Abstract
Poliovirus (PV) mRNA is unusual because it possesses a 5′-terminal monophosphate rather than a 5′-terminal cap. Uncapped mRNAs are typically degraded by the 5′ exonuclease XRN1. A 5′-terminal cloverleaf RNA structure interacts with poly(rC) binding proteins (PCBPs) to protect uncapped PV mRNA from 5′ exonuclease (K. E. Murray, A. W. Roberts, and D. J. Barton, RNA 7:1126-1141, 2001). In this study, we examined de novo polysome formation using HeLa cell-free translation-replication reactions. PV mRNA formed polysomes coordinate with the time needed for ribosomes to traverse the viral open reading frame (ORF). Nascent PV polypeptides cofractionated with viral polysomes, while mature PV proteins were released from the polysomes. Alterations in the size of the PV ORF correlated with alterations in the size of polysomes with ribosomes present every 250 to 500 nucleotides of the ORF. Eukaryotic initiation factor 4GI (eIF4GI) was cleaved rapidly as viral polysomes assembled and the COOH-terminal portion of eIF4GI cofractionated with viral polysomes. Poly(A) binding protein, along with PCBP 1 and 2, also cofractionated with viral polysomes. A C24A mutation that inhibits PCBP-5′-terminal cloverleaf RNA interactions inhibited the formation and stability of nascent PV polysomes. Kinetic analyses indicated that the PCBP-5′ cloverleaf RNA interaction was necessary to protect PV mRNA from 5′ exonuclease immediately as ribosomes initially traversed the viral ORF, before viral proteins could alter translation factors within nascent polysomes or contribute to ribonucleoprotein complexes at the termini of the viral mRNA.
Poliovirus (PV) mRNA has a long 5′ nontranslated region (5′ NTR) without a 5′ cap (22, 40). A cloverleaf RNA structure at the 5′ terminus of PV mRNA forms ribonucleoprotein (RNP) complexes with poly(rC) binding proteins (PCBPs) and with viral protein 3CD (16, 46). A C24A mutation in the 5′ cloverleaf inhibits the binding of PCBP (1) and renders PV mRNA susceptible to degradation by 5′ exonuclease (36). An internal ribosome entry site (IRES) within the 5′ NTR of PV mRNA requires eukaryotic initiation factor 4G (eIF4G) to mediate translation initiation (47, 62), and accumulating evidence indicates that eIF4G likely interacts directly with the IRES (42, 49, 50). PV 2A protease (2APro) expression during the course of PV mRNA translation leads to the cleavage of eIF4GI and eIF4GII (18). Cleavage of eIF4Gs by 2APro prevents cap-dependent host mRNA translation and thereby contributes to the conversion of host mRNA polysomes to predominantly viral mRNA polysomes in infected cells (56). Cleavage of eIF4Gs by 2APro may abrogate some RNA-protein-protein-RNA bridges between the 5′ and 3′ termini of mRNAs (18) while leaving intact other potential RNA-protein-protein-RNA bridges (21, 29). In addition to interactions with the 5′ cloverleaf, PCBPs interact with stem-loop 4 of the PV IRES to mediate the initiation of translation (8, 9). PCBPs may also contribute to RNA-protein-protein-RNA bridges between the 5′ and 3′ termini of PV mRNA (7, 21).
Eukaryotic mRNA translation and mRNA stability are coordinately regulated (38). Because PV mRNA is converted into a template for viral RNA replication following translation (41), it is unlikely that regulated degradation of PV mRNA, like that of host mRNAs, would be advantageous from the perspective of the virus. Interactions between 5′- and 3′-terminal RNP complexes appear to mediate and regulate both mRNA translation and mRNA stability. eIF4GI and eIF4GII are important scaffolding proteins that mediate interactions between translation factors present at the 5′ and 3′ termini of mRNAs (20, 49). eIF4G has domains for protein-protein interactions with eIF4E, the cap binding protein, and poly(A) binding protein (PABP) (23, 34). Deadenylation of mRNAs can lead to 3′-to-5′ mRNA degradation by the exosome (63, 64) or to decapping and degradation of mRNA by a 5′ exonuclease (10). The mechanisms by which translation factors interact with and regulate mRNA degradation machinery coordinate with translation are actively under investigation (13, 14).
Cytoplasmic extracts from uninfected HeLa cells have been used to create cell-free reaction mixtures capable of supporting all of the metabolic steps of PV replication, including the translation and replication of PV mRNA (3, 32). Similar extracts support the translation and replication of coxsackievirus (65), rhinovirus (61), and encephalomyocarditis virus (58). These reactions are advantageous because they faithfully recapitulate the conditions within infected cells, including synergistic interactions between 5′- and 3′-terminal RNPs (59, 60), thereby supporting detailed analyses of the sequential molecular events associated with various steps of PV mRNA translation and replication (4, 6, 33, 35, 37, 53). In this study, we used HeLa cell-free translation reaction mixtures to examine the de novo formation of PV polysomes and the role of PCBP-5′ cloverleaf RNA interactions during polysome formation. We examined de novo polysome formation coincident in time with that needed for ribosomes to traverse the viral mRNA open reading frame (ORF) and compared the kinetics and magnitude of C24A PV mRNA stability, translation, and polysome formation with that of wild-type PV mRNA. The data provide evidence that PV mRNA within polysomes can be degraded by 5′ exonuclease and that PCBP-5′ cloverleaf RNA interactions are important before viral proteins like 2APro or 3CD have an opportunity to modify messenger RNP (mRNP) complexes.
MATERIALS AND METHODS
PV mRNAs.
cDNA clones encoding full-length PV mRNA [pT7-PV1(A)80] (7), DJB1 mRNA with a four-base CUAG insertion at nucleotide 2474 [pT7-PV1(A)80G2474+CTAG] (30), RNA2 mRNA (an efficient RNA replicon) with an in-frame deletion of PV nucleotides 1175 to 2956 (11), and DJB14 mRNA with deletions of nucleotides 630 to 6011 and nucleotides 6061 to 6516 (30) were kindly provided by James B. Flanegan, University of Florida, Gainesville. A C24A mutation was engineered into RNA2 as previously described (36).
PV mRNAs were made from MluI-linearized cDNA templates by T7 transcription (Epicenter, Madison, WI). [α-32P]CTP was included to make radiolabeled RNA when necessary. RNA was precipitated with 2.5 M ammonium acetate, washed with ethanol, and quantified by UV absorption at 260 nm.
Cell-free translation reactions.
PV mRNAs were programmed into reaction mixtures containing cytoplasmic HeLa cell extracts as previously described (3, 5, 35). Reaction mixtures (10 to 300 μl) contained 50% (by volume) S10, 20% (by volume) ribosomal salt wash-derived translation initiation factors, 10% (by volume) 10× nucleotide reaction mix (10 mM ATP, 2.5 mM GTP, 2.5 mM CTP or 2.5 mM UTP, 600 mM KCH3CO2, 300 mM creatine phosphate, 4 mg/ml creatine kinase, and 155 mM HEPES-KOH [pH 7.4]), 2 mM guanidine hydrochloride, and T7 transcripts of viral RNA at 50 μg/ml. Reaction mixtures were incubated at 34°C for the indicated periods of time. When indicated, [35S]methionine (Amersham, Piscataway, NJ) was included in reaction mixtures to radiolabel proteins. Samples (1 μl) of reaction mixtures containing [35S]methionine were precipitated with 5% trichloroacetic acid, collected on nitrocellulose filters, and quantified by scintillation counting to determine the magnitude of protein synthesis. Samples (1 to 4 μl) of reaction mixtures containing [35S]methionine were also solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (2% SDS [Sigma, St. Louis, MO], 62.5 mM Tris-HCl [pH 6.8], 0.5% 2-mercaptoethanol, 0.1% bromophenol blue, 20% glycerol). The samples were heated at 100°C for 5 min and separated by gel electrophoresis in 9 to 18% SDS-polyacrylamide gels. [35S]methionine-labeled proteins were detected by phosphorimaging.
Polysomes.
Sucrose gradients were prepared and samples were treated using conventional protocols with minor modifications (19, 44, 51). Sucrose gradients were prepared in 14- by 89-mm polypropylene tubes (Seton, Los Gatos, CA) by layering 3 to 4 ml of 0.5 M sucrose on top of 7 to 8 ml of 1.5 M sucrose (0.5 or 1.5 M sucrose, 0.3 M NaCl, 10 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 100 μg per ml cycloheximide, and 100 μg per ml heparin) for a total volume of 11 ml. Tubes were capped with parafilm and laid on their sides for 1 h at room temperature to allow for gradient formation.
Cell-free translation reaction mixtures (300 μl) were diluted with an equal volume of solution containing cycloheximide (0.1 M Tris-HCl [pH 7.6], 0.1 M NaCl, 40 mM MgCl2, 0.5 M sucrose, 300 μg per ml cycloheximide, and 100 μg per ml heparin) to stabilize polysomes and 1/10 volume of a 10× nonionic detergent solution (5% Triton X-100, 121 mM sodium deoxycholate). Samples were layered on top of the gradients and subjected to centrifugation at 36,000 rpm (∼160,000 × g) for 3 to 3.5 hours at 4°C using an SW41 rotor (Beckman). Sucrose gradients were fractionated using a BR-184 tube piercer and a syringe pump (Brandel, Gaithersburg, MD), and a UA-6 UV detector (ISCO, Lincoln, NE). Fluorinert FC-40 (ISCO) was pumped into the bottom of sucrose gradients (3.0 pump speed, ∼1 ml per min), while plumbing at the top of the tubes carried the gradient through the UV detector (sensitivity of 0.5, noise filter set at 1.5, peak separator off, and chart speed of 150). Digital data (100 samples per second) were captured during fractionation using a DI-158U USB data acquisition device (DATAQ Instruments, Akron, OH) connecting the UV detector to a personal computer. The digital data were processed using WINDAQ/HS and WINDAQ/XL software (DATAQ Instruments), and UV absorption at 254 nm was plotted versus mobility (from top to bottom of sucrose gradients) using Microsoft Excel.
Twenty 0.5-ml fractions were collected from each gradient. When 32P-labeled viral RNA was used, it was detected by Cerenkov counting sucrose gradient fractions. Total RNA within fractions of sucrose gradients was isolated by phenol-chloroform extraction and ethanol precipitation, fractionated by electrophoresis in 1% agarose Tris-borate-EDTA buffer, stained with ethidium bromide, and detected with UV light or by phosphorimaging for 32P-labeled viral RNA. When [35S]methionine-labeled viral proteins were fractionated, the proteins within sucrose gradient fractions were collected by methanol-chloroform precipitation, solubilized in SDS-PAGE sample buffer, and fractionated by electrophoresis in 9 to 18% polyacrylamide gels.
Western blots.
Proteins fractionated by SDS-PAGE were transferred to Hybond-P membranes (Amersham) using a constant current of 1.6 A for 1.5 h in transfer buffer (25 mM Tris-HCl, 0.2 M glycine, 0.05% SDS, and 20% methanol). Membranes were soaked for 1 h in blocking buffer (20 mM Tris-HCl [pH 7.6], 14 mM NaCl, 0.05% Tween 20, and 5.0% dry powdered milk) and then soaked overnight with 1:1,000 dilutions of eIF4G (NH-terminal portion) rabbit polyclonal antibody (Ab) (AbCAM, Cambridge, MA), eIF4G (COOH-terminal portion) mouse monoclonal Ab (BD Biosciences, San Jose, CA), PABP rabbit polyclonal Ab (Cell Signaling, Danvers, MA), or PCBP1 (hnRNP E1) and PCBP2 (hnRNP E2) goat polyclonal Abs (Santa Cruz Biotechnology, Santa Cruz, CA) (primary Abs) diluted in blocking buffer. Membranes were soaked with secondary Abs (1:10,000 goat anti-mouse Ab conjugated to horseradish peroxidase [HRP; Sigma, St. Louis, MO], 1:25,000 donkey anti-rabbit Ab conjugated to HRP [Jackson ImmunoResearch, West Grove, PA], or 1:5,000 donkey anti-goat Ab conjugated to HRP [Santa Cruz Biotechnology]) diluted in blocking buffer, washed three times for 15 min each time with blocking buffer, and developed using Lightning Plus chemiluminescence (Perkin Elmer, Waltham, MA). Signals were detected using blue-light autoradiography film (ISC BioExpress, Kaysville, UT).
RESULTS
PV mRNA formed polysomes coordinate in time with the elongation of ribosomes across the ORF.
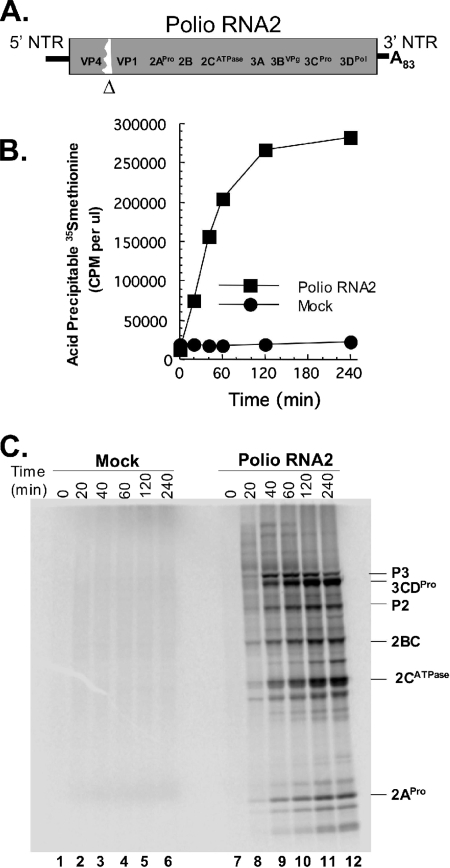
PV RNA2, an efficient RNA replicon (11), was used to examine de novo polysome formation in cell-free reactions (Fig. 1). PV RNA2 has a 4,845-nucleotide-long ORF with an in-frame deletion of two viral capsid proteins, VP2 and VP3 (Fig. 1A). PV RNA2 mRNA translated efficiently when incubated in cell-free translation reaction mixtures (Fig. 1B). Reaction mixtures without viral mRNA did not make proteins (Fig. 1B) because micrococcal nuclease was used to degrade endogenous host mRNAs in the cytoplasmic HeLa cell extracts used for the reaction mixtures (3, 5). [35S]methionine incorporation into PV proteins increased for 2 hours (Fig. 1B). SDS-PAGE revealed a diffuse array of nascent PV polyprotein after 20 min of incubation (Fig. 1C, lane 8). After 40 min of incubation, mature PV proteins were clearly apparent (Fig. 1C, lane 9). 3CDPro, encoded at the 3′ end of the viral ORF, is required for polyprotein processing. The size distribution of nascent PV polyproteins within reaction mixtures incubated for 20 min and the absence of mature viral proteins like 3CDPro within reaction mixtures incubated for 20 min support the conclusion that ribosomes required just over 20 min to traverse the 4,845-nucleotide-long ORF.
FIG. 1.
Kinetics and magnitude of PV mRNA translation in cell-free reaction mixtures. (A) Diagram of poliovirus (Polio) RNA2, a replicon mRNA with an in-frame deletion of capsid proteins VP2 and VP3. (B) Protein synthesis in cell-free reaction mixtures containing PV RNA2 or no mRNA (mock) as measured by the incorporation (in cpm per μl) of [35S]methionine into acid-precipitable material after 0, 20, 40, 60, 120, and 240 min of incubation. (C) Protein synthesis in cell-free reaction mixtures containing [35S]methionine and PV RNA2 (lanes 7 to 12) or no mRNA (lanes 1 to 6) after 0 (lanes 1 and 7), 20 (lanes 2 and 8), 40 (lanes 3 and 9), 60 (lanes 4 and 10), 120 (lanes 5 and 11), and 240 (lanes 6 and 12) minutes of incubation as detected by SDS-PAGE and phosphorimaging. Note that full-length polyproteins corresponding to the ORF are evident by 20 min of incubation but that mature proteins have just begun to appear (lane 8).
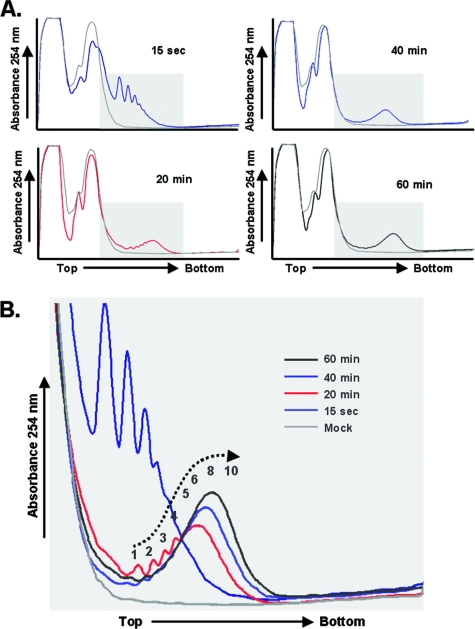
PV mRNA began to form polysomes immediately, and the size of the polysomes increased over time (Fig. 2). The density of ribosomes on each PV mRNA increased from 1 to 4 ribosomes per mRNA after 15 seconds of incubation to 4 to 10 ribosomes per mRNA after 20 min of incubation and then to 8 to 15 ribosomes on most mRNAs after 60 min of incubation (Fig. 2B). As ribosome density increased above six to eight ribosomes per mRNA, the resolution of incrementally larger polysomes was lost. By extrapolating polysome density coordinate with the mobility of polysomes, we estimate that the highest ribosome density on RNA2 was 15 to 20 ribosomes per mRNA. Polysomes were not observed at any time point in reaction mixtures without viral mRNA (Fig. 2B, mock reactions). Rather, in mock reactions, a large amount of free protein and other material was detected by UV absorbance at the top of the sucrose gradients followed by peaks of absorbance for both 40S and 60S/80S ribosomal subunits (Fig. 2A).
FIG. 2.
PV mRNA formed polysomes coordinate in time with the elongation of ribosomes across the ORF. (A) Cell-free reaction mixtures with or without PV RNA2 were incubated at 34°C for 15 seconds or for 20, 40, or 60 minutes as indicated. Reaction products were then separated by centrifugation at 36,000 rpm for 3 h in 0.5 M to 1.5 M sucrose gradients as described in Materials and Methods. UV absorbance was monitored and plotted as the gradients were fractionated. The top and bottom of sucrose gradients are indicated. (B) Polysome traces of indicated time points superimposed to show the maturation of PV polysomes over time. The UV traces from the portion of the gradients with polysomes were cropped and magnified for clarity. Individual peaks corresponding to smaller polysomes were clearly evident. The size of larger polysomes was estimated by natural logarithmic regression and extrapolation based on their mobility.
PV mRNA and nascent polyproteins cofractionated with polysomes.
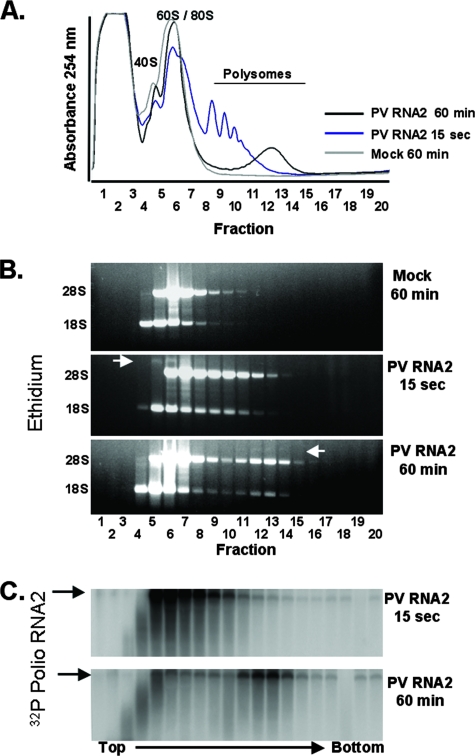
We used agarose gel electrophoresis to examine the distribution of rRNAs (Fig. 3A) and the distribution of PV mRNAs (Fig. 3B and C) in polysome gradients. As expected, 18S rRNA cofractionated with 40S ribosomal subunits and 28S rRNA cofractionated with 60S/80S ribosomal subunits (Fig. 3, mock infections). Proportionate amounts of 28S and 18S rRNAs cofractionated with increasingly larger PV polysomes after 15 seconds and 60 min of incubation (Fig. 3B, 15 sec and 60 min). The distribution and integrity of PV mRNA were assessed by tracking the total amount of 32P in fractions of the gradient (data not shown) and by agarose gel electrophoresis (Fig. 3C). PV mRNA comigrated with free ribosomal subunits and with nascent polysomes immediately after addition to reaction mixtures (Fig. 3A to C, 15 sec). After 60 min of incubation, a large portion of PV mRNA remained intact and comigrated with polysomes (Fig. 3A to C, 60 min). Based on the amounts of 32P in each fraction of the gradient (data not shown), approximately 25% of the PV mRNA programmed into reaction mixtures was intact after 60 min of incubation. The amounts of radiolabel in fractions 1 to 3 increased over time and ran off the bottom of agarose gels, suggesting near complete degradation by exonuclease, while a portion of PV mRNA appears to be partially degraded in fractions 3 and 4 (Fig. 3C). After 60 min of incubation, the majority of intact PV mRNA comigrated with polysomes (Fig. 3B and C).
FIG. 3.
PV mRNA cofractionated with polysomes. Cell-free reactions were programmed with 32P-labeled PV RNA2, and the reaction mixtures were incubated at 34°C for 15 seconds or 60 minutes as indicated. A reaction mixture without mRNA (Mock) was incubated for 60 min. Reaction products were separated by centrifugation at 36,000 rpm for 3 h in 0.5 M to 1.5 M sucrose gradients as described in Materials and Methods. (A) Polysomes were detected by continuous UV absorbance as the gradients were fractionated. (B) RNAs from sucrose gradient fractions 1 to 20 were separated by electrophoresis in 1% agarose, stained with ethidium bromide, and detected by UV light. The mobility of PV RNA2 is marked with a white arrow. (C) 32P-labeled poliovirus (Polio) RNA2 within the agarose gel was detected by phosphorimaging.
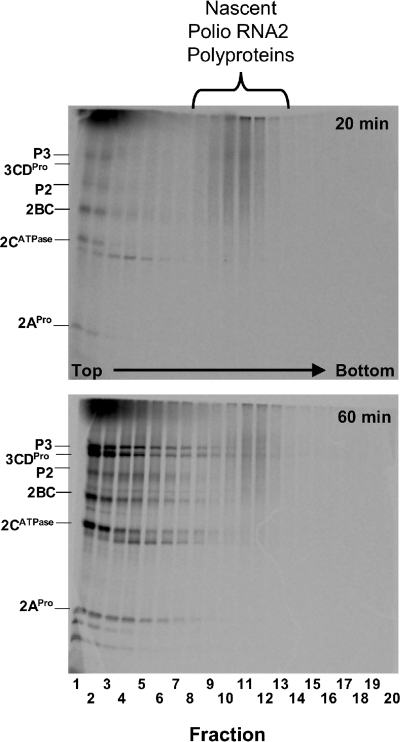
SDS-PAGE revealed that nascent PV polyproteins comigrated with viral polysomes, while mature viral proteins fractionated near the top of the gradients (Fig. 4). It is important to note that reaction products were treated with Triton X-100 before fractionation on sucrose gradients (see Materials and Methods); therefore, the dissociation of mature viral proteins from the polysomes may be due in part to the nonionic detergent treatment. A smear of nascent PV polyproteins comigrated with polysomes after 20 min of incubation, as ribosomes were initially reaching the 3′ terminus of the viral ORF (Fig. 4, 20 min, fractions 8 to 14). Sucrose gradients from mock reaction mixtures had no polysomes (Fig. 2 and 3) and no smear of polyproteins on SDS-polyacrylamide gels (data not shown). Nascent PV polyproteins also comigrated with viral polysomes after 60 min of incubation, while mature viral proteins accumulated at the top of the gradient (Fig. 4).
FIG. 4.
Nascent poliovirus (Polio) polyproteins cofractionated with polysomes. Cell-free reactions containing [35S]methionine were programmed with PV RNA2, and the reaction mixtures were incubated at 34°C for 20 or 60 min as indicated. Reaction products were then separated by ultracentrifugation in sucrose gradients, and the gradients were fractionated (as described in the legends to Fig. 2 and 3). [35S]methionine-labeled proteins from fractions 1 to 20 of the gradients were separated by SDS-PAGE, and radiolabeled proteins were detected by phosphorimaging.
The sizes of the PV polysomes correlated with the sizes of the ORFs.
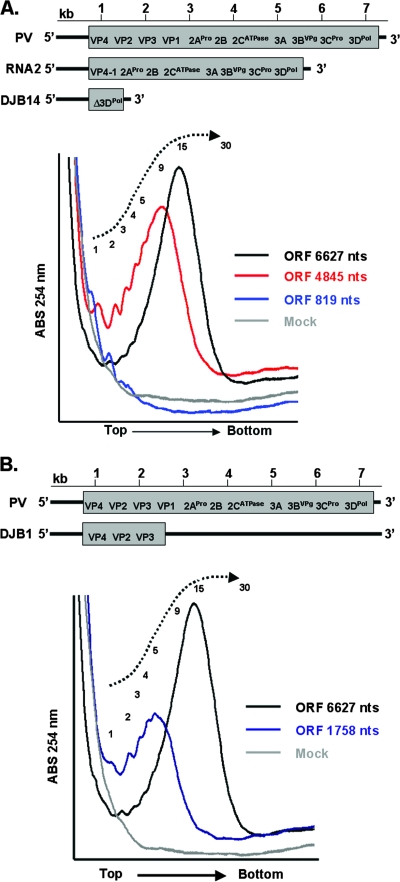
We used PV mRNAs with variable-size ORFs to more carefully examine ribosome density within PV ORFs (Fig. 5). Wild-type PV mRNA has an ORF 6,627 nucleotides long, while PV RNA2 has an ORF 4,845 nucleotides long and DJB14 mRNA has an ORF 819 nucleotides long (Fig. 5A). An out-of-frame stop codon in DJB1 mRNA results in an ORF 1,758 nucleotides long (Fig. 5B). After 60 min of translation, DJB14 mRNAs had a range of 1 to 3 ribosomes per ORF, RNA2 had a range of 1 to ∼20 ribosomes per ORF, and wild-type PV mRNA had a range of 1 to ∼30 ribosomes per ORF (Fig. 5A). The predominant number of ribosomes per ORF was 1 for DJB14 mRNA, ∼9 or 10 for RNA2, and ∼14 or 15 for wild-type PV mRNA (Fig. 5A). DJB1 mRNA had a range of 1 to ∼10 ribosomes per ORF with a peak at 5 ribosomes per ORF (Fig. 5B). Thus, the density of ribosomes on these mRNAs was variable over a limited range; however, a majority of PV mRNAs had a ribosome every 250 to 500 nucleotides of ORF.
FIG. 5.
The size of PV polysomes correlated with the size of ORFs. Four PV mRNAs with variable-length ORFs were incubated for 60 min in cell-free reaction mixtures. A reaction mixture without mRNA (Mock) was incubated for 60 min. Reaction products were then separated by centrifugation in sucrose gradients, and polysomes were detected by continuous UV absorbance (ABS) as the gradients were fractionated (as described in Materials and Methods). The UV trace from the portion of the gradients with polysomes was cropped and magnified for clarity. (A) UV traces for reaction mixtures containing PV mRNA (ORF 6,627 nucleotides [nts] long), RNA2 mRNA (ORF 4,845 nucleotides long), DJB14 mRNA (ORF 819 nucleotides long), and no mRNA (Mock). (B) UV traces for reaction mixtures containing PV mRNA (ORF 6,627 nucleotides long), DJB1 mRNA (ORF 1,758 nucleotides long), and no mRNA (Mock).
eIF4GI was cleaved rapidly after the elongation of ribosomes across PV ORFs.
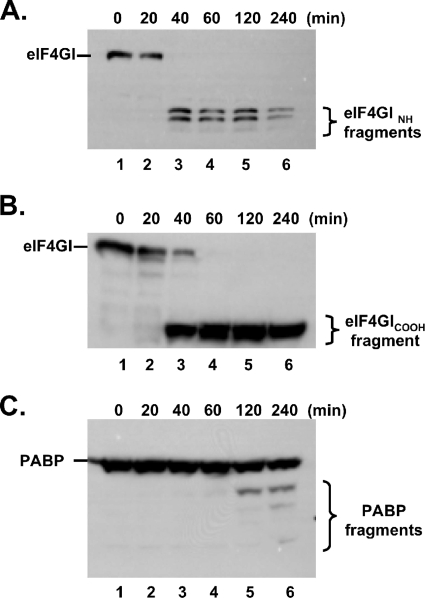
Western blots were used to examine the integrity of eIF4GI during the translation of PV RNA2 (Fig. 6A and B). Antibodies that recognize NH-terminal (Fig. 7A) and COOH-terminal (Fig. 6B) cleavage fragments of eIF4GI were used. eIF4GI was cleaved between 20 and 60 min of incubation (Fig. 6A and B). eIF4GI remained intact in mock reaction mixtures (data not shown). A small amount of 2APro was released from the nascent RNA2 polyprotein by 20 min (Fig. 1B, lane 8). As shown elsewhere (26), 2APro was responsible for degradation of eIF4GI. PABP, a proteolytic target of both 2APro and 3CPro (27, 28), was not cleaved as quickly or completely as eIF4GI in our reaction mixtures (Fig. 6C).
FIG. 6.
eIF4GI was cleaved rapidly as PV polysomes assembled. PV RNA2 was incubated for 0, 20, 40, 60, 120, and 240 min in cell-free reaction mixtures as described in the legend to Fig. 1. Proteins from the reaction mixtures were fractionated by SDS-PAGE and probed by Western blotting with antibodies as described in Materials and Methods. The gels were probed by Western blotting with an eIF4GI polyclonal antibody that recognizes the NH-terminal portion of the eIF4GI protein (eIF4GI NH) (A), eIF4GI monoclonal antibody that recognizes an epitope within the COOH-terminal portion of the eIF4GI protein (eIF4GICOOH) (B), and PABP rabbit polyclonal antibody (C).
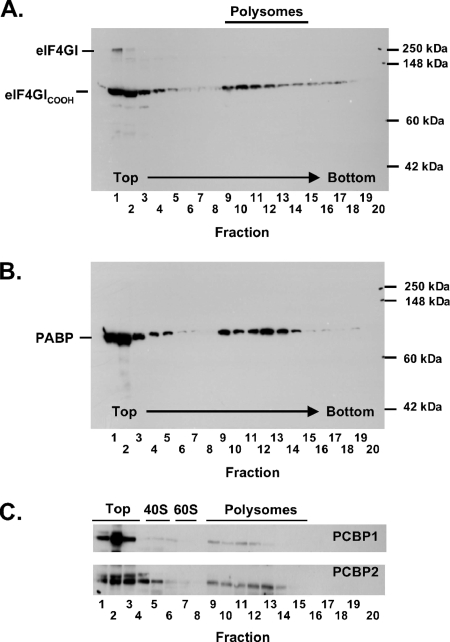
FIG. 7.
PABP, the COOH-terminal portion of eIF4GI, and PCBPs cofractionated with PV polysomes. PV RNA2 was incubated for 60 min in a cell-free reaction mixture as described in the legend to Fig. 5. The reaction products were then separated by centrifugation in sucrose gradients, and the gradients were fractionated (as in Fig. 2 to 4, in 20 0.5-ml fractions per gradient from top to bottom). Proteins from the sucrose gradient fractions were separated by SDS-PAGE and probed by Western blotting with antibodies as described in Materials and Methods. Proteins from sucrose gradient fractions 1 to 8 were diluted 1:10 relative to proteins in sucrose gradient fractions 9 to 20 due to the bulk of material near the top of the gradients. The gels were probed with an eIF4GI monoclonal antibody that recognizes an epitope within the COOH-terminal portion of the protein (eIF4GICOOH) (A), PABP rabbit polyclonal antibody (B), and PCBP 1 antibody or PCBP 2 antibody as indicated (C).
PABP and the COOH-terminal portion of eIF4GI comigrated with PV polysomes.
Because PABP and the COOH-terminal portion of eIF4G are thought to be involved in PV mRNA translation, we examined the distribution of these proteins in polysome gradients (Fig. 7). A portion of both PABP and the COOH terminus of eIF4GI comigrated with PV polysomes (Fig. 7, gradient fractions 9 to 14 correspond to the mobility of polysomes as shown in Fig. 3 and 4). A large amount of these proteins fractionated at the top of polysome gradients (Fig. 7, polysome gradient fractions 1 to 3). eIF4GI remained intact in mock reaction mixtures and fractionated almost exclusively at the top of the polysome gradients (data not shown). Likewise, PABP fractionated at the top of mock reaction mixtures (data not shown). Thus, PABP and the COOH terminus of eIF4GI cofractionated with PV polysomes.
PCBP 1 and 2 cofractionated with PV polysomes.
Because PCBP 1 and 2 interact with the 5′-terminal cloverleaf of PV mRNA and PCBP 2 interacts with the PV IRES (8, 16, 46), we used Western blots to determine whether PCBP 1 and 2 were associated with PV polysomes (Fig. 7C). As would be predicted, a portion of PCBP 1 and PCBP 2 were found to cofractionate with PV polysomes (Fig. 7C).
PV mRNA with a C24A mutation in the 5′ cloverleaf RNA formed unstable polysomes.
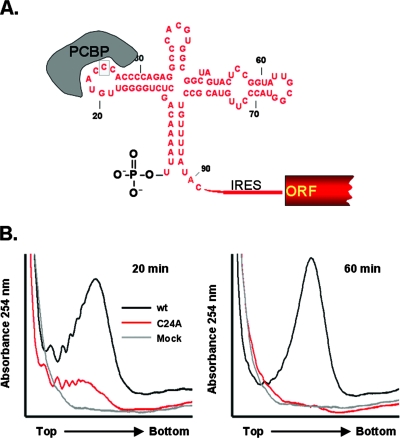
Previous investigations indicated that PCBPs bound to the 5′ terminus of PV mRNA (1, 16, 46) and that a C24A mutation that inhibited this protein-RNA interaction rendered PV mRNA susceptible to 5′ exonuclease (36). To examine how translation machinery interacted with this unstable mRNA, we compared de novo polysome formation of PV mRNA with C24A PV mRNA (Fig. 8). C24A PV mRNA was able to form polysomes in the time needed for ribosomes to traverse the ORF; however, the quality and quantity of C24A polysomes were not like those of wild-type PV mRNA (Fig. 8B, 20 min). Furthermore, after 60 min of incubation, C24A PV mRNA was no longer present in polysomes (Fig. 8B, 60 min). Thus, PV mRNA with a C24A mutation was able to transiently form polysomes coordinate in time with the elongation of ribosomes across the ORF, but C24A mRNA polysomes were smaller than wild-type polysomes and unstable over time. These results indicate that PCBP-5′ cloverleaf RNA interactions are necessary for viral mRNA stability during de novo polysome formation.
FIG. 8.
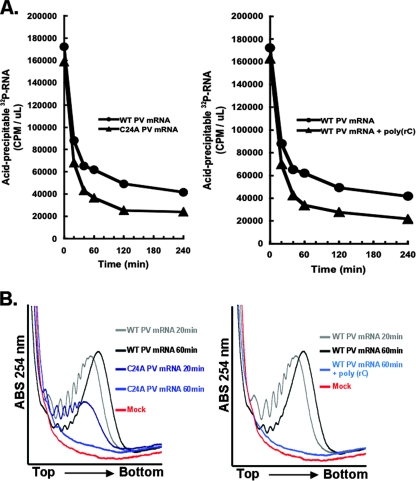
A C24A mutation at the 5′ terminus of PV mRNA inhibited de novo polysome assembly and stability. (A) Diagram of PCBP interaction with the 5′-terminal cloverleaf RNA structure of PV mRNA. (B) Cell-free reaction mixtures containing wild-type (wt) PV RNA2, PV RNA2 with a C24A mutation, or no mRNA (Mock) were incubated for 20 or 60 min as described in the legend to Fig. 1. The reaction products were then separated by centrifugation in sucrose gradients, and the gradients were fractionated (as described in the legends to Fig. 2 to 4). The UV traces from the portion of the gradients with polysomes were cropped and magnified for clarity.
Ribohomopoly(C) competitor RNA rendered wild-type PV mRNA unstable and prevented polysome formation.
PCBP interacts with two RNA elements within the 5′ NTR of PV mRNA, the 5′ cloverleaf and the IRES (8, 15, 16, 46). We previously established that ribohomopoly(C) competitor RNA, like the C24A mutation, prevented PCBP from binding to the PV 5′ cloverleaf RNA and rendered PV mRNA susceptible to 5′ exonuclease. Thus, as predicted, ribohomopoly(C) competitor RNA rendered wild-type PV mRNA unstable [Fig. 9A, note that the kinetics of PV mRNA instability in the presence of ribohomopoly(C) correspond to the kinetics of C24A mRNA instability] and prevented polysome formation by wild-type PV mRNA (Fig. 9B). These data indicate that ribohomopoly(C) competitor RNA can prevent polysome formation by wild-type PV mRNA. Therefore, these data reinforce the important roles of PCBP in both PV mRNA stability and polysome formation.
FIG. 9.
Ribohomopoly(C) competitor RNA inhibited PV mRNA stability and polysome formation. (A) (Left) 32P-labeled wild-type (WT) or C24A PV mRNA were incubated at 34°C for 0 to 240 min in cell-free reaction mixtures as indicated. (Right) 32P-labeled wild-type PV mRNA was incubated in the presence and absence of 25 μg per ml ribohomopoly(C) competitor RNA. The amounts (in cpm per μl) of acid-precipitable PV mRNA were plotted versus time of incubation. Note that ribohomopoly(C) competitor RNA rendered wild-type PV mRNA unstable, similar to the kinetics of C24A mRNA instability. 32P-RNA, 32P-labeled RNA. (B) Cell-free reaction mixtures containing PV RNA2, PV RNA2 with a C24A mutation, or no mRNA (Mock) were incubated for 20 or 60 min as described in the legend to Fig. 8. Ribohomopoly(C) competitor RNA (25 μg per ml) was included in one reaction mixture containing wild-type (WT) PV mRNA. Reaction products were separated by centrifugation at 36,000 rpm for 3.5 h in 0.5 M to 1.5 M sucrose gradients as described in Materials and Methods. UV absorbance (ABS) was monitored and plotted as the gradients were fractionated. The UV traces from the portion of the gradients with polysomes were cropped and magnified for clarity.
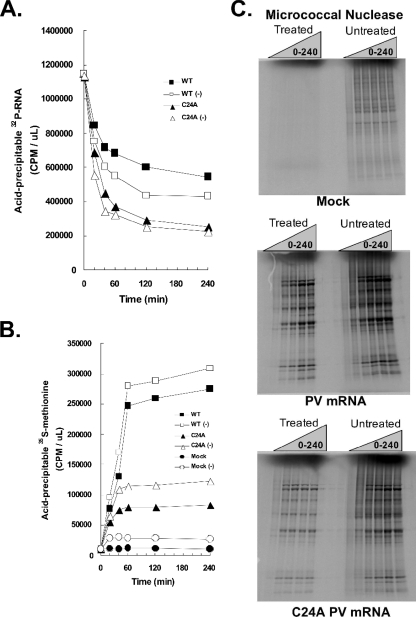
The kinetics and magnitude of PV mRNA stability, translation, and polysome formation were indistinguishable in reaction mixtures containing untreated or micrococcal nuclease-treated HeLa S10 extracts.
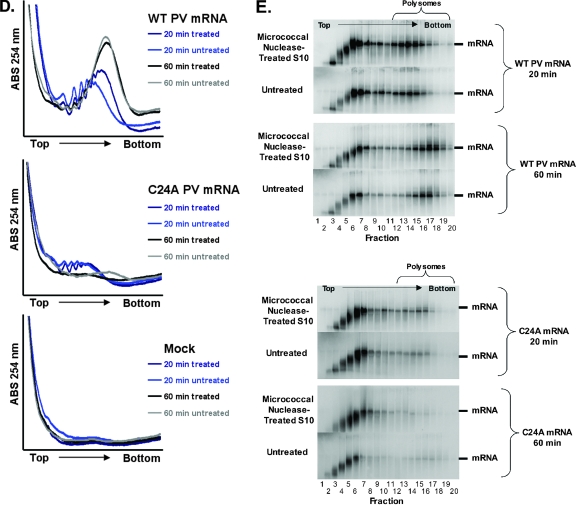
We compared the kinetics of ribosomes loading onto PV mRNA in untreated and micrococcal nuclease-treated cell extracts because competition with host mRNAs in untreated extracts could theoretically affect the ability of PV mRNA to compete for translation and stability factors. Further rationale for this comparison is a recent publication reporting phenotypic differences within reaction mixtures containing micrococcal nuclease-treated or untreated cellular extracts (55). Accordingly, we prepared untreated and micrococcal nuclease-treated HeLa S10 extracts and measured the kinetics and magnitudes of PV mRNA stability (Fig. 10A), PV mRNA translation (Fig. 10B) and polyprotein processing (Fig. 10C), and PV polysome formation (Fig. 10D). Furthermore, we examined the integrity and quantity of wild-type and C24A PV mRNAs within PV polysomes formed in reaction mixtures containing the untreated and treated S10 extracts (Fig. 10E).
FIG. 10.
Comparison of wild-type (WT) and C24A PV mRNA stability, translation, polyprotein processing, and polysome formation in reaction mixtures containing micrococcal nuclease-treated versus untreated HeLa S10 extracts. PV RNA2 (WT), PV RNA2 with a C24A mutation (C24A), or no mRNA (Mock) was incubated for 0 to 240 min in cell-free translation reaction mixtures containing untreated or micrococcal nuclease-treated HeLa S10 extracts as indicated. (A) PV mRNA stability. 32P-labeled wild-type and C24A PV mRNAs were incubated in reaction mixtures containing untreated or micrococcal nuclease-treated HeLa S10 extracts. Acid-precipitable 32P-labeled PV mRNA (32P-RNA) is plotted versus time of incubation. WT mRNA translated in micrococcal nuclease-treated (▪) and untreated (□) HeLa S10, C24A mRNA translated in micrococcal nuclease-treated (▴) and untreated (▵) HeLa S10 are shown. (B) PV mRNA translation. Acid-precipitable [35S]methionine accumulation plotted versus time of incubation. WT mRNA translated in micrococcal nuclease-treated (▪) and untreated (□) HeLa S10, C24A mRNA translated in micrococcal nuclease-treated (▴) and untreated (▵) HeLa S10, and mock reactions without exogenous mRNA in micrococcal nuclease-treated (•) and untreated (○) HeLa S10 are shown. (C) PV polyprotein processing. SDS-PAGE of [35S]methionine-labeled proteins from mock infections, PV mRNA, or C24A mRNA reaction mixtures containing untreated or micrococcal nuclease-treated HeLa S10 extracts after 0, 20, 40, 60, 120, and 240 min of incubation. (D) PV polysome formation. 32P-labeled wild-type PV mRNA, C24A mRNA, or no mRNA (Mock) were incubated for 20 or 60 min in reaction mixtures containing untreated or micrococcal nuclease-treated HeLa S10 extracts. Reaction products were separated by centrifugation at 36,000 rpm for 3.5 h in 0.5 M to 1.5 M sucrose gradients as described in Materials and Methods. UV absorbance (ABS) was monitored and plotted as the gradients were fractionated. The UV traces from the portion of the gradients with polysomes were cropped and magnified for clarity. (E) 32P-labeled wild-type and C24A PV mRNA distribution in polysome gradients. 32P-labeled PV mRNAs from all 20 sucrose gradient fractions in panel D above were separated by electrophoresis in 1% agarose and detected by phosphorimaging (as described in the legend to Fig. 3).
The magnitude of host protein expression from endogenous host mRNAs was greater in reaction mixtures containing untreated S10 extracts than in reaction mixtures containing micrococcal nuclease-treated S10 extracts (Fig. 10B, note the magnitude of [35S]methionine incorporated into acid-precipitable material in mock reactions). [35S]methionine-labeled host proteins were easily detected by SDS-PAGE from reaction mixtures containing untreated S10, whereas host proteins were nearly undetectable from reaction mixtures that had been treated with micrococcal nuclease (Fig. 10C, SDS-PAGE of treated and untreated mock reaction products in the top gel). These data are consistent with the conclusion that micrococcal nuclease treatments of S10 extracts digest host mRNAs, as expected. PV mRNA stability, PV mRNA translation, and PV polysome formation exhibited comparable kinetics and magnitudes in reaction mixtures containing either untreated or micrococcal nuclease-treated S10 (Fig. 10A to D). These data are consistent with the conclusion that competition of PV mRNA for limiting factors with endogenous host mRNAs was minimal in the cell-free reaction mixtures and that endogenous host mRNAs have little impact on the ability of PV mRNA to form functional polysomes in the reaction mixtures. The kinetics and magnitudes of PV C24A mRNA stability, translation, and polysome formation were equally defective relative to wild-type PV mRNA in reaction mixtures containing untreated and micrococcal nuclease-treated HeLa S10 extracts (Fig. 10A to D). These data are consistent with the conclusion that endogenous host mRNAs did not affect the phenotypes associated with the C24A mutation in the 5′ cloverleaf of PV mRNA. Agarose gel electrophoresis revealed that PV mRNA within polysomes was intact, that PV mRNA shifted to larger polysomes between 20 and 60 min of incubation, and that PV mRNA near the top of sucrose gradients was progressively degraded into small RNAs (Fig. 10E). The amounts of C24A PV mRNA within polysomes from reaction mixtures incubated for 20 min were notably less than the amounts of wild-type PV mRNA within polysomes, consistent with the defective stability of C24A PV mRNA (Fig. 10E). Furthermore, vanishingly small amounts of C24A mRNA remained intact relative to the amounts of wild-type mRNA after 60 min of incubation (Fig. 10E). These data indicated that the integrity of PV mRNA correlated with the ability of PV mRNA to form functional polysomes and that the C24A mutation coordinately reduced mRNA stability and polysome formation and stability.
DISCUSSION
In this study, we established experimental conditions to study the de novo formation and maturation of PV polysomes. The PV polysomes formed in HeLa cell-free reaction mixtures were similar to those identified long ago in PV-infected HeLa cells (48, 52). Viral mRNA and nascent viral proteins cofractionated with polysomes (Fig. 3 and 4), and the density of ribosomes on viral mRNA (Fig. 5, ribosomes every 250 to 500 nucleotides of the ORF) was comparable to that on PV mRNA within infected cells (57). A key technical advantage of PV cell-free translation-replication reactions is the ability to synchronize populations of viral RNA within particular steps of replication and to examine the requirements for particular molecules during the progression of metabolic events. Previously, we established experimental conditions to study the synchronous and sequential replication of PV RNA (4) and used such reactions to examine how seemingly distal RNP complexes at the termini of PV mRNA are coordinately required during the replication of viral RNA (7, 35).
Uncapped mRNAs are typically degraded by 5′ exonuclease (45); however, PV mRNA, which is naturally uncapped (22, 39, 40), is able to form stable polysomes (Fig. 8). Our experimental results indicate that uncapped PV mRNA formed polysomes coordinate in time with that needed for ribosomes to traverse the ORF (Fig. 1 and 2), that eIF4GI was cleaved as polysomes matured (Fig. 6), that the COOH terminus of eIF4GI required for translation initiation comigrated with PV polysomes (Fig. 7A), and that a 5′-terminal RNP complex containing PCBP was necessary for the formation and stability of PV polysomes (Fig. 8 to 10). Previous investigations indicated that disruption of the 5′-terminal RNP complex containing PCBP rendered PV mRNA susceptible to degradation by 5′ exonuclease (36). The instability of nascent C24A PV mRNA polysomes supports the conclusion that nucleases, presumably XRN1, can degrade PV mRNA within nascent polysomes and that PCBP-5′ cloverleaf RNA interactions prevent this degradation. The PCBP-5′ cloverleaf RNA interaction was required during the formation of polysomes (0 to 20 min of incubation), before ribosomes traversed the PV ORF, before the synthesis of viral protein 3CD, and before cleavage of eIF4GI by 2APro. Therefore, PCBP-5′ cloverleaf interactions are important during de novo polysome formation and function independently of viral protein 3CD, which forms RNP complexes associated with the cloverleaf RNA that are required for steps of replication long after polysome formation (7, 30, 35). Because PCBP-5′ cloverleaf interactions are required to mediate viral mRNA stability before the synthesis of viral protein 3CD, our experimental observations are not compatible with the idea that accumulating viral protein 3CD functions to recruit PCBP to the 5′ cloverleaf RNA from another binding site within the IRES as suggested by others (15).
PV mRNA is converted into a template for viral RNA replication following translation (41); therefore, it would be advantageous for the viral mRNA to avoid the cellular mRNA turnover machinery. Recently, discrete foci containing mRNA degradation enzymes were identified in yeast (54) and in HeLa cells (12, 24). These so-called P-bodies (mRNA processing bodies) are thought to be localized sites for degradation of cellular mRNAs. Furthermore, specific mRNA binding proteins may target cellular mRNAs to P-bodies for degradation (17), while other specific mRNA binding proteins like PCBP stabilize mRNAs. PCBP is known to interact with CU-rich sequences in the 3′ NTR of globin mRNA, and these interactions make globin mRNA one of the most stable mRNAs identified (31, 66). PV mRNA is uncapped, localized within endoplasmic reticulum membrane-associated polysomes, bound by PCBP, and present within 2APro-modified mRNP complexes containing the COOH-terminal portion of eIF4GI. Cofractionation of the COOH-terminal eIF4G fragment with PV mRNA polysomes (Fig. 7) is consistent with evidence that this portion of eIF4GI mediates IRES-dependent translation initiation (43). The NH-terminal portion of eIF4GI is dispensable for IRES-dependent translation (43) and is not associated with PV polysomes (data not shown). The NH-terminal portion of eIF4G interacts with eIF4E, the cap binding protein of eIF4F, and eIF4E in concert with eIF4E-T may be required for targeting 5′ capped mRNAs to P-bodies (2). Intriguingly, 2APro activity contributes to increased stability of PV mRNA (25). Ironically, the absence of a 5′ cap on PV mRNA in conjunction with 2APro activity may be part of the concerted mechanisms that uncouple PV mRNPs from the cellular mRNA turnover machinery.
Acknowledgments
Kevin Durand and Megan Filbin provided technical assistance. We thank Jerry Schaack and Richard Davis for their critical evaluation of the manuscript.
This work was supported by Public Health Service grants AI42189 (D.J.B.) and T32 AI07537 (B.J.K.) from the National Institutes of Health.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 123587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrei, M. A., D. Ingelfinger, R. Heintzmann, T. Achsel, R. Rivera-Pomar, and R. Luhrmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., and J. B. Flanegan. 1993. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 67822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 718482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 27535-57. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 7310104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 201439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 9311115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 716243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73861-890. [DOI] [PubMed] [Google Scholar]
- 11.Collis, P. S., B. J. O'Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 666480-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 16531-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenger-Gron, M., C. Fillman, B. Norrild, and J. Lykke-Andersen. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20905-915. [DOI] [PubMed] [Google Scholar]
- 14.Fillman, C., and J. Lykke-Andersen. 2005. RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol. 17326-331. [DOI] [PubMed] [Google Scholar]
- 15.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 742219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3882-892. [PMC free article] [PubMed] [Google Scholar]
- 17.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14571-583. [DOI] [PubMed] [Google Scholar]
- 18.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 9511089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond, M. L., and L. H. Bowman. 1988. Insulin stimulates the translation of ribosomal proteins and the transcription of rDNA in mouse myoblasts. J. Biol. Chem. 26317785-17791. [PubMed] [Google Scholar]
- 20.Hentze, M. W. 1997. eIF4G: a multipurpose ribosome adapter? Science 275500-501. [DOI] [PubMed] [Google Scholar]
- 21.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hewlett, M. J., J. K. Rose, and D. Baltimore. 1976. 5′-terminal structure of poliovirus polyribosomal RNA is pUp. Proc. Natl. Acad. Sci. USA 73327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 177480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingelfinger, D., D. J. Arndt-Jovin, R. Luhrmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 81489-1501. [PMC free article] [PubMed] [Google Scholar]
- 25.Jurgens, C. K., D. J. Barton, N. Sharma, B. J. Morasco, S. A. Ogram, and J. B. Flanegan. 2006. 2Apro is a multifunctional protein that regulates the stability, translation and replication of poliovirus RNA. Virology 345346-357. [DOI] [PubMed] [Google Scholar]
- 26.Kempf, B. J., and D. J. Barton. 2008. Poliovirus 2APro increases viral mRNA stability and polysome stability coordinately in time with cleavage of eIF4G. J. Virol. 825847-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuyumcu-Martinez, N. M., M. Joachims, and R. E. Lloyd. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 762062-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 241779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd, R. E. 2006. Translational control by viral proteinases. Virus Res. 11976-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 7510696-106708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makeyev, A. V., and S. A. Liebhaber. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8265-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 2541647-1651. [DOI] [PubMed] [Google Scholar]
- 33.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 775136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 774739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray, K. E., A. W. Roberts, and D. J. Barton. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 71126-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray, K. E., B. P. Steil, A. W. Roberts, and D. J. Barton. 2004. Replication of poliovirus RNA with complete internal ribosome entry site deletions. J. Virol. 781393-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newbury, S. F. 2006. Control of mRNA stability in eukaryotes. Biochem. Soc. Trans. 3430-34. [DOI] [PubMed] [Google Scholar]
- 39.Nomoto, A., N. Kitamura, F. Golini, and E. Wimmer. 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. USA 745345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomoto, A., Y. F. Lee, and E. Wimmer. 1976. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc. Natl. Acad. Sci. USA 73375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 81726-1737. [DOI] [PubMed] [Google Scholar]
- 42.Ochs, K., A. Zeller, L. Saleh, G. Bassili, Y. Song, A. Sonntag, and M. Niepmann. 2003. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 77115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohlmann, T., M. Rau, V. M. Pain, and S. J. Morley. 1996. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 151371-1382. [PMC free article] [PubMed] [Google Scholar]
- 44.Palacios, R., R. D. Palmiter, and R. T. Schimke. 1972. Identification and isolation of ovalbumin-synthesizing polysomes. I. Specific binding of 125 I-anti-ovalbumin to polysomes. J. Biol. Chem. 2472316-2321. [PubMed] [Google Scholar]
- 45.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11121-127. [DOI] [PubMed] [Google Scholar]
- 46.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 31124-1134. [PMC free article] [PubMed] [Google Scholar]
- 47.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 81103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Penman, S., K. Scherrer, Y. Becker, and J. E. Darnell. 1963. Polyribosomes in normal and poliovirus-infected HeLa cells and their relationship to messenger-RNA. Proc. Natl. Acad. Sci. USA 49654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prevot, D., J. L. Darlix, and T. Ohlmann. 2003. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell 95141-156. [DOI] [PubMed] [Google Scholar]
- 50.Prevot, D., D. Decimo, C. H. Herbreteau, F. Roux, J. Garin, J. L. Darlix, and T. Ohlmann. 2003. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 221909-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan, H., C. Y. Brown, and D. R. Morris. 1997. Analysis of ribosome loading onto mRNA species: implications for translational control, p. 305-321. In J. D. Richter (ed.), mRNA formation and function. Academic Press, New York, NY.
- 52.Scharff, M. D., A. J. Shatkin, and L. Levintow. 1963. Association of newly formed viral protein with specific polyribosomes. Proc. Natl. Acad. Sci. USA 50686-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma, N., B. J. O'Donnell, and J. B. Flanegan. 2005. 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J. Virol. 793565-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soto Rifo, R., E. P. Ricci, D. Decimo, O. Moncorge, and T. Ohlmann. 2007. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 35e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Summers, D. F., and J. V. Maizel, Jr. 1967. Disaggregation of HeLa cell polysomes after infection with poliovirus. Virology 31550-552. [DOI] [PubMed] [Google Scholar]
- 57.Summers, D. F., J. V. Maizel, Jr., and J. E. Darnell, Jr. 1967. The decrease in size and synthetic activity of poliovirus polysomes late in the infectious cycle. Virology 31427-435. [DOI] [PubMed] [Google Scholar]
- 58.Svitkin, Y. V., and N. Sonenberg. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 776551-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svitkin, Y. V., and N. Sonenberg. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 257155-170. [DOI] [PubMed] [Google Scholar]
- 60.Thoma, C., A. Ostareck-Lederer, and M. W. Hentze. 2004. A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. Methods Mol. Biol. 257171-180. [DOI] [PubMed] [Google Scholar]
- 61.Todd, S., J. S. Towner, and B. L. Semler. 1997. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology 22990-97. [DOI] [PubMed] [Google Scholar]
- 62.Trono, D., R. Andino, and D. Baltimore. 1988. An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol. 622291-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Hoof, A., P. A. Frischmeyer, H. C. Dietz, and R. Parker. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 2952262-2264. [DOI] [PubMed] [Google Scholar]
- 64.van Hoof, A., and R. Parker. 2002. Messenger RNA degradation: beginning at the end. Curr. Biol. 12R285-R287. [DOI] [PubMed] [Google Scholar]
- 65.van Ooij, M. J., D. A. Vogt, A. Paul, C. Castro, J. Kuijpers, F. J. van Kuppeveld, C. E. Cameron, E. Wimmer, R. Andino, and W. J. Melchers. 2006. Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative- and positive-strand RNA synthesis. J. Gen. Virol. 87103-113. [DOI] [PubMed] [Google Scholar]
- 66.Wang, X., and S. A. Liebhaber. 1996. Complementary change in cis determinants and trans factors in the evolution of an mRNP stability complex. EMBO J. 155040-5051. [PMC free article] [PubMed] [Google Scholar]