpUL25 but not vice versa; and (iv) pUL25(HSV-1) is also able to rescue plaque formation of PrV-ΔpUL25, albeit not to wild-type levels. Thus, our data indicate a functional overlap between the HSV-1 and PrV pUL25 proteins.
Proteins homologous to pUL25 of HSV-1 are conserved throughout the Herpesviridae and are essential for viral replication in cultured cells as studied thus far (1, 6, 16, 22, 42). However, their exact role has not yet been elucidated. pUL25 deletion mutants of HSV-1 are deficient in the formation of mature capsids by an impairment of packaging of viral DNA into preformed capsids (22, 42). However, in contrast to other proteins involved in cleavage/packaging, concatemeric viral DNA is cleaved into unit length genomes in the absence of pUL25 in HSV-1 and bovine herpesvirus 1 (BHV-1 [6, 22, 42]) but is either less efficiently packaged or less stable within the capsid (42). A direct association of pUL25(HSV-1) with DNA has been suggested as the basis for its packaging function (32). However, pUL25 also plays a role in a step after DNA encapsidation, since even those mature C capsids which form in the absence of pUL25, are unable to leave the nucleus for continuing virion maturation (6, 16, 42). This phenotype is even more striking in PrV: in the absence of pUL25(PrV) DNA-containing C capsids are formed and can also be found in close association with the inner nuclear membrane. However, they are apparently unable to bud, implicating a function for pUL25 as a trigger for mature capsids to undergo primary envelopment (16).
pUL25 homologs of PrV and HSV-1 are 54% identical at the amino acid level, a homology that spans the whole protein. This significant conservation coincides with the observed cross-reactivity of our anti-pUL25(PrV) serum with pUL25(HSV-1). Since this high degree of sequence conservation might be indicative of common functions in the replication cycle of the respective virus, we followed on experiments started previously to uncover functional similarity of homologous herpesvirus proteins (18, 25). To this end, we isolated a HSV-1 UL25 expressing rabbit kidney cell line and generated a HSV-1 UL25 deletion mutant paralleling the experimental design as described recently for PrV (16).
Interestingly, in our virus-cell system the ultrastructural analyses of cells infected with either PrV-ΔUL25 or HSV1-ΔUL25 showed similar phenotypes. Electron-dense C capsids were present in high numbers in the nuclei of cells infected with either virus. Thus, in the absence of pUL25 in PrV or HSV-1, genome packaging apparently proceeds. However, the C capsids did not leave the nucleus since no stages of nuclear egress or viral capsids in the cytosol were observed. This does not correlate with a defect in the primary envelopment machinery, since vesicles (termed “primary L-particles” [17]) were present in the perinuclear cleft of cells infected by either virus. Thus, in both viruses pUL25 is apparently required for the inclusion of mature capsids into the primary envelope, which correlates with the proposal that pUL25(HSV-1) plays an important role during later stages of capsid maturation, prior to the release of capsids into the cytoplasm (42, 43), and our demonstration that pUL25(PrV) is not required for cleavage and/or encapsidation of genomic DNA but for egress of DNA-containing intranuclear C capsids from the nucleus (16). The role pUL25 homologs may play for nuclear egress primarily of C capsids correlates with its exclusive (for PrV [16]) or predominant (for HSV-1 [32, 40]) localization at the outside of C capsids, whereas immature capsid forms either lack pUL25 altogether (for PrV [16]) or carry it in significantly reduced amounts (for HSV-1 [32, 40]). pUL25(HSV-1) has been shown to be located on the outside of the mature capsid as part of the penton and hexon structures (32, 44), also suggesting that it may play a role in tethering mature capsids to the future primary envelope for nuclear egress.
Since the pUL25 proteins of PrV and HSV-1 proved to be antigenically related and since the phenotypes of the deletion mutants were similar, we tested in heterologous transcomplementation assays whether the proteins also shared functional homology. Whereas neither mutant was able to replicate to a significant extent in noncomplementing cells, titers of PrV-ΔUL25 propagated on RK13-UL25(HSV-1) cells reached 4 × 104 PFU/ml, which was only ∼25-fold lower than titers obtained on cells expressing the homologous pUL25(PrV). This demonstrates that pUL25(HSV-1) can exert functions of pUL25(PrV). The formation and release of infectious herpesvirus particles requires transfer of intranuclear DNA-containing C capsids into the cytosol for final maturation. This step is blocked in PrV-ΔUL25, essentially abolishing infectious virus production. Since pUL25(HSV-1) can partially rescue this defect, it is reasonable to assume that pUL25(HSV-1) also functions during nuclear egress. Thus, our data indicate that at least one of the functions of pUL25(HSV-1) is similar to that of pUL25(PrV), which is supposed to trigger nuclear egress. In fact, nuclear egress of PrV-ΔUL25 from pUL25(HSV-1)-expressing cells could also be observed directly in electron microscopic analyses (Fig. 7). In contrast, PrV pUL25 was unable to detectably complement the HSV-1 UL25 deletion mutant in either one-step replication or plaque formation, which indicates that infectious progeny did not form in the cytosol, presumably since nuclear egress was blocked. Taken together, these results demonstrate that pUL25 of PrV and HSV-1 share functions required for the primary envelopment of intranuclear capsids. However, pUL25 of HSV-1 may exert additional effects that are needed prior to nuclear egress, presumably during capsid maturation or DNA encapsidation, which PrV pUL25 cannot provide. Alternatively, due to the sequence divergence, pUL25(PrV) may not be able to engage one or more of the corresponding partner proteins in HSV.
Unidirectional complementation between homologous herpesviral proteins is not uncommon. It has been reported for the gB homologs of PrV and BHV-1 (18), as well as for gB of PrV and HSV-1 (25). Although BHV-1 gB could complement the defect of the PrV gB deletion mutant, the HSV-1 homolog did not. In contrast, the BHV-1 gB deletion mutant was unable to replicate with the assistance of PrV gB, whereas HSV-1 was complemented by PrV gB, indicating that, although the proteins share common functions, certain domains or protein interaction faces might be unique.
Our data demonstrate that HSV-1 pUL25 is incorporated into PrV virions and is able to partially complement the defect of PrV-ΔUL25. This would also mean that, if the pUL17/pUL25 complex as described for HSV-1 (44) is involved in this interaction, pUL25(HSV-1) would have to interact with pUL17(PrV) to execute its function. Thus, an inefficient interaction between the PrV and HSV-1 partners of the complex could explain the one-way partial complementation. Thus far, heterologous transcomplementation of pUL17 mutants of either virus has not been attempted, which, however, would be most interesting since pUL17 of PrV (13) and HSV-1 (43) have been localized to different subviral structures. It remains also to be analyzed whether transcomplementation is improved by the coexpression of pUL17 and pUL25 derived from the same virus.
Although pUL25 is required for primary envelopment, the transport of capsids to the inner nuclear membrane appears not to be impaired without pUL25, at least in PrV. In the absence of pUL25(PrV), DNA-containing C capsids are able to access the inner nuclear membrane but unable to undergo primary envelopment (16). Thus, pUL25 is not required for the contact of capsids with the inner nuclear membrane. In cells infected with HSV1-ΔUL25, capsids were only rarely observed in intimate contact with the inner nuclear membrane, indicating that HSV-1 pUL25 may also be involved in directing capsids to the inner nuclear membrane. This difference in properties may contribute to the observed failure of pUL25(PrV) to complement HSV1-ΔUL25.
In summary, we show that pUL25 of PrV and HSV-1 have overlapping, although not identical functions. Comparative studies such as these are important to elucidate common features and subtle differences between herpesvirus proteins that cannot be uncovered by analyses of single virus-cell systems.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG Me 854/8-2).
We thank Charlotte Ehrlich, Petra Meyer, and Diana Werner for expert technical assistance and Mandy Jörn for photographic help.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Ali, M. A., B. Forghani, and E. M. Cantin. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216278-283. [DOI] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. G. Barell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310207-211. [DOI] [PubMed] [Google Scholar]
- 3.Bjerke, S. L., and R. J. Roller. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154125-169. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desloges, N., and C. Simard. 2003. Implication of the product of the bovine herpesvirus type 1 UL25 gene in capsid assembly. J. Gen. Virol. 842485-2490. [DOI] [PubMed] [Google Scholar]
- 7.Dezelee, S., F. Bras, P. Vende, B. Simonet, X. Nguyen, A. Flamand, and M. J. Masse. 1996. The BamHI fragment 9 of pseudorabies virus contains genes homologous to the UL24, UL25, UL26, and UL 26.5 genes of herpes simplex virus type 1. Virus Res. 4227-39. [DOI] [PubMed] [Google Scholar]
- 8.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. Torrisi, A. Faggioni, and H.-J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 793703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7107-122. [DOI] [PubMed] [Google Scholar]
- 11.Kaelin, K., S. Dezelee, M. J. Masse, F. Bras, and A. Flamand. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7394-407. [DOI] [PubMed] [Google Scholar]
- 13.Klupp, B. G., A. Karger, H. Granzow, and T. C. Mettenleiter. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J. Virol. 7913442-13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 7410063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 806235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klupp, B. G., H. Granzow, W. Fuchs, G. M. Keil, S. Finke, and T. C. Mettenleiter. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA 1047241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp, A., and T. C. Mettenleiter. 1992. Stable rescue of a glycoprotein gII deletion mutant of pseudorabies virus by glycoprotein gI of bovine herpesvirus 1. J. Virol. 662754-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BRFR1 and BFLF2 interact and coexpression alters their cellular localization. Virology 32099-106. [DOI] [PubMed] [Google Scholar]
- 20.Lubinski, J., T. Nagashunmugam, and H. M. Friedman. 1998. Viral interference with antibody and complement. Semin. Cell Dev. Biol. 9329-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGeoch, D. J., C. Cunningham, G. McIntyre, and A. Dolan. 1991. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J. Gen. Virol. 723057-3075. [DOI] [PubMed] [Google Scholar]
- 22.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 721060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171623-625. [DOI] [PubMed] [Google Scholar]
- 24.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113163-169. [DOI] [PubMed] [Google Scholar]
- 25.Mettenleiter, T. C., and P. G. Spear. 1994. Glycoprotein gB (gII) of pseudorabies virus can functionally substitute for glycoprotein gB in herpes simplex virus type 1. J. Virol. 68500-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 27.Milbradt, J., S. Auerochs, and M. Marschall. 2007. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 882642-2650. [DOI] [PubMed] [Google Scholar]
- 28.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297854-857. [DOI] [PubMed] [Google Scholar]
- 29.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 806286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 747137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connor, M., M. Peifer, and W. Bender. 1989. Construction of large DNA segments in Escherichia coli. Science 2441307-1312. [DOI] [PubMed] [Google Scholar]
- 32.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 751427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purves, F. C., D. Spector, and B. Roizman. 1992. UL34, the target of the herpes simplex virus U(S)3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 664295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 785564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, PA.
- 39.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenny. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 812397-2405. [DOI] [PubMed] [Google Scholar]
- 42.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 7510755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 821423-1428. [DOI] [PubMed] [Google Scholar]