Abstract
Cell entry by paramyxoviruses requires fusion of the viral envelope with the target cell membrane. Fusion is mediated by the viral fusion (F) glycoprotein and usually requires the aid of the attachment glycoprotein (G, H or HN, depending on the virus). Human respiratory syncytial virus F protein (FRSV) is able to mediate membrane fusion in the absence of the attachment G protein and is unique in possessing two multibasic furin cleavage sites, separated by a region of 27 amino acids (pep27). Cleavage at both sites is required for cell-cell fusion. We have investigated the significance of the two cleavage sites and pep27 in the context of Sendai virus F protein (FSeV), which possesses a single monobasic cleavage site and requires both coexpression of the HN attachment protein and trypsin in order to fuse cells. Inclusion of both FRSV cleavage sites in FSeV resulted in a dramatic increase in cell-cell fusion activity in the presence of HN. Furthermore, chimeric FSeV mutants containing both FRSV cleavage sites demonstrated cell-cell fusion in the absence of HN. The presence of two multibasic cleavage sites may therefore represent a strategy to regulate activation of a paramyxovirus F protein for cell-cell fusion in the absence of an attachment protein.
Human respiratory syncytial virus (RSV) and Sendai virus (SeV) are members of the Pneumovirinae and Paramyxovirinae subfamilies, respectively, within the Paramyxoviridae family of negative-stranded RNA viruses (Mononegavirales) (5). Cell entry by paramyxoviruses requires fusion of the viral envelope with the target cell membrane at the cell surface. Paramyxovirus-infected cells are also capable of fusing with adjacent cells to form syncytia (multinuclear, giant cells). Fusion in both cases is mediated by viral envelope glycoproteins, namely, the fusion (F) protein, a trimeric type I integral membrane glycoprotein.
The F polypeptide is synthesized as an inactive precursor (F0), which requires proteolytic cleavage to yield fusion-competent, disulfide-linked F2-F1 chains (for a review see reference 21). Cleavage takes place at either a mono- or multibasic (RXK/RR) cleavage site, recognized by trypsin- or furin-like proteases, respectively, immediately upstream of a hydrophobic fusion peptide that is inserted into the cell membrane during fusion (30). Cleavage is an absolute requirement for fusion, and indeed the nature of the cleavage motif has been shown to play a role in the pathogenicity of some paramyxoviruses (17).
Whereas for most paramyxoviruses, fusion mediated by the F glycoprotein requires participation of the homotypic attachment protein (G, H, or HN, depending on the virus), the F protein of human respiratory syncytial virus is able to fuse membranes in the absence of the attachment G protein. Spontaneous mutants (15) or genetically engineered recombinant RSVs expressing F as the only surface glycoprotein (38, 39) are able to infect cells and form syncytia. Furthermore, expression of the RSV F protein as the only viral protein in transfected cells is sufficient to induce syncytium formation (12, 45).
RSV F protein (FRSV) is unique among the Paramyxoviridae, since FRSV possesses two furin cleavage sites (site I, RARR109, and site II, KKRKRR136), which are separated by a region of 27 amino acids (pep27), as shown in Fig. 1A, below. The two cleavage sites and the length, but not the sequence, of pep27 are highly conserved in all bovine and human RSV strains. Proteolytic processing at both cleavage sites is required by FRSV to form syncytia in transfected cells (12, 45). Thus, while membrane fusion by FRSV does not depend on coexpression of an attachment protein, there is a requirement for double cleavage of the F protein and removal of the intervening pep27, which is secreted into the medium (28). The function of the double cleavage and subsequent pep27 removal are currently unknown, although completion of cleavage is associated with a change in shape of the RSV F protein (25, 28). An immunomodulatory role has been suggested for bovine RSV pep27 (48); however, no such role has been attributed to the human RSV intervening peptide, which does not share sequence similarity with its bovine counterpart.
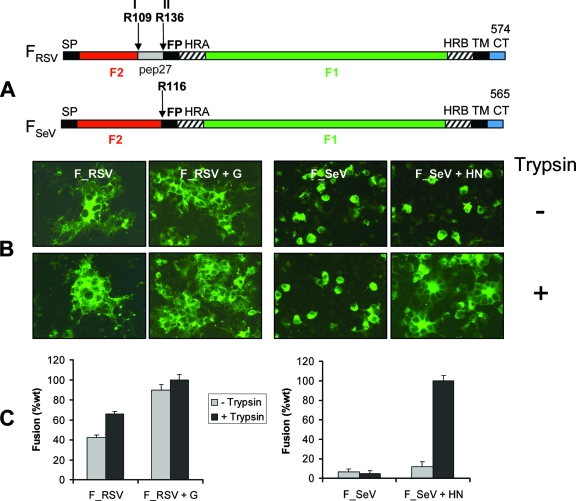
FIG. 1.
RSV and SeV fusion proteins have different requirements for cell-cell fusion. (A) Schematic diagram of RSV fusion (FRSV) and SeV fusion (FSeV) proteins. The three principal hydrophobic regions found in paramyxovirus F proteins are shown in black: the N-terminal signal peptide (SP), the fusion peptide (FP), located adjacent to the cleavage site at the N terminus of F1, and the C-terminal transmembrane region (TM). Heptad repeat sequences HRA and HRB (hatched boxes) are located adjacent to the FP and TM regions, respectively. Cleavage sites of FRSV (site I, R109, and site II, R136) and FSeV (R116) are indicated by arrows. Cleavage results in the formation of disulfide-linked F2 and F1 polypeptides, and in the case of FRSV, cleavage at both sites results in the removal of the intervening segment pep27, shown in light gray. (B) BSR-T7/5 cells were cotransfected in microchamber culture slides with 0.25 μg of pTM1 plasmids encoding RSV or SeV F genes and 0.25 μg pTM1-G or pTM1-HN plasmids, as indicated, or 0.25 μg empty vector pTM1 (not indicated). The transfection mixture was removed 7 h posttransfection and cells were incubated in serum-free medium with (+) or without (−) trypsin. Cells were fixed and immunostained as indicated in Materials and Methods. Immunofluorescence of cells transfected with the empty pTM1 plasmid was conducted in parallel, and the absence of fluorescence confirmed specific antibody staining (data not shown). (C) BSR-T7/5 cells were transfected in 48-well plates with plasmids encoding RSV or SeV genes. BHK cells were simultaneously transfected with a plasmid encoding the luciferase gene (pTM1-Luc). At 7 h posttransfection, BHK cells were detached using 1 mM EDTA, resuspended at a density of 1 × 106 cells/ml in serum-free medium, and overlaid at a 1:1 ratio onto BSR cells, either in the presence or absence of trypsin. At 30 h posttransfection (or 48 h posttransfection for experiments involving FRSV), cells were lysed and analyzed for luciferase activity. Results (relative light units) are expressed as a percentage of the wild-type (wt) fusion (FSeV plus HN plus trypsin, or FRSV plus G plus trypsin), with background activity subtracted (determined by overlaying BHK cells transfected with pTM1-Luc onto BSR cells transfected with the empty pTM1 plasmid). Mean values from three independent experiments are shown.
Despite low sequence identity, paramyxovirus F proteins share structural elements, including the location of cysteine residues, hydrophobic domains, and heptad repeats (HR) A and B, as shown in Fig. 1A, below. Crystallographic data indicate that refolding of the F protein during fusion results in the assembly of HRA and HRB into a six-helix bundle, consisting of an internal core of three HRA helices surrounded by three antiparallel HRB helices (2, 44). Such an arrangement positions the fusion peptide and transmembrane domain at the same end of the F protein and is thought to be directly linked to merging of viral and cell membranes, with the free energy released on six-helix bundle formation driving membrane fusion (26, 30). The crystal structures of the pre- and postfusion forms of two paramyxovirus F proteins have recently been solved, revealing substantial conformational differences between the two structures (42, 43). The trigger responsible for activation of the prefusion F protein for fusion is currently unknown. However, in the case of viruses belonging to the Paramyxovirinae subfamily, binding of the attachment protein to the corresponding cell surface receptor is thought to play a key role in F activation (for a review see reference 20).
Paramyxovirus attachment proteins are type II integral membrane proteins that differ widely in their structure and receptor usage. An interaction between the F and HN proteins has been demonstrated on the surface of transfected (8) or infected cells (36). This interaction has been shown to decrease under conditions that favor HN attachment (24), suggesting that F and HN interact before the binding of HN to its receptor takes place. The current model of membrane fusion hypothesizes that the HN protein forms a prefusion, metastable complex with F in the virion. Following attachment of the HN protein to the target cell receptor, conformational changes that take place in HN may be transduced to the F protein to activate it for fusion “at the right time and in the right place” (20). However, the fusion protein of human metapneumovirus (HMPV) and human and bovine RSV is sufficient to mediate attachment and fusion in the absence of other surface glycoproteins (3, 15, 33, 34, 38, 39). Thus, the precise role in fusion of attachment proteins belonging to members of the Pneumovirinae subfamily is less clear.
In order to explore the significance of the double cleavage of FRSV for fusion, we decided to investigate the effect of the FRSV proteolytic processing region in the context of the Sendai virus F protein (FSeV). Sendai virus is a member of the Paramyxovirinae subfamily, and in contrast to the RSV F protein, FSeV is cleaved at a monobasic cleavage site (R116) and requires coexpression of both the HN attachment protein and trypsin in order to fuse cells in culture (16, 32). A series of chimeric mutants of FSeV were constructed by inserting one or both FRSV cleavage sites and various regions of pep27 into FSeV. The ability of these mutants to cause cell-cell fusion was evaluated in the absence and presence of the HN protein and trypsin. Inclusion of both FRSV cleavage sites in FSeV resulted in a dramatic increase in cell-cell fusion activity in the presence of HN. Furthermore, chimeric mutants containing both FRSV cleavage sites also demonstrated cell-cell fusion in the absence of HN. These results suggest that the presence of two multibasic cleavage sites may represent an alternative strategy to regulate the activation of a paramyxovirus F protein for cell-cell fusion in the absence of an attachment protein.
MATERIALS AND METHODS
Cell lines.
BSR-T7/5 cells (4) (a gift from K.-K. Conzelmann, Munich, Germany) and BHK-21 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; BioWhittaker), supplemented with 10% fetal calf serum (FCS). Alternate passages of BSR cells were supplemented with 1 mg/ml G-418 sulfate (Sigma-Aldrich), in order to select for T7 polymerase expression.
Plasmids and mutagenesis.
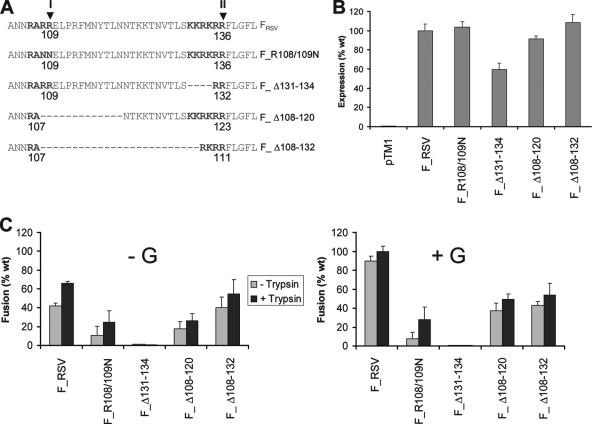
The pTM1 plasmid (9), carrying the full-length cDNA insert of the RSV F gene under transcriptional control of the T7 promoter (pTM1-FRSV), has been described previously (12). Plasmids carrying the mutations in FRSV shown in Fig. 7A, below, have also been reported (28, 29). The mutant F genes were subsequently subcloned in pTM1 between NcoI and BamHI restriction sites.
FIG. 7.
Coexpression of the attachment G protein fails to restore the reduced fusogenic capacity of RSV F cleavage site mutants. (A) The amino acid sequence surrounding the FRSV proteolytic processing region (amino acids 103 to 141) is shown. Cleavage sites of FRSV (site I, RARR109, and site II, KKRKRR136) are shown in bold. Deleted residues are indicated by a dashed line. (B) The levels of cell surface expression of wild-type (wt) and mutant FRSV proteins were determined by flow cytometry of transfected BSR-T7/5 cells 24 h posttransfection, using the monoclonal antibody 2F. The mean fluorescent intensity of cells is represented as a percentage of wt FRSV expression, and mean values from three independent experiments are shown. (C) BSR-T7/5 cells were cotransfected in 48-well plates with 0.25 μg pTM1 plasmids encoding wt or mutant FRSV genes and 0.25 μg pTM1 (−G) or pTM1-G (+G). BHK cells were simultaneously transfected with 30 μg of pTM1-Luc and overlaid at a 1:1 ratio onto BSR cells, as described in the legend to Fig. 1C. Cells were lysed and analyzed for luciferase activity at 48 h posttransfection. Results (relative light units) are expressed as a percentage of wt fusion (FRSV plus G plus trypsin), with mean values from three independent experiments shown.
The genes encoding Sendai virus F and HN proteins (Harris strain) were amplified from pGEM-F and pGEM-HN plasmids (a gift from L. Roux, Geneva, Switzerland), by PCR, and cloned into the pTM1 plasmid downstream of the T7 promoter, between NcoI and SpeI restriction sites. The resulting pTM1-FSeV plasmid was subjected to mutagenesis, using the QuikChange site-directed mutagenesis kit (Stratagene) to generate the mutants shown in Fig. 2, below. Three (RKR) or five (KKRKR) basic residues from the FRSV cleavage site II were inserted into FSeV using the forward oligonucleotides pTM1-F_RKR, 5′GCCGGTGTTCCACAGTCGAGGAAAAGAAGATTCTTCGGTGCTGTG, and pTM1-F_KKRKR (Fc), 5′GCCGGTGTTCCACAGTCGAAGAAAAGGAAAAGAAGATTCTTCGGTGCTG, with complementary reverse oligonucleotides. Subsequent mutagenesis of pTM1-F_KKRKR (termed F cleavage, or Fc) was carried out in order to insert FRSV cleavage site I, producing the pTM1-Fc_103-110 mutant (where the numbers refer to inserted FRSV residues), using the following forward oligonucleotide: pTM1-Fc_103-110, 5′GATCTTCAGGAGGCTCTGATAACTGCAAACAATCGAGCCAGAAGAGAAGTCACCAATGATACGACACAAAATGCC.
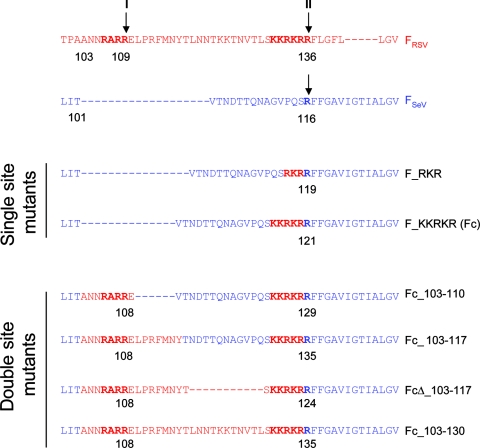
FIG. 2.
Design of SeV F protein cleavage site mutants. An alignment of FSeV (amino acids 99 to 129 [blue]) and FRSV (amino acids 100 to 144 [red]) is shown. Cleavage sites of FRSV (site I, RARR109, and site II, KKRKRR136) and FSeV (R116) are shown in bold and indicated by arrows. Residues from FRSV that were inserted into a backbone of FSeV are shown in red. The numbering of FSeV mutants produced by mutation of Fc refers to the inserted FRSV residues.
Further mutagenesis of pTM1-Fc_103-110 was performed as detailed in Fig. 2, below, using the following forward oligonucleotides to insert various regions of the intervening segment between the two cleavage sites (pep27): pTM1-Fc_103-117, 5′CTGCAAACAATCGAGCCAGAAGAGAACTACCAAGGTTTATGAATTACACCAATGATACGACACAAAATGCCGGTG; pTM1-FcΔ_103-117, 5′GCCAGAAGAGAACTACCAAGGTTTATGAATTACACCTCGAAGAAAAGGAAAAGAAGATTCTTCGGTGC; pTM1-Fc_103-130, 5′GCCAGAAGAGAACTACCAAGGTTTATGAATTACACCCTCAACAATACCAAAAAAACCAATGTAACATTATCGAAGAAAAGGAAAAGAAGATTCTTCGGTGC.
All mutations were confirmed by automated sequencing of the complete F gene. The RSV G protein was similarly cloned into the pTM1 plasmid between NcoI and SpeI restriction sites (pTM1-G), and the gene encoding luciferase was cloned into pTM1 between NcoI and PstI restriction sites (pTM1-Luc).
Syncytium formation assay.
BSR-T7/5 cells (a BHK-derived cell line that constitutively expresses the T7 RNA polymerase) were grown in microchamber culture slides to 90 to 100% confluence and transfected in DMEM (2.5% FCS) with 0.5 μg total DNA, using FuGENE HD (Roche Biochemicals, Inc.). The transfection mixture was removed 7 h posttransfection, and cells were incubated in serum-free medium with or without 0.25 μg/ml trypsin (Sigma-Aldrich). Cells were fixed for 32 h (FSeV) or 48 h posttransfection (FRSV) with cold methanol for 5 min, followed by cold acetone for 30 s. Fixed cells were subsequently immunostained using monoclonal antibodies directed against FSeV (GB5; a gift from S. Wharton and J. J. Skehel, London, United Kingdom) or FRSV (2F) (11), followed by incubation with anti-mouse fluorescein-linked antibody (GE Healthcare UK Ltd.). Cells were examined using a Zeiss microscope and photographed using an AxioCam HRC digital camera and Axiovision 3.1 software.
Luciferase reporter gene cell-cell fusion assay.
BSR-T7/5 cells were transfected in 48-well plates with 0.5 μg total DNA, as described above. BHK cells in 100-cm dishes were simultaneously transfected with 30 μg of a plasmid encoding the luciferase gene (pTM1-Luc). For the cell-cell fusion assay, BHK cells were detached 7 h posttransfection using 1 mM EDTA in Ca2+- and Mg2+-free phosphate-buffered saline (PBS), resuspended at a density of 1 × 106 cells/ml in serum-free medium, and overlaid at a 1:1 ratio onto BSR cells, either in the presence or absence of 0.25 μg/ml trypsin (Sigma-Aldrich). The mixed donor and target cells were subsequently incubated for 22 h to allow fusion. At 30 h posttransfection (or 48 h posttransfection for experiments involving FRSV), cells were washed with PBS and lysed in 1× passive lysis buffer (Promega, Inc.), according to the manufacturer's instructions. Ten to 20 μl of clarified cell extract was mixed with 100 μl luciferase assay substrate (Promega) and immediately assayed for luciferase activity over a 10-second measurement using a Turner Biosystems 20/20n luminometer instrument. Background activity (subtracted from all results shown) was determined by overlaying BHK cells transfected with pTM1-Luc onto BSR cells, which had previously been transfected with the empty pTM1 plasmid.
Flow cytometry.
Transfected BSR-T7/5 cells were detached 24 h posttransfection using 1 mM EDTA in Ca2+- and Mg2+-free PBS and resuspended in DMEM (2% FCS). A total of 3 × 105 cells were incubated for 30 min at 4°C with monoclonal antibody GB5 (FSeV) or 2F (FRSV). After pelleting and washing the cells two times with PBS, cells were stained by subsequent 30-min incubations with biotinylated anti-mouse immunoglobulin (Amersham Biosciences UK Ltd.) and streptavidin-R-phycoerythrin (Southern Biotechnology Associates). Finally, cells were fixed in 1% paraformaldehyde and the fluorescence of 2 × 104 cells was determined using a Becton Dickinson FACSCalibur instrument with CellQuest software. Data were analyzed using FlowJo software (Tree Star, Inc.).
Immunoprecipitation and Western blotting.
BSR-T7/5 cells were transfected as previously described. Twenty-four hours posttransfection, cells were washed with PBS and extracts of membrane proteins prepared using the ProteoExtract native membrane protein extraction kit (Calbiochem). For the immunoprecipitation experiments, purified GB5 antibody was bound to CNBr-activated Sepharose (Pharmacia), according to the manufacturer's instructions. One ml of membrane protein extract was incubated at 4°C with 50 μl of GB5-conjugated Sepharose. Following an overnight incubation, Sepharose beads were washed four times with PBS, resuspended in 50 μl sample buffer (80 mM Tris, 2% sodium dodecyl sulfate [SDS], 10% glycerol, and 0.01% bromophenol blue), and boiled for 6 min. Following centrifugation to remove beads from the eluted proteins, 5% β-mercaptoethanol was added and the immunoprecipitated proteins were separated on a 10% acrylamide SDS-polyacrylamide gel electrophoresis gel. Proteins were transferred to an Immobilon membrane (Millipore) and subjected to Western blotting in order to detect FSeV proteins using biotinylated GB5 monoclonal antibody and streptavidin-horseradish peroxidase conjugate (Amersham Biosciences UK Ltd.). Bands were visualized using the Amersham ECL Advance Western blotting detection kit (GE Healthcare UK Ltd.) and imaged using a VersaDoc camera and Quantity One 1D analysis software (Bio-Rad Laboratories).
RESULTS
RSV and SeV fusion proteins have different requirements for cell-cell fusion.
Qualitative and quantitative assays of cell-cell fusion were carried out in order to compare the requirements for fusion of RSV and SeV F proteins (comparative primary structure diagrams of both proteins are shown in Fig. 1A) and to confirm previous findings. BSR-T7/5 cells, which constitutively express T7 RNA polymerase, were transfected with pTM1 plasmids encoding viral glycoproteins under transcriptional control of the T7 promoter. Cells were fixed 32 h (FSeV) or 48 h (FRSV) posttransfection and immunostained using monoclonal antibodies directed against FSeV or FRSV proteins (Fig. 1B). Formation of syncytia (multinuclear, fused cells) was observed in transfected cells that expressed FRSV as the only viral protein, both in the presence and absence of trypsin. The number and size of syncytia was enhanced by coexpression of FRSV with G and by the presence of trypsin in the culture medium. In contrast, FSeV displayed a strict requirement for both HN coexpression and trypsin in order to form syncytia. Thus, in the absence of HN, only individual, unfused cells expressing FSeV were observed.
These results were confirmed by a luciferase reporter gene assay (Fig. 1C). BSR cells transfected with the genes of interest were mixed with BHK cells, which had been previously transfected with the pTM1-Luc plasmid, and left to fuse in the absence or presence of trypsin. Since pTM1-Luc contains the luciferase reporter gene under transcriptional control of the T7 promoter, expression of luciferase only takes place in BHK cells that have fused with BSR cells, permitting quantitative measurement of cell-cell fusion. While trypsin produced only a moderate enhancement in the level of luciferase activity induced by FRSV, coexpression of G increased cell-cell fusion by approximately twofold. Therefore, while not a strict requirement, G enhances FRSV-mediated cell-cell fusion. In accordance with the syncytium formation assay, FSeV required both HN coexpression and trypsin in order to produce significant luciferase activity. These results are in good agreement with previously published findings, both for FSeV, expressed with or without the HN protein (37), and for FRSV, expressed in the absence of the G protein (12, 45).
Design and expression of SeV F protein cleavage site mutants.
As indicated in Fig. 1A, RSV F protein is cleaved by furin-like proteases at two cleavage sites (site I R109 and site II R136), which are separated by a region of 27 amino acids (pep27). In contrast, SeV F protein contains a single arginine residue at its cleavage site (R116) and requires the addition of trypsin to the cell culture medium for cleavage. Alignment of FRSV and FSeV (Fig. 2) revealed that although both proteins possess homologous fusion peptides, the two multibasic cleavage sites and the intervening segment of FRSV cannot be aligned with FSeV. We decided to investigate if the unique characteristics of the FRSV proteolytic processing region play a role in the distinct fusion requirements by FRSV and FSeV. To this aim, mutagenesis was carried out on the FSeV gene (cloned in the pTM1 plasmid) in order to reproduce partially, or totally, the sequences that determine cleavage of FRSV. Initially, part of the second cleavage site of FRSV was inserted directly preceding the single cleavage site of FSeV (R116), producing F_RKR or F_KKRKR (Fc) mutants, containing a minimal furin recognition sequence (RKRR) or the complete FRSV cleavage site II (KKRKRR), respectively (Fig. 2). Further mutation of the Fc construct was carried out in order to introduce FRSV cleavage site I, producing the Fc_103-110 mutant (where the numbering refers to FRSV residues). As detailed in Fig. 2, regions of pep27 were subsequently inserted to produce additional mutants, including Fc_103-117, which contains both FRSV cleavage sites I and II separated by 27 intervening amino acids derived from both FRSV and FSeV. FcΔ_103-117 was produced from the Fc_103-117 mutant by a deletion N-terminal to the second cleavage site, resulting in a shortened intervening segment composed entirely of FRSV residues. Finally, the remaining residues of FRSV pep27 were inserted into FcΔ_103-117 to produce the mutant Fc_103-130, which contains the complete FRSV pep27 sequence.
In order to confirm cell surface expression, BSR-T7/5 cells were transfected with the chimeric FSeV mutants, and flow cytometry of the cells was performed using a monoclonal antibody directed against FSeV. As shown in Fig. 3A, all FSeV mutants were expressed at the cell surface at levels comparable to the wild-type F protein, although a slight decrease in expression was noted for mutants containing both FRSV cleavage sites.
FIG. 3.
Cell surface expression and proteolytic cleavage of chimeric SeV F mutants. (A) The cell surface expression levels of wild-type (wt) and mutant FSeV proteins were determined by flow cytometry analysis of transfected BSR-T7/5 cells 24 h posttransfection, using the monoclonal antibody GB5. The mean fluorescent intensity of cells is represented as a percentage of wt FSeV expression, and mean values from three independent experiments are shown. (B) Extracts of membrane proteins were prepared from transfected BSR-T7/5 cells incubated in the absence (upper panel) or presence (lower panel) of trypsin and subjected to immunoprecipitation using purified GB5 antibody bound to Sepharose beads. Following an overnight incubation, immunoprecipitated FSeV proteins were eluted, fractionated by SDS-polyacrylamide gel electrophoresis under reducing conditions, and analyzed by Western blotting using biotinylated GB5 antibody and a streptavidin-horseradish peroxidase conjugate.
With the aim of determining whether the FSeV mutants could be cleaved by cellular proteases, GB5-conjugated Sepharose beads were used to immunoprecipitate SeV F proteins from membrane extracts of transfected BSR cells, which were subsequently visualized by Western blotting. As shown in Fig. 3B (upper panel), the SeV wild-type F0 protein precursor remained essentially uncleaved in the absence of trypsin, whereas some cleavage of F0 to F1 was observed for the mutant F_RKR. Insertion of the complete FRSV cleavage site II into FSeV led to a greater extent of cleavage, as observed for the Fc mutant. Similarly, mutants Fc_103-110, Fc_103-117, and Fc_103-130, which contain both FRSV cleavage sites I and II, were also present as uncleaved F0 and fully cleaved F1 forms in approximately equal proportions. A band migrating at a lower molecular weight than F0 was observed for the mutant FcΔ_103-117. Although the identity of this band has not been thoroughly investigated, it may represent the F0 precursor of the FcΔ_103-117 mutant, which lacks an N-glycosylation site that has previously been shown to be used in the Z strain of SeV F protein (N104) (35).
When trypsin was added to the medium of the transfected cells (Fig. 3B, lower panel), cleavage of the F0 precursor of both wild-type FSeV and F_RKR to F1 was clearly enhanced. In contrast, only a slight increase in F0 processing was detected when trypsin was added to cells expressing the Fc mutant. However, the extent of F0-to-F1 cleavage was enhanced by trypsin in all four FSeV mutants containing both FRSV cleavage sites, and Fc_103-130 appeared fully cleaved to F1 in the presence of trypsin. Thus, insertion of an FRSV cleavage site II in the SeV F protein increases the extent of cleavage at this site in the absence of trypsin. This cleavage is further enhanced by trypsin, following the insertion of FRSV cleavage site I upstream of site II.
Insertion of RSV F cleavage site II in SeV F leads to trypsin-independent syncytium formation.
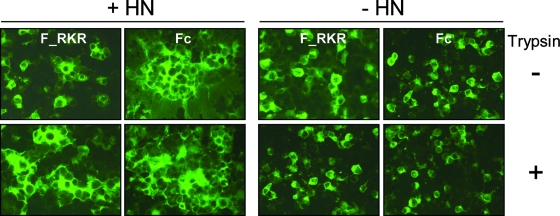
Cell-cell fusion by the SeV F protein mutants F_RKR or Fc, containing part (RKRR) or the complete FRSV cleavage site II (KKRKRR), respectively, was tested in the syncytium formation assay. In the presence of the HN attachment protein, only small syncytia were observed for F_RKR when trypsin was excluded from the culture medium (Fig. 4). This reflects the poor cleavage of F_RKR in the absence of trypsin (Fig. 3B). In contrast, the Fc mutant formed very large syncytia even in the absence of trypsin (Fig. 4), consistent with the enhanced cleavage of F0 to F1 that was observed for this mutant (Fig. 3B). Fc was also capable of forming more extensive syncytia than wild-type FSeV (compare with Fig. 1B). However, both F_RKR and Fc mutants continued to require HN coexpression in order to form syncytia (Fig. 4). Thus, in spite of the large syncytia formed by Fc in the presence of HN, insertion of the complete FRSV cleavage site II in FSeV does not lead to HN-independent fusion. Similar results were obtained with F_RKR and Fc mutants in the quantitative luciferase assay, presented comparatively with other mutants in subsequent sections.
FIG. 4.
Insertion of the RSV F cleavage site II in SeV F leads to trypsin-independent syncytium formation. BSR-T7/5 cells growing in microchamber wells were cotransfected as previously described with 0.25 μg pTM1-F_RKR or pTM1-Fc mutants and 0.25 μg pTM1-HN plasmid (+HN) or 0.25 μg empty pTM1 vector (−HN). The transfection mixture was removed 7 h posttransfection, and cells were incubated in serum-free medium with (+) or without (−) trypsin. Cells were fixed 32 h posttransfection and subjected to immunostaining as described in Materials and Methods.
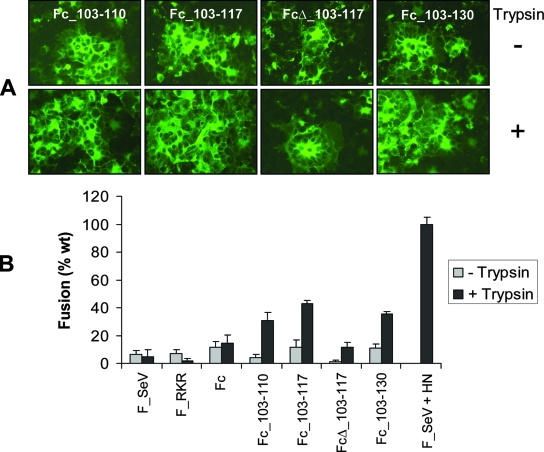
Insertion of both RSV F cleavage sites I and II in SeV F protein leads to enhanced membrane fusion in the presence of HN.
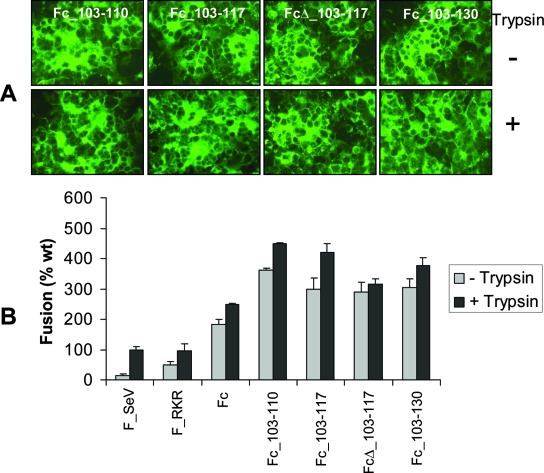
Mutants containing both FRSV cleavage sites I and II (Fig. 2) were also tested for their ability to fuse cells in the syncytium formation assay. As shown in Fig. 5A, all FSeV chimeras containing the two FRSV cleavage sites formed extremely large syncytia in the presence of HN, without a requirement for trypsin. In order to confirm the results of the syncytium formation assay, all of the mutants detailed in Fig. 2 were coexpressed with HN and analyzed for cell-cell content mixing using the luciferase reporter gene assay (Fig. 5B). Insertion of the complete FRSV cleavage site II (KKRKRR) in FSeV to form the Fc mutant led to a marked increase in cell-cell fusion compared to wild-type FSeV. In the absence of trypsin, Fc resulted in 181% of wild-type FSeV fusion (FSeV coexpressed with HN in the presence of trypsin represents 100% fusion). In contrast, the F_RKR mutant containing the partial FRSV cleavage site II produced only 50% of wild-type fusion in the absence of trypsin and fused cells to the same extent as the wild-type F protein on addition of trypsin. Thus, the presence of six basic residues at the single cleavage site of FSeV enhances the fusogenic potential of this protein when coexpressed with HN, even in the absence of trypsin. In the presence of HN, all mutants containing both FRSV cleavage sites I and II dramatically increased fusion with respect to the wild-type F protein (by up to 364% or 450% in the absence or presence of trypsin, respectively). The intervening sequence between the two cleavage sites appeared to exert a minor modulatory role on fusion in the presence of HN.
FIG. 5.
Insertion of both RSV F cleavage sites I and II in SeV F protein enhances membrane fusion in the presence of HN. (A) BSR-T7/5 cells growing in microchamber wells were cotransfected with 0.25 μg pTM1-Fc_103-110, pTM1-Fc_103-117, pTM1-FcΔ_103-117, or pTM-Fc_103-130 mutants and 0.25 μg pTM1-HN plasmid and processed for syncytium formation and immunostaining as described in the legend for Fig. 1. (B) BSR-T7/5 cells were cotransfected in 48-well plates with 0.25 μg pTM1 plasmids encoding wild-type (wt) or mutant FSeV genes and 0.25 μg pTM1-HN, as previously described. BHK cells were simultaneously transfected with 30 μg of pTM1-Luc and overlaid at a 1:1 ratio onto BSR cells, as described in the legend to Fig. 1C. Cells were mixed either in the presence or absence of trypsin and incubated for 22 h to allow fusion. Cells were subsequently lysed and analyzed for luciferase activity at 30 h posttransfection. Results (relative light units) are expressed as a percentage of wt fusion (FSeV plus HN plus trypsin), with mean values from three independent experiments shown.
No strict correlation was observed between the extent of F0 cleavage to F1 in the absence or presence of trypsin (Fig. 3B) and the relative differences in fusion observed between the mutants (Fig. 5B). However, the enhanced fusogenicity of double cleavage site mutants reflects their increased susceptibility to cleavage when trypsin was added to the culture medium (Fig. 3B, lower panel).
Insertion of both RSV F cleavage sites I and II in SeV F protein leads to a decreased dependence on HN for membrane fusion.
In contrast to the Fc mutant that contains only the second FRSV cleavage site, all of the FSeV mutants containing both FRSV cleavage sites were also able to form syncytia in the absence of HN coexpression (Fig. 6A). Therefore, inclusion of the two multibasic FRSV cleavage sites in FSeV facilitates syncytium formation, even in the absence of an attachment protein. Although HN-independent cell-cell fusion was observed in the absence of trypsin, trypsin inclusion did enhance the size and number of syncytia formed. The insertion of both FRSV cleavage sites and not the intervening pep27 sequence per se conveys HN independence for cell-cell fusion, since the Fc_103-110 mutant forms syncytia in the absence of HN. However, the nature and/or length of the intervening residues between the two cleavage site does appear to influence fusion to a greater extent in the absence of HN, since Fc_103-117 formed large syncytia whereas FcΔ_103-117 formed only small syncytia without HN.
FIG. 6.
Insertion of both RSV F cleavage sites I and II in SeV F protein decreases dependency on HN for membrane fusion. (A) BSR-T7/5 cells growing in microchamber wells were cotransfected with 0.25 μg pTM1-Fc_103-110, pTM1-Fc_103-117, pTM1-FcΔ_103-117, or pTM-Fc_103-130 mutants and 0.25 μg empty pTM1 plasmid and processed in parallel to the cultures shown in Fig. 5A. (B) BSR-T7/5 cells were cotransfected in 48-well plates with 0.25 μg pTM1 plasmids encoding wild-type (wt) or mutant FSeV genes and 0.25 μg empty pTM1 plasmid. BHK cells were simultaneously transfected with 30 μg of pTM1-Luc, overlaid at a 1:1 ratio onto BSR cells to allow fusion, and processed in parallel to those in Fig. 5B.
In the absence of HN coexpression, three mutants containing both FRSV cleavage sites (Fc_103-110, Fc_103-117, and Fc_103-130) produced fusion activity in the luciferase reporter gene assay in the presence of trypsin (Fig. 6B). As observed in the syncytium formation assay, Fc did not produce significant fusion in the absence of HN. FcΔ_103-117 also failed to produce HN-independent fusion, reflecting the small syncytia produced by this mutant in the absence of an attachment protein, in spite of the fact that it contains both FRSV cleavage sites.
Enhanced membrane fusion in the absence of HN by the double cleavage site mutants paralleled the increase in F0-to-F1 cleavage by trypsin, compared with single site mutants (Fig. 3B). However, no precise quantitative correlation between F0 processing and membrane fusion activity in the absence of HN could be found. For instance, while FcΔ_103-117 was highly susceptible to trypsin cleavage, this mutant was poorly active in both membrane fusion assays (Fig. 6). Furthermore, although Fc_103-130 was cleaved by trypsin more efficiently than Fc_103-110 or Fc_103-117 (Fig. 3B), it did not produce larger syncytia or induce greater luciferase activity in the absence of HN (Fig. 6).
The Fc_103-117 mutant produced the greatest fusion activity in the absence of the HN protein and was subjected to a dose-response fusion assay in the presence of trypsin and compared with Fc (Table 1). Increasing quantities of DNA were used to transfect BSR cells, and the corresponding increase in cell surface expression was determined by flow cytometry. An average fusion index was subsequently calculated from the fusion and expression data. While increasing expression of Fc did not enhance fusion, a dose response of fusion was seen for Fc_103-117, confirming the ability of this double cleavage site mutant to fuse cells in the absence of HN. The fusion index estimated for Fc_103-117 was 0.72, compared to 0.08 for the Fc mutant and 1.0 for wild-type F (FSeV coexpressed with HN in the presence of trypsin). However, higher expression levels of Fc_103-117 were required in the luciferase reporter gene assay, compared to the syncytium formation assay, in order to observe a level of HN-independent cell-cell fusion comparable to the wild type. This may reflect differences in the requirements of the two assays. For instance, the presence of F protein in both target and donor cells in the syncytium formation assay may facilitate cell-cell fusion with respect to the reporter gene assay, in which F protein is present only in the donor BSR cells. Furthermore, the reporter gene assay involves the interaction between two distinct populations of cells and may depend to a greater extent on the attachment ability of the mutated F proteins.
TABLE 1.
Dose effect of Fc103-117 and Fc on membrane fusion
| Plasmid | F DNA (ng)a | Fusion (% of wt)b | Cell surface expression (% of wt)c | Fusion index (fusion/cell surface expression) | Avg fusion indexd |
|---|---|---|---|---|---|
| FSeV + HN | 250 | 100.0 ± 10.8 | 100.0 ± 9.1 | 1.00 | 1.0 |
| Fc_103-117 | 250 | 45.7 ± 9.2 | 78.6 ± 7.7 | 0.58 | 0.72 ± 0.13 |
| 375 | 72.1 ± 8.0 | 85.7 ± 13.0 | 0.84 | ||
| 500 | 88.1 ± 7.0 | 119.5 ± 12.6 | 0.74 | ||
| Fc | 250 | 10.7 ± 9.9 | 112.0 ± 6.4 | 0.10 | 0.08 ± 0.04 |
| 375 | 14.3 ± 0.4 | 124.3 ± 10.9 | 0.11 | ||
| 500 | 6.0 ± 1.4 | 136.8 ± 2.2 | 0.04 |
The total amount of transfected DNA was maintained constant at 500 ng by using empty pTM1 plasmid (Fc_103-117 and Fc) or pTM1-HN (FSeV).
Wild-type (wt) fusion was determined in a luciferase reporter gene assay using 250 ng FSeV coexpressed with HN in the presence of trypsin. Fusion data represent the means of three independent experiments in the presence of trypsin.
Wild-type (wt) cell surface expression was determined by flow cytometry of BSR cells transfected with 250 ng FSeV, as previously described. Flow cytometry results represent the means of at least two independent experiments.
An average fusion index was calculated for each F protein by averaging the fusion index (fusion/cell surface expression) for each quantity of transfected DNA.
Coexpression of the attachment G protein fails to restore the reduced fusogenic capacity of RSV F cleavage site mutants.
It has previously been shown that cleavage at both sites I and II is a requirement for fusion of the FRSV protein, expressed in transfected cells in the absence of the attachment G protein. Mutations of FRSV cleavage site I abrogated syncytium formation in the absence of trypsin, whereas deletion of FRSV cleavage site II prevented syncytium formation, even in the presence of trypsin (12, 45). In light of our findings that insertion of FRSV cleavage site I in FSeV led to syncytium formation by FSeV in the absence of HN, we aimed to determine whether coexpression of RSV G protein with FRSV cleavage site mutants would restore cell-cell fusion.
A series of FRSV mutants (Fig. 7A), which have been previously described (12, 28, 29), were subcloned in the pTM1 plasmid. These mutations included changes in cleavage site I (RARR) to produce the F_R108/109N mutant (RANN) or deletion of four basic residues in cleavage site II (KKRKRR), resulting in the F_Δ131-134 mutant (12). Further deletions in F_R108/109N resulted in the RSV F_Δ108-120 and F_Δ108-132 mutants (28, 29). All mutant proteins were expressed at the cell surface at a level comparable to FRSV (Fig. 7B), with the exception of the cleavage site II mutant F_Δ131-134, which was expressed at a slightly reduced level (60% of wild-type FRSV). This is consistent with previous findings that mutation of FRSV cleavage site II increases the susceptibility of the protein to degradation (45).
FRSV cleavage mutants were subjected to the luciferase fusion assay described in Fig. 1C. F_R108/109N produced a low level of fusion, which was increased by the presence of trypsin but not by coexpression of the G protein. F_Δ131-134 failed to produce fusion above background levels, irrespective of the inclusion of trypsin or G protein. Further mutation of F_R108/109N by deletion of pep27 to produce F_Δ108-120 and F_Δ108-132 mutants led to an increase in cell-cell fusion activity, compared to F_R108/109N. However, the fusion activity of both F_Δ108-120 and F_Δ108-132 in the presence of G was approximately half that of the wild type (54% and 49% of wild-type F coexpressed with G in the presence of trypsin, respectively). Therefore, coexpression of the G protein failed to rescue the reduced fusogenic capacity of the RSV F cleavage site mutants.
DISCUSSION
In the current study, the differences between cell-cell fusion mediated by RSV and SeV fusion proteins were initially highlighted. In addition to confirming previously reported qualitative measurements of cell-cell fusion mediated by FRSV, it was observed that the G protein increased cell-cell fusion by approximately twofold in a quantitative assay (Fig. 1C). Therefore, while not a strict requirement, G enhances FRSV-mediated cell-cell fusion by a mechanism that remains essentially unknown. These results are in good agreement with those of Techaarpornkul et al. (38), who reported a similar level of fusion enhancement by the G protein in recombinant RSV that expressed either F protein or both F and G proteins.
In contrast, FSeV displayed an absolute requirement for both HN coexpression and trypsin for cell-cell fusion, confirming previously published results (37). FSeV protein contains a single basic cleavage site (R116), whereas FRSV requires cleavage of two furin-dependent cleavage sites (RARR109 and KKRKRR136) for cell-cell fusion (12, 45). A number of chimeras between RSV and SeV fusion proteins were constructed in order to determine whether the differential requirements for fusion exhibited by FRSV and FSeV are related to the distinct proteolytic processing pathways of the two proteins.
Insertion of the complete FRSV cleavage site II (KKRKRR) in FSeV augmented cell-cell fusion in an HN-dependent manner and decreased the dependence on trypsin for fusion. The presence of six basic residues at the cleavage site of Fc increased cleavage and cell-cell fusion over the minimal furin recognition sequence present in F_RKR. However, the Fc mutant was still dependent on the coexpression of HN for cell-cell fusion.
Insertion of the FRSV cleavage site I (RARR) in the backbone of the Fc mutant further augmented trypsin-independent cell-cell fusion in the presence of HN. Although no clear correlation was found between the extent of F0 cleavage in the absence of trypsin and the ability of the differing mutants to mediate cell-cell fusion, mutants with double cleavage sites were more susceptible to proteolytic processing when trypsin was added to the culture medium. The presence of two cleavage sites may increase the local level of F protein processing by proteases in the vicinity of the target membrane, increasing the number of F protein molecules available for engagement in membrane fusion. It has previously been shown that cleavage of FRSV at site I, in the absence of cleavage at site II, results in a partially cleaved protein, F_Δ1-109, seen as an intermediate band between F0 and F1 (12, 45). In the present study, we did not observe a partially cleaved intermediate for FSeV mutants containing both cleavage sites. This suggests that site I is either not used in the chimeric FSeV mutants or that if cleavage at site I does take place, it is immediately proceeded by cleavage at site II to form F1. Interestingly, other studies of FRSV cleavage in transfected (18) or infected (47) cells also failed to observe F_Δ1-109, suggesting that the ability to observe this partially cleaved FRSV intermediate is dependent on cell type and/or the expression system.
Insertion of both FRSV cleavage sites in FSeV also resulted in syncytium formation in the absence of the HN protein. All four mutants that possessed two FRSV cleavage sites formed syncytia without a requirement for HN coexpression. The intervening segment between the two cleavage sites slightly modulated the capacity of the FSeV protein to fuse membranes, and this modulation appeared greater in the absence of HN. In particular, while the Fc_103-117 mutant formed large syncytia without HN, FcΔ_103-117 (containing a shortened intervening segment) produced only small syncytia. FcΔ_103-117 is related to Fc_103-117 by a deletion N-terminal to the second cleavage site, which results in removal of an N-glycosylation motif that has previously been shown to be glycosylated in the Z strain of SeV F protein (35). There are three N-glycosylation sites present in pep27, and it has been estimated that at least two of the three sites are glycosylated (45). Mutation of one site (N126) resulted in increased FRSV cell-cell fusion, suggesting that differential glycosylation of pep27 may modulate fusion (46, 47).
While the exact mechanism by which introduction of the two FRSV cleavage sites in FSeV leads to HN-independent membrane fusion is not known, there are several possibilities which deserve consideration. The presence of two multibasic cleavage sites may influence F protein activation by altering the energy threshold required to trigger the conformational changes for fusion. Mutations in the parainfluenza virus 5 F protein that resulted in HN-independent syncytium formation are thought to destabilize the prefusion conformation of the F protein, lowering the activation energy barrier required to convert the high-energy prefusion form into the more thermostable postfusion form (14, 27, 31, 41).
The introduction of basic residues at the FSeV cleavage site may also facilitate attachment of the SeV F protein to the target cell. FRSV has previously been shown to bind to cells in an interaction that is largely dependent on glycosaminoglycans (GAGs) (10, 39). Studies using overlapping peptides of FRSV revealed that a heparin-binding peptide, corresponding to FRSV cleavage site II and part of the fusion peptide, was one of only two FRSV peptides capable of inhibiting both attachment and infection of Vero cells by the RSV A2 strain (7). Since binding of the peptide to cells was dependent on GAGs, it was hypothesized that this region of FRSV is involved in interactions with cell surface proteoglycans. An attractive hypothesis is that the insertion of FRSV cleavage site I directly mediates attachment to cells. Alternatively, the insertion of cleavage site I may alter the local structure of the FSeV proteolytic processing region, thereby facilitating the projection of cleavage site II (KKRKRR) from the surface of the F prefusion structure to bind to target cell GAGs. This hypothesis is supported by previous findings which suggest that the sequence surrounding the FSeV cleavage site is important for fusion (13).
It is currently unknown how activation of the F protein for fusion could be coordinated with attachment so that triggering of the F protein would only occur following binding of the F protein to the target membrane. In support of a role for cellular attachment in F protein activation, FSeV has previously been shown to mediate fusion of virosomes with red blood cells in the absence of the HN protein, via an interaction with the asialoglycoprotein receptor (1). In addition, virus-like particles that expressed SeV protein, but that lacked HN, were able to infect cells via the asialoglycoprotein pathway (23). The ability of FSeV to bind to cells may reduce the requirement of an attachment protein for fusion, as has been observed for FRSV (38, 39). Correct timing of F protein activation is crucial to ensure activation for fusion “at the right time and in the right place,” (19). As shown by Western blotting of transfected cell extracts, intracellular cleavage of F0 to F1 in the FSeV double cleavage site mutants was incomplete. Previous studies have also confirmed incomplete cleavage of FRSV in both transfected and infected cell extracts (12, 45). It is possible that partial cleavage of some, but not all, of the F protein monomers in the trimeric F protein prevents conformational changes to the postfusion form before the F protein reaches the cell surface. The presence of uncleaved FRSV monomers at the cell surface would permit attachment of the proposed heparin binding region at cleavage site II to cell surface glycosaminoglycans. Following attachment, cleavage of all three monomers and removal of the intervening segment (pep27) may be completed by trypsin added to the culture medium, or by furin present at the cell surface as it recycles between the plasma membrane and Golgi network (for a review see reference 40). Completion of cleavage could, in turn, facilitate activation of the F protein for membrane fusion.
We have shown that the RSV F proteolytic processing region may be involved in attachment protein-independent fusion, at least in the context of chimeric SeV F. However, the failure of the RSV G protein to rescue membrane fusion when coexpressed with site I mutants RSV F_R108/109N and F_Δ108-120 (Fig. 7) contrasts with the high fusion activity displayed by SeV Fc in the presence of HN (Fig. 5), given that all three F proteins share the same polybasic sequence at site II. It therefore appears that the cooperative effect of HN on SeV F-mediated fusion cannot be reproduced by the G protein when coexpressed with RSV F proteins that contain only a single furin recognition sequence (site II).
Interestingly, despite displaying drastically reduced syncytium formation, recombinant bovine RSV expressing F protein with an intact cleavage site II, but lacking both site I and the intervening segment (Δ106-130), replicated with kinetics similar to that of the parental virus (47). Recombinant bovine RSV expressing F_R108/109N also replicated in cells, albeit with a growth retardation. Thus, while mutations of FRSV cleavage site I and the intervening peptide affect cell-cell fusion, they still permit virus replication in cell culture. It is possible that the structural requirements for virus-cell and cell-cell fusion are different, as recently suggested for parainfluenza virus 5 (6). Similarly, while certain mutations that destabilize the envelope (Env) glycoprotein of a murine leukemia virus enhanced syncytium formation in transfected cells, the same mutations were detrimental when incorporated into virus particles (22). It is possible that unstable Env proteins at the cell membrane that are continuously being replaced by new molecules facilitate cell-cell fusion. In contrast, such unstable proteins may be prematurely activated in the virus particle before virus-cell contact is able to take place, leading to virus inactivation. It remains to be tested whether the chimeric SeV F proteins reported here could replace SeV F in infectious virus particles.
Further studies are currently under way to investigate the mechanism by which insertion of the FRSV proteolytic processing region in FSeV is able to confer decreased dependence on HN for cell fusion. Although the RSV F protein possesses unique properties among the Paramyxoviridae family of viruses, studies of such features may shed light on the mechanism of F protein activation, particularly in the absence of an attachment protein. HMPV F protein has also been shown to mediate cell-cell fusion (34) and virus-cell fusion (3) in the absence of the attachment G protein. Interestingly, syncytium formation by the HMPV F protein expressed alone in transfected cells was dependent on low pH (34). It is therefore possible that members of the Pneumovirinae subfamily have evolved distinct mechanisms of F protein activation compared to those of the Paramyxovirinae subfamily of paramyxoviruses.
Acknowledgments
This work was supported by grant SAF2006-07805, awarded to J.A.M., from the Ministerio de Educación y Ciencia. This laboratory participates in the “Virus-host” framework funded by the Comunidad de Madrid and in CIBER de Enfermedades Respiratorias, an initiative of ISCIII.
We thank L. Roux (Geneva, Switzerland) for the pGEM-F and pGEM-HN plasmids, K.-K. Conzelmann (Munich, Germany) for the BSR-T7/5 cells, and S. Wharton and J. J. Skehel (London, United Kingdom) for the GB5 monoclonal antibody. We are also grateful for advice and comments from Teresa Corral, Paulino Gómez-Puertas, and Concepción Palomo.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Bagai, S., A. Puri, R. Blumenthal, and D. P. Sarkar. 1993. Hemagglutinin-neuraminidase enhances F protein-mediated membrane fusion of reconstituted Sendai virus envelope with cells. J. Virol. 673312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3309-319. [DOI] [PubMed] [Google Scholar]
- 3.Biacchesi, S., M. H. Skiadopoulos, L. Yang, E. W. Lamirande, K. C. Tran, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2004. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J. Virol. 7812877-12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K.-K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, P. L., and J. E. Crowe. 2007. Respiratory syncytial virus and metapneumovirus, p. 1601-1646. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 6.Connolly, S. A., and R. A. Lamb. 2007. Paramyxovirus fusion: real-time measurement of parainfluenza virus 5 virus-cell fusion. Virology 355203-212. [DOI] [PubMed] [Google Scholar]
- 7.Crim, R. L., S. A. Audet, S. A. Feldman, H. S. Mostowski, and J. A. Beeler. 2007. Identification of linear heparin-binding peptides derived from human respiratory syncytial virus fusion glycoprotein that inhibit infectivity. J. Virol. 81261-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, R., Z. Wang, P. J. Mahon, M. Marinello, A. Mirza, and R. M. Iorio. 1999. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 25343-54. [DOI] [PubMed] [Google Scholar]
- 9.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. USA 866126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman, S. A., S. Audet, and J. A. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulphate. J. Virol. 746442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Barreno, B., C. Palomo, C. Peñas, T. Delgado, P. Perez-Breña, and J. A. Melero. 1989. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J. Virol. 63925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González-Reyes, L., M. B. Ruiz-Argüello, B. García-Barreno, L. Calder, J. A. López, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 989859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heminway, B. R., Y. Yang, Y. Tanaka, M. Panin, Y. T. Huang, and M. S. Galinski. 1995. Role of basic residues in the proteolytic activation of Sendai virus fusion glycoprotein. Virus Res. 3615-35. [DOI] [PubMed] [Google Scholar]
- 14.Ito, M., M. Nishio, H. Komada, Y. Ito, and M. Tsurudome. 2000. An amino acid in the heptad repeat domain 1 is important for the hemagglutinin-neuraminidase-independent fusing activity of simian virus 5 fusion protein. J. Gen. Virol. 81719-727. [DOI] [PubMed] [Google Scholar]
- 15.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 9413961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kido, H., M. Murakami, K. Oba, Y. Chen, and T. Towatari. 1999. Cellular proteinases trigger the infectivity of the influenza A and Sendai virus. Mol. Cell 9235-244. [PubMed] [Google Scholar]
- 17.Klenk, H.-D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 239-43. [DOI] [PubMed] [Google Scholar]
- 18.König, P., K. Giesow, K. Schuldt, U. J. Buchholz, and G. M. Keil. 2004. A novel protein expression strategy using recombinant bovine respiratory syncytial virus (BRSV): modifications of the peptide sequence between the two furin cleavage sites of the BRSV fusion protein yield secreted proteins, but affect processing and function of the BRSV fusion protein. J. Gen. Virol. 851815-1824. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 1971-11. [DOI] [PubMed] [Google Scholar]
- 20.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 34430-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb, R. A., and T. S. Jardetzky. 2007. Structural basis of virus invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette, D., M. Maurice, C. Roche, S. J. Russell, M. Sitbon, and F.-L. Cosset. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J. Virol. 729955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leyrer, S., M. Bitzer, U. Lauer, J. Kramer, W. J. Neubert, and R. Sedlmeier. 1998. Sendai virus-like particles devoid of haemagglutinin-neuraminidase protein infect cells via the human asialoglycoprotein receptor. J. Gen. Virol. 79683-687. [DOI] [PubMed] [Google Scholar]
- 24.McGuinnes, L. W., and T. G. Morrison. 2006. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J. Virol. 802894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melero, J. A. 2007. Molecular biology of human respiratory syncytial virus, p. 1-42. In P. A. Cane (ed.), Respiratory syncytial virus. Perspectives in medical virology, vol. 14. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 26.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 27017-30. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Argüello, M. B., L. González-Reyes, L. J. Calder, C. Palomo, D. Martin, M. J. Saiz, B. García-Barreno, J. J. Skehel, and J. A. Melero. 2002. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology 298317-326. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Argüello, M. B., D. Martín, S. A. Wharton, L. J. Calder, S. R. Martin, O. Cano, M. Calero, B. García-Barreno, J. J. Skehel, and J. A. Melero. 2004. Thermostability of the human respiratory syncytial virus fusion protein before and after activation: implications for the membrane-fusion mechanism. J. Gen. Virol. 853677-3687. [DOI] [PubMed] [Google Scholar]
- 30.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 204024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell, C. J., K. L. Kantor, T. S. Jardetzky, and R. A. Lamb. 2003. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 163363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57475-490. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt, U., J. Beyer, U. Polster, L. J. Gershwin, and U. J. Buchholz. 2002. Mucosal immunization with live recombinant bovine respiratory syncytial virus (BRSV) and recombinant BRSV lacking the envelope glycoprotein G protects against challenge with wild-type BRSV. J. Virol. 7612355-12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schowalter, R. M., S. E. Smith, and R. E. Dutch. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 8010931-10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segawa, H., T. Yamashita, M. Kawakita, and H. Taira. 2000. Functional analysis of the individual oligosaccharide chains of Sendai virus fusion protein. J. Biochem. 12865-72. [DOI] [PubMed] [Google Scholar]
- 36.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 716287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 706112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 756825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Techaarpornkul, S., P. L. Collins, and M. E. Peeples. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294296-304. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsurudome, M., M. Ito, M. Nishio, M. Kawano, H. Komada, and Y. Ito. 2001. Hemagglutinin-neuraminidase-independent fusion activity of Simian Virus 5 fusion (F) protein: difference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J. Virol. 758999-9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 1029288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 43938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 9714172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. J. Biol. Chem. 27631642-31650. [DOI] [PubMed] [Google Scholar]
- 46.Zimmer, G., I. Trotz, and G. Herrler. 2001. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J. Virol. 754744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmer, G., K. K. Conzelmann, and G. Herrler. 2002. Cleavage at the furin consensus sequence RAR/KR109 and presence of the intervening peptide of the respiratory syncytial virus fusion protein are dispensable for virus replication in cell culture. J. Virol. 769218-9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmer, G., M. Rohn, G. P. McGregor, M. Schemann, K. K. Conzelman, and G. Herrler. 2003. Virokinin, a bioactive peptide of the tachykinin family, is released from the fusion protein of bovine respiratory syncytial virus. J. Biol. Chem. 27846854-46861. [DOI] [PubMed] [Google Scholar]