Abstract
The human nuclear envelope proteins emerin and lamina-associated polypeptide 2α (LAP2α) have been proposed to aid in the early replication steps of human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MLV). However, whether these factors are essential for HIV-1 or MLV infection has been questioned. Prior studies in which conflicting results were obtained were highly dependent on RNA interference-mediated gene silencing. To shed light on these contradictory results, we examined whether HIV-1 or MLV could infect primary cells from mice deficient for emerin, LAP2α, or both emerin and LAP2α. We observed HIV-1 and MLV infectivity in mouse embryonic fibroblasts (MEFs) from emerin knockout, LAP2α knockout, or emerin and LAP2α double knockout mice to be comparable in infectivity to wild-type littermate-derived MEFs, indicating that both emerin and LAP2α were dispensable for HIV-1 and MLV infection of dividing, primary mouse cells. Because emerin has been suggested to be important for infection of human macrophages by HIV-1, we also examined HIV-1 transduction of macrophages from wild-type mice or knockout mice, but again we did not observe a difference in susceptibility. These findings prompted us to reexamine the role of human emerin in supporting HIV-1 and MLV infection. Notably, both viruses efficiently infected human cells expressing high levels of dominant-negative emerin. We thus conclude that emerin and LAP2α are not required for the early replication of HIV-1 and MLV in mouse or human cells.
Viruses that enter or egress from the host cell nucleus are influenced by proteins in the nuclear milieu and have devised strategies to overcome and exploit innate barriers in order to sustain and spread infection (36). The nuclear architecture of eukaryotes is maintained by a network of intermediate filament proteins, the lamins, located beneath the inner nuclear membrane known as the lamina (5). In addition to providing mechanical strength and shape to nuclei, lamins support both the nuclear pores and other lamin-associated proteins. Some of these other lamina-associated proteins include a family defined by a ∼40-residue motif called the LEM domain (9, 15). The LEM domain family consists of several proteins, including emerin (3), lamina-associated polypeptide 2 (LAP2) (7), MAN1 (18), otefin (8), Lem3 (13), and Lem2 (4). The LEM domain is essential for interaction of these laminar proteins with the host chromatin protein barrier-to-autointegration factor (BAF) (14, 22, 26). BAF is believed to be a component of human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MLV) preintegration complexes (PICs) and to facilitate productive infection by preventing intramolecular integration of the viral DNA into itself (autointegration) and promoting intermolecular integration into extraneous (host) DNA (6, 16, 17, 29).
Recent studies have suggested that two LEM domain proteins, emerin and LAP2α, promote HIV-1 and MLV infectivity by enhancing chromatin association and integration of viral cDNA, likely through BAF interactions (11, 30). Depletion of emerin from human cells impaired infection by HIV-1 displaying wild-type Env or the G protein of vesicular stomatitis virus (VSV-G), whereas depletion of LAP2α reduced infection by HIV-1 displaying wild-type Env, MLV bearing wild-type Env, or MLV pseudotyped with VSV-G (11). Emerin, in particular, appeared to be critical for HIV-1 infection of primary macrophages. In addition, it has been reported that mouse cells depleted of LAP2α are diminished in their capacity to support MLV replication (30). However, the requirement of these factors for HIV-1 or MLV infection has been questioned (27).
Because these discordant results were generated through similar RNA interference (RNAi)-based techniques, we sought to investigate the roles of emerin and LAP2α in retroviral infection using alternative approaches. Studies of the HIV-1 IN binding protein, LEDGF, revealed that even minute quantities of a viral cofactor remaining after RNAi-mediated depletion are sufficient to permit efficient virus integration (19). We therefore examined whether HIV-1 or MLV could infect primary cells from mice whose genes that encode emerin, LAP2α, or both emerin and LAP2α were ablated. In contrast to studies implicating roles for emerin and LAP2α in retroviral infection, we did not observe either protein to be essential for HIV-1 or MLV infection of mouse cells. In addition, a dominant-negative form of emerin did not impair HIV-1 or MLV infectivity in human cells. While these studies do not rule out an interaction of HIV-1 or MLV with nuclear lamin proteins during infection, they suggest that these interactions may not be critical for provirus establishment.
MATERIALS AND METHODS
Generation of knockout mice.
The emerin-deficient (Emd−/−) mice have been described previously (24). The LAP2α-deficient (Tmpo−/−) mice have also been described elsewhere (N. Naetar et al., submitted for publication). The Emd−/− and Tmpo−/− mice were intercrossed to obtain Emd−/− Tmpo−/− double homozygous mice. All mice were maintained in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals of the NCI-Frederick ACUC Guidelines and Policies Committee.
Isolation of mouse embryonic fibroblasts and immunofluorescence analysis.
Mouse embryos were harvested at day 13 postconception, eviscerated, and homogenized in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 0.1 mg/ml DNase I (Sigma), and 0.5 mg/ml collagenase (Sigma). After 30 min of incubation, cells were washed in DMEM containing 10% FBS and 100 U/ml penicillin and 0.1 mg/ml streptomycin and plated to establish the primary fibroblast lines. Individual mouse embryonic fibroblast (MEF) lines were expanded and genotyped by Southern blot analysis as described elsewhere (24; Naetar et al., submitted). Cells used in infection experiments were at passage 4 to 5. For immunofluorescence analysis, MEFs were grown on microscope slide coverslips, fixed with 4% paraformaldehyde, stained with antibodies specific for either emerin (Novocastra, United Kingdom) or LAP2α (35) and then with secondary antibodies conjugated to Alexa 568 (Molecular Probes, Eugene, OR), and counterstained with 4′,6′-diamidino-2-phenylindole. After immunolabeling, coverslips were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and visualized using a Zeiss Axiophot inverted microscope.
Genotyping (RT-PCR).
Mice of known genetic backgrounds were used to isolate macrophages or used as breeding pairs to obtain MEFs. Emerin and LAP2α genotypes were further characterized in isolated cells by reverse transcription-PCR (RT-PCR). Total cellular RNA from the mouse macrophages used in Fig. 4, below, and the MEFs used in Fig. 2 and 3, below, were extracted with the RNeasy kit (Qiagen). Each sample was normalized for total RNA by spectrophotometric analysis and was subjected to a single round of RT-PCR using oligo(dT) oligonucleotides and the First Strand cDNA synthesis kit (Roche) to generate cDNA. The cDNA was subjected to 30 rounds of PCR using the following specific primers: for Emd (NM_000117), forward primer 5′-TTGTCTGCCATGGACGACTATGC-3′ and reverse primer 5′-TAAGAGCTGCTCTAAAACCAATACC-3′, which amplify a 914-bp product containing sequence from exon 1 through exon 6; for Tmpo (U09086), forward primer 5′-TGGAGGGAAGAGTAGAGCTCAG-3′ and reverse primer 5′-GTTCGGATCCAGGTGTATTCTG-3′, which amplify a 373-bp product within exon 4; for Actb (actin, beta; NM_007393), forward primer 5′-TGAACCCTAAGGCCAACCGTG-3′ and reverse primer 5′-GCTCATAGCTCTTCTCCAGGG-3′; for Hprt1 (hypoxanthine guanine phosphoribosyl transferase 1; NM_013556), forward primer 5′-CCTAAGATGAGCGCAAGTTGAA-3′ and reverse primer 5′-CCACAGGACTAGAACACCTGCTAA-3′. Following PCR amplification, the samples were run on 1.0% agarose to confirm genotype, i.e., the absence or presence of mRNA.
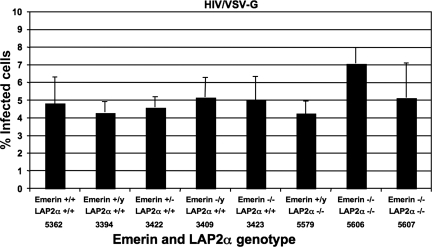
FIG. 4.
HIV-1 can infect primary mouse macrophages from emerin and LAP2α knockout mice. Infection efficiency of HIV/VSV-G was examined in macrophages from emerin knockout, LAP2α knockout, and emerin and LAP2α double knockout mice compared to wild-type littermate controls. At 60 h postinfection, the cells were stained for the F4/80 macrophage-specific marker. The percent infectivity represents the percentage of GFP-positive cells in the F4/80-positive population. Each data set represents the mean of four separate wells of infected cells, and error bars indicate standard deviations.
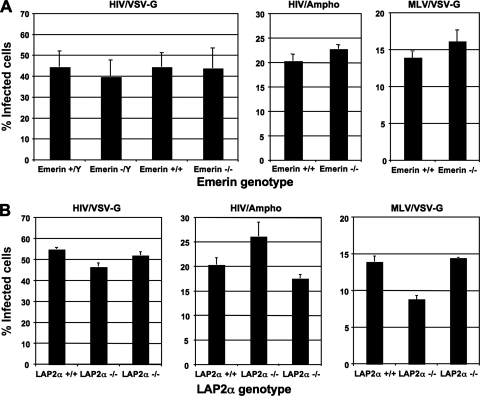
FIG. 2.
HIV-1 and MLV can infect MEFs that are knocked out for either emerin or LAP2α expression. (A) Infection efficiency of HIV/VSV-G, HIV/Ampho, or MLV/VSV-G was examined in Emd (emerin) knockout MEFs compared to sex-matched, wild-type littermate control MEFs. The percent infectivity represents the percentage of GFP-expressing MEFs 48 h postinfection with HIV-1 or MLV vectors. Each data set represents the mean of three independent experiments, and error bars indicate standard deviations. (B) Infection efficiency of HIV/VSV-G, HIV/Ampho, or MLV/VSV-G was also examined in LAP2α knockout MEFs compared to wild-type littermate control MEFs. The percent infectivity represents the percentage of GFP-expressing MEFs 48 h postinfection with HIV-1 or MLV vectors. Each data set represents the mean of three separate wells of infected cells, and error bars indicate standard deviations.
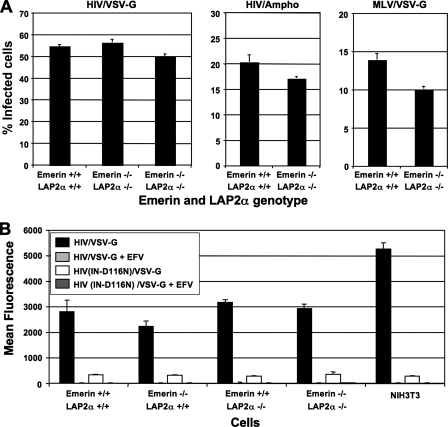
FIG. 3.
HIV-1 and MLV can infect MEFs that are knocked out for genes encoding both emerin and LAP2α. (A) Infection efficiency of HIV/VSV-G, HIV/Ampho, or MLV/VSV-G was examined in emerin and LAP2α double knockout MEFs compared to wild-type littermate control MEFs. The percent infectivity represents the percentage of GFP-expressing MEFs 48 h postinfection with HIV-1 or MLV vectors. Each data set represents the mean of three separate wells of infected cells, and error bars indicate standard deviations. (B) The infectivity of HIV/VSV-G and HIVIN-D116N/VSV-G was examined in wild-type, emerin knockout, LAP2α knockout, or emerin and LAP2α double knockout MEFs and NIH 3T3 cells. The cells were infected with an equal dose (200 ng p24 antigen) of virus. Infection was measured in the absence and presence of the nonnucleoside reverse transcriptase inhibitor EFV (100 nM). Infectivity was measured as the mean fluorescence intensity of GFP expression from infected cells to increase sensitivity for differences in gene expression. Each data set represents the mean of three separate wells of infected cells, and error bars indicate standard deviations.
Cell culture and antiviral drugs.
The 293T and NIH 3T3 cell lines were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. The MEFs were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 0.1 mM β-mercaptoethanol. The primary mouse macrophages were maintained in RPMI 1640 medium supplemented with 10% FBS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 0.1 mM β-mercaptoethanol, 0.3% sodium bicarbonate, and 0.25% conditioned medium containing granulocyte-macrophage colony-stimulating factor (gift of Derya Unutmaz, NYU). The nonnucleoside reverse transcriptase inhibitor efavirenz (EFV) was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Plasmids.
The HIV-CMV-GFP (HCG) plasmid was constructed from the HIV-EGFP plasmid (32) using the unique 5′ NotI and 3′ XhoI at the ends of the enhanced green fluorescent protein (EGFP) coding sequence, thus replacing EGFP with cytomegalovirus (CMV)-EGFP. This plasmid contains deletions in vif, vpr, env, and nef. The NLdE-CMV-GFP (NLdECG) plasmid was constructed from wild-type pNL4-3 to contain the same env gene deletion as the pLai3ΔenvGFP3 plasmid (38). The NLdECG plasmid contains deletions in env and nef. The integrase catalytic site mutant, IN-D116N, was generated by PCR mutagenesis and confirmed by sequencing.
The cDNA for full-length human Emd (GenBank accession number NM_000117) was obtained from Open Biosystems. The two dominant-negative LEM mutants were made by site-directed mutagenesis. The delta-LEM mutant is deleted for the entire LEM domain coding sequence (coding for amino acids 3 to 44) as described by Jacque and Stevenson (11), while the m24-LEM mutant described by Lee and colleagues has alanine substitutions at amino acid positions 24, 25, 26, and 27 (14). Both wild-type and LEM mutant emerin were inserted into the pIRES-puro3 expression vector (Clontech) using the AgeI and BamHI sites.
Virus stocks.
HIV-1 virus stocks were produced by DNA transfection on monolayer cultures of 293T cells grown in 10-cm culture dishes using Lipofectamine 2000 transfection reagent (Invitrogen). Each 10-cm dish was cotransfected with 18 μg of viral vector and 6 μg of p-L-VSV-G (1) or pSV-A-MLV (amphotropic) env (12) expression plasmids. Moloney MLV stocks were produced by DNA transfections of 293T cells grown in six-well plates using the calcium phosphate DNA precipitation method. Each cell monolayer (well) was cotransfected with 5 μg of pMIGR1 (25), 2.5 μg of pJK3 (1), 1 μg of pCMV-Tat, and 2 μg of p-L-VSV-G plasmids (1). Culture supernatants from the 293T cells were collected 48 h posttransfection, clarified by low-speed centrifugation (1,000 × g, 10 min), and filtered through 0.2-μm-pore-size sterile filters. For the HIV-1 vectors, the clarified supernatants were analyzed for p24 antigen concentration by enzyme-linked immunosorbent assay (Beckman-Coulter Inc.).
Infection assays.
Infectivity for all virus stocks was initially determined by titration on NIH 3T3 cells. Briefly, virus-containing supernatants were serially diluted (threefold dilutions) and used to infect monolayer cultures of NIH 3T3 cells plated on the previous day at 20,000 cells/well in 12-well tissue culture plates (Corning). At 48 h postinfection, the cells were trypsinized and resuspended in phosphate-buffered saline (PBS) containing 2% FBS. The expression of GFP following infection by the different HIV-1 and Moloney MLV vectors was measured by fluorescence-activated cell sorter (FACS) analysis (FACSCalibur; Becton Dickinson). Using the titration curves obtained from the NIH 3T3 infections, set doses of VSV-G- or Ampho Env-pseudotyped HCG vector (HIV/VSV-G and HIV/Ampho, respectively) or VSV-G Env-pseudotyped MIGR1 vector (MLV/VSV-G) were used to infect the MEFs that were plated on the previous day at 20,000 cells/well in 12-well tissue culture plates (Corning). The infection of MEFs by the HIV-1 and MLV vectors was measured by GFP expression at 48 h postinfection using FACS analysis, as described above. The percent infected cells represents the percentage of GFP-positive cells in the cell population.
The infection efficiencies of HIV/VSV-G on macrophages from Emd−/− or Tmpo−/− knockout and Emd−/− Tmpo−/− double knockout mice compared to wild-type littermate controls were examined. The peritoneal cavity of mice was injected with 2 ml of 3% thioglycolate in PBS. At 3 days after thioglycolate injection, the thioglycolate-responsive macrophage populations were harvested in 5 ml ice-cold Hanks' balanced salt solution and washed once with macrophage culture medium. The macrophages were plated at a cell density of 100,000 cells/well in 24-well low-attachment plates (Corning). After overnight incubation at 37°C, the cells were infected with a set dose of VSV-G Env-pseudotyped NLdECG vector (HIV/VSV-G). At 60 h postinfection, the cells were taken off the plate, washed once with 10 ml PBS containing 2% FBS, and kept on ice for subsequent steps. The cells were resuspended in 200 μl of PBS containing 2% FBS. The rat anti-mouse CD16/CD32 (FcγIII/II receptor) monoclonal antibody (mouse BD Fc block) was used to block the Fc-mediated binding of antibodies to mouse Fcγ receptor-bearing macrophages as recommended by the manufacturer (BD Pharmingen). After blocking the Fcγ receptors, the cells were stained with either the mouse macrophage-specific anti-F4/80-allophycocyanin or the isotype control antibody (Caltag Laboratories). F4/80 is a macrophage-restricted cell surface glycoprotein (23). The cells were washed twice after staining for 30 min, resuspended in 0.5 ml PBS containing 2% FBS, and analyzed by FACS. The typical purity of isolated macrophages by F4/80 staining is shown under the M1 gate in Fig. S1 in the supplemental material. The percent infected cells was measured as the percentage of GFP-positive cells in the F4/80-positive population.
The effect of dominant-negative emerin was examined using cells expressing wild-type and mutant forms of emerin. To measure infection by HIV-1 and MLV, 293T cells were transiently transfected with pIRES-puro3, pEmerin(WT)-IRES-puro, pEmerin(delta-LEM)-IRES-puro, or pEmerin(m24-LEM)-IRES-puro in a six-well plate. Cells were cotransfected with pDsRed-monomer-N1 (Clontech) at a threefold-lower molar amount. The cells were replated 1 day posttransfection at 1 × 106 cells/well in a fresh 6-well plate for obtaining Western blot cell lysates and at 4 × 104 cells/well in a 24-well plate for infection with HIV/VSV-G or MLV/VSV-G virus. On day 2 posttransfection, the cells from the six-well plate were collected for Western blotting. The cells in the 24-well plate were infected with a set dose of previously titrated HIV/VSV-G or MLV/VSV-G virus. At 48 h from the time of infection the cells were trypsinized, resuspended in PBS containing 2% FBS, and analyzed by FACS. The percent infected cells was measured as the percentage of GFP-positive cells in the DsRed-positive population.
RESULTS
Analysis of cells from knockout mice.
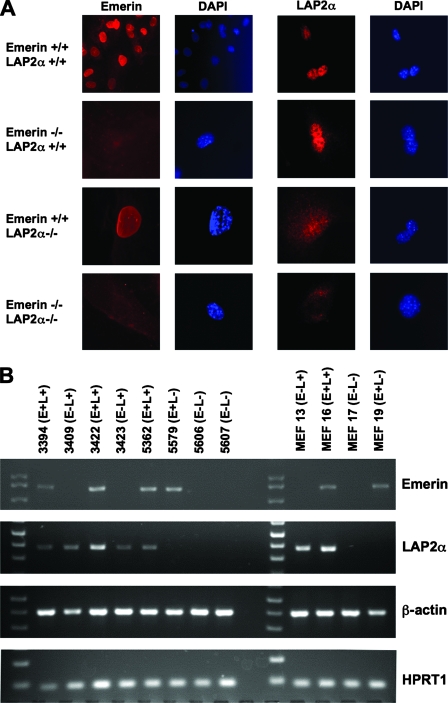
The emerin- and LAP2α-deficient mice have been described previously (24; Naetar et al., submitted). Deletion of Emd exons 2 to 5 eliminates all but the first exon of the X-linked gene, resulting in a complete loss of emerin synthesis (24). By contrast, the Tmpo knockout used in this study was restricted to exon 4, which enables expression of all LAP2 isoforms except for LAP2α (Naetar et al., submitted). Removal of emerin did not impair LAP2α production or subcellular distribution in MEFs, and the elimination of LAP2α had no effect on emerin levels or distribution (Fig. 1A and data not shown). In addition, the overall nuclear morphologies in the single and double knockout cells were grossly similar to wild-type cells.
FIG. 1.
Emerin- and LAP2α-deficient mouse cells. (A) The expression of emerin and LAP2α in fibroblasts derived from the single and double homozygote mice was analyzed by immunofluorescence. Staining with an emerin antibody showed the absence of the perinuclear staining pattern in all Emd−/− cells. Staining with LAP2α antibody showed a nucleoplasmic staining pattern that was absent in Lap2α−/− cells, although a low level of background staining was evident with this antibody (35). (B) Genotyping of primary macrophages and MEFs by RT-PCR. Total cellular RNA from mouse macrophages used in Fig. 4, below, and MEFs used in Fig. 2 and 3, below, was subjected to RT-PCR to generate cDNA. Using primers specific for emerin, LAP2α, β-actin, and the HPRT-encoding mRNA sequence, the cDNA was subjected to 30 rounds of PCR. After PCR amplification, the samples were run on 1.0% agarose to confirm genotypes. β-Actin and HPRT were amplified as controls to demonstrate the presence of RNA in the cell extracts.
For subsequent infection experiments, control cells expressing wild-type proteins were obtained from littermates of knockout mice to minimize possible maternal effects and ensure matched developmental times. Mouse macrophage and MEF genotypes were verified by RT-PCR. The macrophages and MEFs from wild-type littermate controls showed the presence of both Emd and Tmpo mRNA, and single knockout mice for either Emd or Tmpo showed an absence of the respective mRNA, while those from the double knockout mice showed an absence of both mRNAs (Fig. 1B). The RT-PCR for β-actin and HPRT mRNA confirmed the presence of cellular RNA in all the samples.
HIV-1 and MLV can infect MEFs lacking either emerin or LAP2α.
RNAi studies by Jacque and Stevenson suggested that emerin regulated HIV-1 infection of immortalized (dividing) human cells after nuclear entry (11). These results have been questioned by Shun and colleagues employing both RNAi in human cells and one Emd knockout male MEF line that we provided to assay sensitivity to HIV-1 or MLV infection (27). The underlying reasons for the different experimental outcomes are unclear. In the studies with the mouse cells, we examined whether other Emd knockout lines were also susceptible to retroviral infection or if other LEM-domain containing proteins played a more prominent role in the infection of mouse cells. To test whether emerin is required for HIV-1 or MLV infection of dividing mouse cells, we infected MEFs that do not have a functional Emd gene with HIV/VSV-G, HIV/Ampho, or MLV/VSV-G. HIV/VSV-G was used to infect emerin-deficient MEFs from both male and female mice (−/Y and −/−, respectively) along with MEFs from sex-matched wild-type littermate controls (+/Y and +/+, respectively). Infection of these MEF lines with an equal dose of HIV/VSV-G did not result in significantly different infection levels (Fig. 2A, left panel). Also, the HIV/Ampho and MLV/VSV-G viruses showed similar infectivity on emerin-deficient (−/−) and wild-type (+/+) MEFs (middle and right panels, respectively). Infections with greater or lesser titers of virus did not result in any observable differences in susceptibility between the different wild-type and knockout lines (data not shown).
LAP2α has been associated with HIV-1 and MLV infections of immortalized human and mouse cell lines (11, 30). It is possible that the presence of LAP2α in the emerin-deficient MEFs enabled efficient retroviral infection. To examine the role of LAP2α in both HIV-1 and MLV infection, we infected MEFs from mice lacking exon 4 of the Tmpo gene with HIV/VSV-G, HIV/Ampho, or MLV/VSV-G. Infection of the LAP2α-deficient MEFs (−/−) with HIV/VSV-G and HIV/Ampho was similar to that of MEFs from wild-type littermate controls (+/+) (Fig. 2B, left and middle panels, respectively). In MLV/VSV-G infections, while some LAP2α-deficient MEF lines showed wild-type-like infection, other LAP2α-deficient MEF lines showed an approximately 30% reduced infection (right panel).
HIV-1 and MLV can infect MEFs lacking both emerin and LAP2α.
Because both emerin and LAP2α are LEM domain proteins, we determined whether these proteins could be functionally redundant in supporting retroviral infection. To evaluate whether emerin and LAP2α are important for HIV-1 and MLV infection, we examined the infectivity of HIV/VSV-G, HIV/Ampho, or MLV/VSV-G on MEFs from Emd and Tmpo double knockout mice. Infection of the emerin- and LAP2α-deficient MEFs (−/−) with HIV/VSV-G and HIV/Ampho was comparable to that of MEFs from wild-type littermate controls (+/+) (Fig. 3A, left and middle panels, respectively). MLV/VSV-G infectivity of the double knockout MEFs showed an approximately 30% reduced infection compared to wild-type MEFs (right panel). Although the susceptibility of this line was consistently lower, another double knockout line showed similar infectivity to wild-type cells in other experiments (data not shown).
Because it has been suggested that emerin and LAP2α regulate the fate of viral DNA (vDNA) in the nucleus, potentially facilitating interactions with chromatin (11), it was conceivable that the positive infections scored in the absence of the LEM domain-containing proteins actually reflected stable forms of vDNA that were transcriptionally active but failed to integrate. To test whether Emd knockout, Tmpo knockout, or Emd and Tmpo double knockout MEFs enriched unintegrated vDNAs to a greater extent than wild-type control MEFs, we infected these MEFs with wild-type HIV/VSV-G or HIVIN-D116N/VSV-G (IN catalytic site mutation, D116N). As previously seen, infecting Emd or Tmpo knockout, Emd and Tmpo double knockout, and wild-type MEFs with an equal dose of HIV/VSV-G (200 ng p24/CA) resulted in similar infection levels (Fig. 3B). Notably, although the infectivity of HIVIN-D116N/VSV-G virus in the MEFs was reduced approximately 100-fold compared to wild-type HIV/VSV-G, these infection levels were similar on Emd or Tmpo knockout, Emd and Tmpo double knockout, and wild-type MEFs. Because increased reporter gene expression by IN-defective HIV-1 was not apparent in knockout MEFs, wild-type HIV-1 infection detected in the absence of emerin or LAP2α remained integration dependent. The inability to detect infection of MEFs by wild-type HIV/VSV-G or HIVIN-D116N/VSV-G in the presence of the nonnucleoside reverse transcriptase inhibitor EFV further confirmed that the infection observed was real (and not pseudo-infection by VSV-G vesicles containing GFP).
HIV-1 can infect macrophages from mice lacking emerin or LAP2α.
The LEM domain-containing nuclear envelope proteins emerin and LAP2α have also been implicated in HIV-1 transduction of nondividing primary human macrophages (11). To test the role of emerin and LAP2α in HIV-1 infection, we obtained macrophages from Emd or Tmpo knockout, Emd and Tmpo double knockout, and wild-type littermate control mice. The genotypes of these mice were confirmed by RT-PCR from lysates of the macrophages used in the infectivity studies (Fig. 1B).
The ability of HIV/VSV-G to infect macrophages from these Emd or Tmpo knockout, Emd and Tmpo double knockout, and wild-type littermate control mice was tested. The percentage of GFP-positive cells in the population expressing the mouse macrophage-specific F4/80 glycoprotein was used to determine the percent infected cells. The macrophages from the wild-type control mice numbers 5362 (female; Emd+/+ LAP2α+/+), 3394 (male; Emd+/y LAP2α+/+), and 3422 (female; Emd+/− LAP2α+/+) showed an approximate infectivity of 4.5% (Fig. 4B, lanes 1, 2, and 3). The macrophages from the Emd-deficient mice, numbers 3409 (male; Emd−/y LAP2α+/+) and 3423 (female; Emd−/− LAP2α+/+), showed an infectivity of 5.0% (lanes 4 and 5). The macrophages from the Tmpo-deficient mouse, number 5579 (male; Emd+/y LAP2α−/−), had an infectivity of approximately 4.2% (lane 6). The macrophages from the Emd and Tmpo double knockout mice, numbers 5606 and 5607 (female; Emd−/− LAP2α−/−), showed infectivities of approximately 7.0% and 5.0% (lanes 7 and 8). The mouse macrophages were inherently more resistant to HIV-1 infection. Despite being infected with 5.2 × 105 infectious units (based on titration on NIH 3T3 cells) of virus per 1.0 × 105 cells, the infectivity on the mouse macrophages ranged between 4.5 and 7.0%.
Analysis of HIV-1 infection in the presence of dominant-interfering human emerin.
The N-terminal LEM domain of emerin is necessary for interaction with BAF (14). Mutant emerin that fails to interact with BAF has been reported to impair HIV-1 infection (11). Because our prior analyses did not provide evidence for an essential role of emerin in retroviral infection of mouse cells, we sought to examine whether such a role was confined to human cells. HeLa cells stably expressing wild-type and LEM mutated forms of emerin were derived and tested for sensitivity to HIV-1 infection. However, no differences were observed in susceptibility relative to control lines (data not shown).
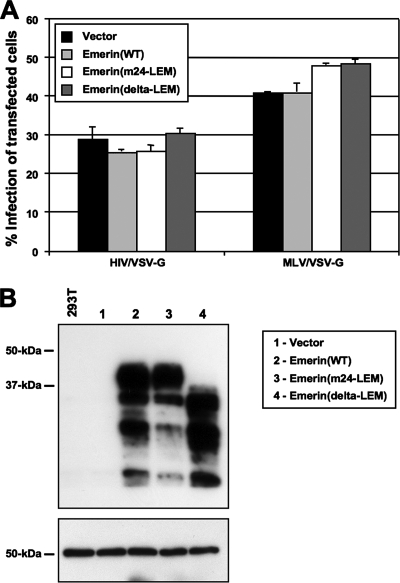
Because expression levels of mutant emerin might have been inadequate to interfere with infection in the stable cell lines, we reexamined the effect of LEM mutated forms of emerin via transient overexpression in HEK 293T cells. Two different LEM mutants were used: (i) the m24 LEM mutant characterized by Lee and colleagues as a mutant with four alanine substitution mutations in the LEM that is unable to bind BAF (14), and (ii) the ΔLEM mutant described by Jacque and Stevenson (11). Human 293T cells were transiently cotransfected with pDsRed-monomer-N1 and either empty vector, pEmerin(wild-type), pEmerin(m24-LEM), or pEmerin(ΔLEM) expression constructs. These transiently transfected cells were subsequently infected with HIV/VSV-G or MLV/VSV-G, and the percentage of cells expressing GFP was examined after 2 days. In individual samples, no differences between the infection rate of positively transfected cells (DsRed positive) and untransfected cells (DsRed negative) were observed (data not shown). Significantly, DsRed-positive cells expressing emerin with LEM mutations were as permissive to HIV-1 infection as cells expressing empty vector or high levels of wild-type emerin (Fig. 5A, left panel). Infection by MLV appeared to increase slightly in the presence of emerin mutants (Fig. 5A, right panel). A fraction of the transfected cells was lysed and analyzed by Western blotting for emerin levels (Fig. 5B). Exogenously expressed wild-type and mutant emerin were present in vast excess relative to endogenous emerin, such that endogenous protein in control samples was undetectable under the short exposure time needed to detect the transfected proteins (Fig. 5B, top panel). The blot was reprobed with monoclonal antibody to α-tubulin (Sigma) to confirm cell lysate loading consistency (bottom panel).
FIG. 5.
HIV-1 infection in cells expressing wild-type and dominant-negative emerin. (A) 293T cells transiently transfected with pIRES-puro3, pEmerin(WT)-IRES-puro, pEmerin(m24-LEM)-IRES-puro, or pEmerin(delta-LEM)-IRES-puro were infected with HIV/VSV-G or MLV/VSV-G at similar efficiency. Cells were cotransfected with pDsRed-monomer-N1 (Clontech) at a threefold-lower molar amount as a transfection control. The percent infectivity was measured as the percentage of GFP-positive cells in the DsRed-positive population as measured by FACS. (B) Top panel: expression of wild-type emerin or mutant emerin proteins in 293T cells used in the above infections was checked using Western blotting by probing with an anti-emerin polyclonal antiserum (Santa Cruz Biotechnology, Inc.). Bottom panel: the blot was reprobed with monoclonal antibody to α-tubulin (Sigma) to confirm cell lysate loading consistency.
DISCUSSION
The ability of viruses to coopt host cell proteins and machinery is fundamental to their propagation and spread. Productive infection by retroviruses like HIV-1 and MLV requires integration of the virus into the host genome. The nuclear chromatin-associated protein BAF and nuclear lamina-associated LEM domain proteins emerin and LAP2α have been suggested to assist in targeting the viral PICs to chromatin during integration. Prior investigation of BAF demonstrated its presence in viral PICs and suggested its requirement for integration in vitro (6, 16, 17, 29). The association of emerin and LAP2α with PICs is thought to be indirect and via LEM domain interactions with BAF (11, 30). Functional evaluation of emerin and LAP2α in retroviral infection has relied heavily on RNAi experiments but has resulted in conflicting data regarding the role of these proteins in HIV-1 and MLV infection (11, 27, 30). While MEFs from emerin-deficient mice had also been previously tested (27), this analysis was limited to one knockout line and did not include emerin-deficient macrophages from adult mice. Moreover, LAP2α knockout mice were only recently derived, allowing investigation of retroviral permissivity in MEFs or macrophages singly and doubly ablated for LEM domain proteins. With these tools, we demonstrate that both HIV-1 and MLV can infect dividing MEFs and that HIV can infect nondividing mouse macrophages from emerin knockout, LAP2α knockout, or emerin and LAP2α double knockout mice as efficiently as cells from wild-type littermate mice. Moreover, human cell lines expressing wild-type or dominant-negative mutant emerin were found to be equally susceptible to HIV-1. Based on these findings, we conclude that these two LEM domain-containing proteins are not required for infection of mouse cells by HIV-1 and MLV.
Because Jacque and Stevenson (11) conducted experiments in human cells, it could be argued that cross-species differences between mouse and human emerin or LAP2α orthologs prevent productive interaction with HIV-1 and MLV PICs within the murine cells. However, at least with regards to MLV infection, murine cells are the most relevant targets. Thus, because emerin or LAP2α removal from mouse cells has no clear effect on MLV vector transduction, these proteins are probably irrelevant to this process. Prior analysis in mouse cells by others appeared to demonstrate that MLV replication was impaired after depletion of LAP2α by RNAi, but these effects manifested only after cycles of replication, and it was not determined if they occurred early in the viral life cycle (30).
The differences observed in the essential nature of emerin or LAP2α for HIV-1 infection by Shun and colleagues (27) and what we have presented here are more difficult to reconcile with the findings of Jacque and Stevenson (11). In our work, we have primarily relied on mouse knockout cells to avoid the variability that is sometimes observed when using RNAi-based assays. In prior studies, we observed HIV-1 vector transduction to be comparable in mouse and human fibroblasts, indicating efficiency in early replication steps (2). It thus seems unlikely that HIV-1 utilizes entirely different pathways when infecting mouse versus human cells, as this would suggest that contacts with putative cofactors are simply incidental. Indeed, essential cofactors such as LEDGF/p75 are required by HIV-1 both in mouse and human cell types (19, 28, 33). Interestingly, analysis of LEDGF/p75 indicates that it may direct HIV-1 PICs to the host cell chromatin after nuclear entry, a function that would seemingly be redundant to the proposed role of emerin in HIV-1 infection (10, 20, 21, 31, 34).
Although beyond the scope of our analyses, it is possible that our assay systems fail to reveal subtle or cell-specific phenotypes in the absence of emerin or LAP2α. For example, integration site specificity could be altered in cells lacking either factor without impairing infection efficiency. Alternatively, MLV or HIV-1 infection of specific cell types or under different cell growth conditions may elicit a greater requirement for these factors. However, even with these limitations, retroviral infection efficiency in highly permissive or less permissive primary cell types, MEFs and macrophages, respectively, is strikingly consistent in the presence or absence of the LEM domain proteins. In addition, the emerin and LAP2α deficiencies did not appear to change the biology of HIV-1 infection by permitting accumulation of vDNA in the absence of integration. Our data indicate that HIV-1 IN function was required to establish infection in the absence of either or both LEM domain proteins.
Analysis of protein functional relevance in cells from gene-ablated mice can present other concerns. During the development of cell lines from the knockout animals, it is conceivable that there could be some compensatory upregulation of other nuclear envelope proteins, which would restore HIV/MLV infectivity to levels observed in wild-type cells. Because both emerin and LAP2α had been proposed to play similar roles in retroviral infection, our experiments with double knockout cells ruled out the possibility that these proteins were functionally redundant, thus masking effects in single knockout cells. While expression of other LEM domain-containing proteins is not significantly increased in emerin or LAP2α knockout lines (data not shown), another LEM domain protein could, in theory, productively interact with PICs in the absence of competing factors. Nonetheless, even if a third factor supported retroviral infection and/or enhanced BAF interactions with chromatin in the double knockout cells, it would again imply that emerin and LAP2α are not essential for HIV/MLV infection and are easily replaced by other proteins.
Despite caveats, an advantage of using knockout cells is the complete gene-level ablation of the factor being characterized. While RNAi-based techniques permit the rapid analysis of gene function in a variety of cell types, it is difficult to deplete all the mRNA for a targeted factor. It is also conceivable that a rapid depletion of emerin or LAP2α by RNAi could have collateral effects on BAF stability and/or distribution that contribute to the reduced permissivity of these cells. Because of the clear discordance in RNAi-dependent analyses of the role of emerin and LAP2α in retroviral infection in preceding studies, we also employed a third strategy to investigate emerin function in human cells by expressing mutant forms of emerin previously argued to have dominant-negative effects on HIV-1 infection (11). However, using two different LEM domain mutated forms of emerin, we could not reproduce an inhibition of HIV-1 infection even in the presence of extremely high levels of mutant proteins. These results were consistent with our observations using knockout mouse cells and further undercut an essential role for emerin in human cells.
Interestingly, Shun and colleagues have also failed to detect an effect on retroviral infection after small interfering RNA-dependent depletion of BAF in human target cells (27). An essential role for BAF in retroviral infection in vivo is integral to models that consider the potential influence of LEM domain proteins. BAF is proposed to act as the linchpin that connects these proteins to the retroviral PIC. Future studies with BAF-deficient mouse cells may be revealing in this regard. Recent studies with poxviruses suggest that BAF may, in some circumstances, act as an antiviral factor (37). Further examination of the role of BAF during retroviral infection, both alone and together with other nuclear lamin proteins, will be of great interest given the clear interaction of BAF with retroviral PICs.
Supplementary Material
Acknowledgments
We thank Zandrea Ambrose and Derya Unutmaz for reagents and advice and Michael Emerman, Alan Engelman, and Mario Stevenson for discussions.
This research was supported by the Intramural Research Program of the National Cancer Institute.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 9 April 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12337-342. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, J. G., D. Unutmaz, M. D. Miller, S. K. Breun, S. M. Grill, J. Mirro, D. R. Littman, A. Rein, and V. N. KewalRamani. 2004. Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 7812537-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bione, S., E. Maestrini, S. Rivella, M. Mancini, S. Regis, G. Romeo, and D. Toniolo. 1994. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 8323-327. [DOI] [PubMed] [Google Scholar]
- 4.Brachner, A., S. Reipert, R. Foisner, and J. Gotzmann. 2005. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell Sci. 1185797-5810. [DOI] [PubMed] [Google Scholar]
- 5.Burke, B., and C. L. Stewart. 2006. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu. Rev. Genomics Hum. Genet. 7369-405. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 9515270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foisner, R., and L. Gerace. 1993. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 731267-1279. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg, M., H. Lu, N. Stuurman, R. Ashery-Padan, A. M. Weiss, J. Yu, D. Bhattacharyya, P. A. Fisher, Y. Gruenbaum, and M. F. Wolfner. 1998. Interactions among Drosophila nuclear envelope proteins lamin, otefin, and YA. Mol. Cell. Biol. 184315-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruenbaum, Y., A. Margalit, R. D. Goldman, D. K. Shumaker, and K. L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 621-31. [DOI] [PubMed] [Google Scholar]
- 10.Hombrouck, A., J. De Rijck, J. Hendrix, L. Vandekerckhove, A. Voet, M. De Maeyer, M. Witvrouw, Y. Engelborghs, F. Christ, R. Gijsbers, and Z. Debyser. 2007. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 3e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacque, J. M., and M. Stevenson. 2006. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature 441641-645. [DOI] [PubMed] [Google Scholar]
- 12.Landau, N. R., and D. R. Littman. 1992. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J. Virol. 665110-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, K. K., Y. Gruenbaum, P. Spann, J. Liu, and K. L. Wilson. 2000. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell 113089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, K. K., T. Haraguchi, R. S. Lee, T. Koujin, Y. Hiraoka, and K. L. Wilson. 2001. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 1144567-4573. [DOI] [PubMed] [Google Scholar]
- 15.Lee, K. K., and K. L. Wilson. 2004. All in the family: evidence for four new LEM-domain proteins Lem2 (NET-25), Lem3, Lem4 and Lem5 in the human genome. Symp. Soc. Exp. Biol. 2004329-339. [PubMed] [Google Scholar]
- 16.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 951528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, C. W., and A. Engelman. 2003. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 775030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, F., D. L. Blake, I. Callebaut, I. S. Skerjanc, L. Holmer, M. W. McBurney, M. Paulin-Levasseur, and H. J. Worman. 2000. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 2754840-4847. [DOI] [PubMed] [Google Scholar]
- 19.Llano, M., D. T. Saenz, A. Meehan, P. Wongthida, M. Peretz, W. H. Walker, W. Teo, and E. M. Poeschla. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314461-464. [DOI] [PubMed] [Google Scholar]
- 20.Llano, M., M. Vanegas, N. Hutchins, D. Thompson, S. Delgado, and E. M. Poeschla. 2006. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 360760-773. [DOI] [PubMed] [Google Scholar]
- 21.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 27833528-33539. [DOI] [PubMed] [Google Scholar]
- 22.Mansharamani, M., and K. L. Wilson. 2005. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J. Biol. Chem. 28013863-13870. [DOI] [PubMed] [Google Scholar]
- 23.McKnight, A. J., A. J. Macfarlane, P. Dri, L. Turley, A. C. Willis, and S. Gordon. 1996. Molecular cloning of F4/80, a murine macrophage-restricted cell surface glycoprotein with homology to the G-protein-linked transmembrane 7 hormone receptor family. J. Biol. Chem. 271486-489. [DOI] [PubMed] [Google Scholar]
- 24.Melcon, G., S. Kozlov, D. A. Cutler, T. Sullivan, L. Hernandez, P. Zhao, S. Mitchell, G. Nader, M. Bakay, J. N. Rottman, E. P. Hoffman, and C. L. Stewart. 2006. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet. 15637-651. [DOI] [PubMed] [Google Scholar]
- 25.Pear, W. S., J. P. Miller, L. Xu, J. C. Pui, B. Soffer, R. C. Quackenbush, A. M. Pendergast, R. Bronson, J. C. Aster, M. L. Scott, and D. Baltimore. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 923780-3792. [PubMed] [Google Scholar]
- 26.Shumaker, D. K., K. K. Lee, Y. C. Tanhehco, R. Craigie, and K. L. Wilson. 2001. LAP2 binds to BAF. DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 201754-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shun, M. C., J. E. Daigle, N. Vandegraaff, and A. Engelman. 2007. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 81166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shun, M. C., N. K. Raghavendra, N. Vandegraaff, J. E. Daigle, S. Hughes, P. Kellam, P. Cherepanov, and A. Engelman. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 211767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, Y., and R. Craigie. 2002. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J. Virol. 7612376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki, Y., H. Yang, and R. Craigie. 2004. LAP2α and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 234670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turlure, F., G. Maertens, S. Rahman, P. Cherepanov, and A. Engelman. 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 341653-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 1891735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandekerckhove, L., F. Christ, B. Van Maele, J. De Rijck, R. Gijsbers, C. Van den Haute, M. Witvrouw, and Z. Debyser. 2006. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 801886-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanegas, M., M. Llano, S. Delgado, D. Thompson, M. Peretz, and E. Poeschla. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 1181733-1743. [DOI] [PubMed] [Google Scholar]
- 35.Vlcek, S., B. Korbei, and R. Foisner. 2002. Distinct functions of the unique C terminus of LAP2alpha in cell proliferation and nuclear assembly. J. Biol. Chem. 27718898-18907. [DOI] [PubMed] [Google Scholar]
- 36.Whittaker, G. R. 2003. Virus nuclear import. Adv. Drug Deliv. Rev. 55733-747. [DOI] [PubMed] [Google Scholar]
- 37.Wiebe, M. S., and P. Traktman. 2007. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe 1187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 785670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.