Abstract
In the φX174 procapsid crystal structure, 240 external scaffolding protein D subunits form 60 pairs of asymmetric dimers, D1D2 and D3D4, in a non-quasi-equivalent structure. To achieve this arrangement, α-helix 3 assumes two different conformations: (i) kinked 30° at glycine residue 61 in subunits D1 and D3 and (ii) straight in subunits D2 and D4. Substitutions for G61 may inhibit viral assembly by preventing the protein from achieving its fully kinked conformation while still allowing it to interact with other scaffolding and structural proteins. Mutations designed to inhibit conformational switching in α-helix 3 were introduced into a cloned gene, and expression was demonstrated to inhibit wild-type morphogenesis. The severity of inhibition appears to be related to the size of the substituted amino acid. For infections in which only the mutant protein is present, morphogenesis does not proceed past the first step that requires the wild-type external scaffolding protein. Thus, mutant subunits alone appear to have little or no morphogenetic function. In contrast, assembly in the presence of wild-type and mutant subunits is blocked prematurely, before D protein is required in a wild-type infection, or channeled into an off-pathway reaction. These data suggest that the wild-type protein transports the inhibitory protein to the pathway. Viruses resistant to the lethal dominant proteins were isolated, and mutations were mapped to the coat and internal scaffolding proteins. The affected amino acids cluster in the atomic structure and may act to exclude mutant subunits from occupying particular positions atop pentamers of the viral coat protein.
Proper virion assembly requires numerous macromolecular interactions proceeding along an ordered morphogenetic pathway. While structural proteins are incorporated into the final product, morphogenesis is equally dependent upon scaffolding proteins, which are not included in the mature virion. During morphogenesis, these proteins mediate conformational switches to ensure the efficiency and fidelity of assembly (13). Although many large DNA viruses rely on a single internal scaffolding protein, the small microviruses (canonical species φX174, G4, and α3) and the satellite P4-like viruses require both internal and external scaffolding proteins (10, 14, 21). Although the internal scaffolding proteins in these dual systems are absolutely essential, several lines of evidence suggest that the external scaffolding proteins may be more critical. Successive targeted genetic selections have led to the isolation of a φX174 multiple mutant that can assemble without its internal scaffolding protein in vivo (5). Moreover, P4 procapsid-like particles can be assembled in vitro using only the capsid and external scaffolding proteins (26).
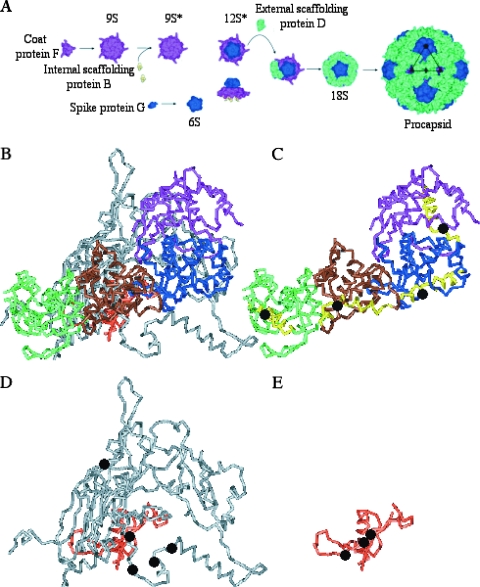
The morphogenetic roles of the φX174 internal and external scaffolding proteins are illustrated in Fig. 1A. The first identifiable assembly intermediates are pentamers of the viral coat F and major spike G proteins, the 9S and 6S particles, respectively, which can form in the absence of both scaffolding proteins (23). Five internal scaffolding B proteins bind to the underside of the 9S particle, yielding the 9S* intermediate. This interaction induces a conformational change that allows 9S*-6S particle associations, forming the 12S* intermediate. This particle also includes the DNA pilot protein H (10, 16). Twenty external scaffolding D proteins, most likely in the form of five tetramers or ten asymmetric dimers (18), interact with the 12S* particle to form the short-lived 18S intermediate (4). D-D contacts mediate the construction of the procapsid (108S), an immature virus particle, presumably by allowing 18S pentamers to interact across twofold axes of symmetry (6, 8, 18).
FIG. 1.
(A) The φX174 morphogenetic pathway. (B) The four external scaffolding subunits (D1 [purple], D2 [blue], D3 [brown], and D4 [green]), coat protein (gray), and internal scaffolding protein (peach) in the asymmetric unit. (C) The four external scaffolding proteins. The position of glycine reside 61 within α-helix 3, which is highlighted in yellow, is indicated by a black dot. (D) Location of the resistance mutations (black dots) in the coat protein. Amino acid F426 is not depicted, since the very C terminus is unordered in the atomic structure. (E) Location of the resistance mutations (black dots) in the internal scaffolding protein. The structures in panels B, C, D, and E are rendered at the same scale and orientation.
In the atomic structure of the viral procapsid (6, 8), there are four structurally distinct external scaffolding proteins (D1 to D4), arranged as dimers of dimers (D1D2, D3D4), per viral coat protein (Fig. 1B and C). To achieve this unique arrangement, one monomer in each asymmetric dimer, D1 and D3, must be bent 30°; this occurs at glycine residue 61 in α-helix 3 (Fig. 1C). This kink is also present in the crystal structure of the assembly naive dimer DADB (18). A mutation at this site may allow the protein to fold into only one of these two conformations. Although the mutant protein would retain the ability to interact with other D and structural proteins, the product of these interactions may inhibit lattice formation and subsequent virus assembly. The putative dominant lethal phenotypes of some glycine 61 substitutions have been reported (3). However, the molecular mechanism of inhibition could not be characterized due to difficulties associated with propagating the mutant strains.
To determine the mechanism of inhibition and circumvent the problems associated with propagating dominant lethal viral mutants, genetically engineered D genes were constructed, and the proteins were expressed in wild-type φX174-infected cells. This alternate approach also allowed mutants resistant to the expression of the inhibitory proteins to be isolated, elucidating possible resistance mechanisms that may be anticipated for antiviral therapies that target virus-specific assembly processes. The results of this analysis suggest that the presence of the wild-type external scaffolding protein is required for the mutant inhibitory D protein to enter the morphogenetic pathway. This may lead to premature D protein interactions with structural proteins or off-pathway reactions involving later intermediates. The location of the resistance mutations within the procapsid crystal structure suggest a mechanism in which inhibitory D protein subunits are excluded from occupying particular positions atop 12S* pentamers.
MATERIALS AND METHODS
Phage plating, media, buffers, and stock preparation.
The reagents, media, buffers, and protocols have been previously described (11).
Bacterial strains, phage strains, and plasmids.
Escherichia coli C strains C122 (sup°) and BAF30 (recA) have been previously described (11, 12). The host slyD mutation confers resistance to E-protein-mediated lysis (20). The φX174 nullD mutant and the complementing plasmid pφXDJ have been previously reported (2).
Generation of cloned genes with substitution for the glycine 61 codon.
The 5′ ends of the genes, which contained the mutated codons, were amplified by PCR, using a cloned wild-type D gene as the template. The downstream primer concurrently mutated the G61 codon, created amber mutations in gene E and introduced a SacII restriction site. The amber E and SacII substitutions do not alter the primary structure of the D protein. The upstream primer annealed to the multiple cloning site of the plasmid. The 5′ end of the gene was then cloned into the pSE420 expression vector, between the BglII and SacII sites. The 3′ end of the gene was amplified by using primers that introduced the SacII site and a downstream primer that annealed to the multiple cloning site. It was then cloned behind the 5′ end of the gene. Plasmid names reflect the expressed mutant protein. For example, pG61D refers to a φX174 D protein with a glycine→aspartic acid substitution at amino acid 61. Likewise, G61D refers to the mutant D protein expressed from the plasmid.
Isolation of resistant mutants.
Wild-type φX174 was plated on cells expressing the mutated protein, BAF30/pG61V, BAF30/pG61K, and BAF30/pG61D. Plates were incubated at 37°C until plaques appeared. Plaques were then purified, and subsequent plating assays confirmed the resistant phenotype bred true.
Detection of virion and intermediate particles from infected cells.
A total of 100 ml of cells was infected with wild-type φX174 at a multiplicity of infection of 4.0, followed by incubation for 3 h at 37°C. Afterward, the cells were concentrated and resuspended in 2.0 ml of BE buffer (11) and lysed with T4 lysozyme as previously described (15). Material between 1.3 and 1.4 gm/cm3 was isolated from CsCl gradients (2). After dialysis to remove the CsCl, extracts were concentrated to 200 μl. For the detection of particles with S values larger than 70S, samples were loaded atop 5 to 30% sucrose gradients and spun at 45,000 × g for 1.0 h in a Beckman SW50.1 rotor. After fractionation into approximately 50 100-μl fractions, virus-derived protein complexes were detected by spectrophotometry (λ = 280 nm). The positions of infectious virions (114S) were further determined by plating assays. To detect particles with S values of less than 50S, no CsCl gradients were performed. Extracts were analyzed in either 5 to 30% or 5 to 20% sucrose gradients spun at 108,000 × g for 16 h. After fractionation, material was detected by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE).
RESULTS
Biological activity of the φX174 mutant external scaffolding proteins.
Proteins with substitutions for glycine 61 were expressed from cloned genes under lac induction as described in Materials and Methods. To determine whether the mutant proteins displayed activity, they were assayed for the ability to complement a nullD mutant and to inhibit wild-type φX174 (Table 1). With the exception of the protein with a substitution of alanine (referred to as G61A), the mutant proteins were unable to complement the nullD strain. Complementation by the G61A protein was only observed at 42°C. Although plating efficiency was 0.6, plaque size was extremely small compared to that obtained by complementing with the wild-type protein.
TABLE 1.
Plating efficiency of wild-type φX174
| Strain | Plating efficiencya of protein with substitution for G61 expressed in vivo
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gb | V | D | K | P | S | A | T | |
| Wild type | 1.0 (100) | 10−2 (0.1) | <10−4 (0.3) | <10−4 (0.1) | <10−4 (0.2) | 1.0 (14) | 1.0 (5.0) | 1.0 (6.0) |
| nullD mutant | 1.0 | <10−4 | <10−4 | <10−4 | <10−4 | <10−4 | <10−4 | <10−4 |
The plating efficiency for wild-type φX174 is the titer on a cell line expressing a mutant external scaffolding protein/titer on a cell line expressing the wild-type external scaffolding protein. Burst sizes are given in parentheses as the percentage of the control infection, which was 128 phage/cell.
That is, the wild-type protein.
The expression of the proteins with a valine, aspartic acid, lysine, or proline substitution (G61V, G61D, G61K, or G61P, respectively) inhibited wild-type plaque formation (Table 1). In liquid culture, burst sizes were reduced 2 orders of magnitude compared to control infections in cells expressing the wild-type protein (Table 1). Although the expression of the mutant G61S, G61A, and G61T proteins did not reduce plating efficiency, the plaque size was reduced at 37°C. Accordingly, the expression of these proteins resulted in lower bursts (Table 1). These dominant lethal and codominant phenotypes indicate that the mutant proteins retain a near-wild-type fold, permitting them to interact with wild-type D protein subunits and/or structural proteins.
Biochemical analysis of particles produced by the external scaffolding protein with G61 substitutions.
In an effort to elucidate the molecular basis of inhibition, high-molecular-weight intermediates synthesized in wild-type infected cells expressing the inhibitory G61D external scaffolding protein were analyzed. The analysis was conducted under two induction conditions. For one condition (high induction), the concentration of the IPTG (isopropyl-β-d-thiogalactopyranoside) inducer yields plating efficiencies of <10−4, and the wild-type/mutant D-protein ratios are approximately 1:1 as previously determined (24). For the other condition (low induction), the inducer concentration does not affect plating efficiencies, but the plaque size is significantly reduced. Under these conditions, the level of the mutant protein was below detection by SDS-PAGE analysis. Thus, it was not possible to calculate a wild-type/mutant D-protein ratio. After incubation, infected cells were concentrated by centrifugation to remove unabsorbed virions. After chemically induced lysis, soluble proteins and particles were purified in CsCl gradients and then analyzed by 5 to 30% sucrose gradient sedimentation. After rate-zonal sedimentation, samples were separated into approximately 50 0.1-ml fractions. Particles, regardless of infectivity, were detected by UV spectroscopy. The presence of infectious virions was determined by plating assays.
Low levels of the G61D protein result in the production of virions and defective procapsids.
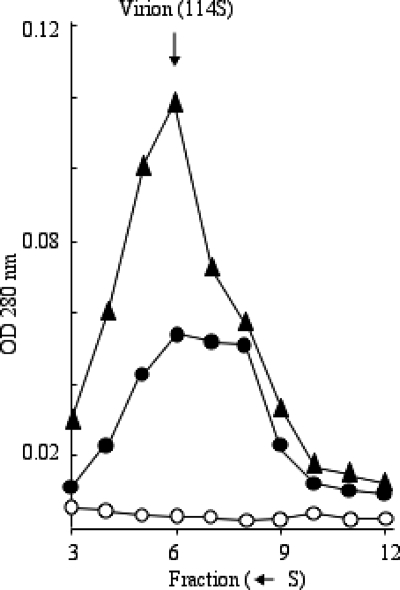
Although low induction conditions resulted in the production of infectious virions, approximately one-half of the large particles isolated from cells expressing the G61D protein sedimented more slowly than virions (Fig. 2), which was determined by direct plating assays. This was not observed in the control experiment. In both gradients, the highest concentration of infectious particles and the optical density at 280 nm (OD280) were found in fraction 6. To calculate the specific infectivity, the PFU of each fraction was divided by its OD reading (Table 2). To compensate for differences in particle yields resulting from sample preparation and possible discrepancies in the OD readings, each gradient was normalized to its most infectious fraction, fraction 6. This value was set to 1.0. In the control infection, the specific infectivities of each fraction did not differ significantly, ranging from 0.41 to 1.0. This indicates that the vast majority of particles in every fraction is the virions. However, the material present in fractions 7 and 8, which was only abundant in extracts from cells expressing the G61D protein, have a specific infectivity 2 orders of magnitude lower than fraction 6 of either infection.
FIG. 2.
Large particles synthesized in wild-type infected cells. Symbols: ▴, no inhibitory protein expression; •, low induction of the inhibitory protein; ○, high induction of the inhibitory protein. See the text for the definition of low- and high-induction conditions.
TABLE 2.
Specific infectivity of large particles synthesized in the presence of the G61D inhibitory protein
| Infection | Specific infectivitya of fraction from the bottom of the gradient (S←):
|
||||
|---|---|---|---|---|---|
| Fraction 4 | Fraction 5 | Fraction 6 | Fraction 7 | Fraction 8 | |
| Wild-type expression | 0.68 | 0.41 | 1.0 | 0.66 | 0.71 |
| G61D expression | 0.05 | 0.25 | 1.0 | 0.07 | 0.01 |
The specific infectivity is defined as the number of infectious particles/OD280. Values of the most infectious fractions were set to 1.0.
These fractions were also analyzed by SDS-PAGE. Both internal and external scaffolding proteins were detected (data not shown), suggesting that low levels of the mutant protein can be incorporated into procapsids, which cannot be filled.
High levels of the G61D protein inhibit large particle formation.
The yield of infectious particles (114S) in cells expressing the G61D protein under high induction was very low compared to the control infection (Fig. 2). Moreover, empty and degraded procapsids, which sediment at 108S and 70S, respectively, were not detected. Similar results were obtained with wild-type-infected cells expressing the G61V and G61P external scaffolding proteins (data not shown). These data indicate that these mutant external scaffolding proteins inhibit wild-type φX174 assembly early in the pathway, before procapsid morphogenesis.
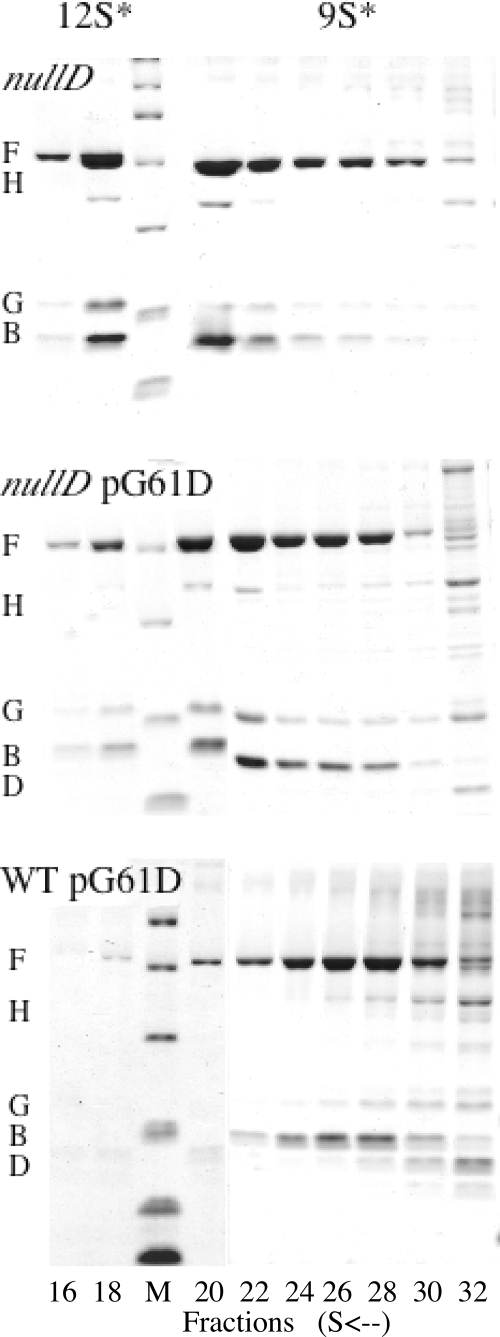
To determine the exact step in early assembly inhibited by the mutant scaffolding protein when present at approximately the same concentration as the wild-type protein, high induction lysates of wild-type-infected cells were examined for low-molecular-weight intermediates (Fig. 3). After rate zonal sedimentation, the fractions were analyzed by SDS-PAGE. Analysis of a nullD mutant infection demonstrated which assembly intermediates accumulate in the complete absence of D protein. As can be seen in Fig. 3, this results in the accumulation of 12S* particles, which contain the coat F, the internal scaffolding B, the major spike G, and the DNA pilot H proteins. The 12S* particle is the last assembly intermediately prior to the D protein entering the morphogenetic pathway (Fig. 1A). A nullD mutant infection in cells expressing the lethal G61D external scaffolding protein produced a similar result (Fig. 3), suggesting that the mutant protein alone does not interfere with early assembly. Due to the properties of rate zonal sedimentation, the small amounts of G protein observed in the 9S* fractions result from the presence of G protein in the surrounding 12S* and 6S peaks. The entire 6S regions of the gradients are not depicted in the figure. In cells infected with wild-type φX174, both the wild-type D protein and the lethal dominant G61D protein would be expressed. This results in the accumulation of 9S* particles, containing only the coat and internal scaffolding proteins (Fig. 3). Thus, the wild-type D protein is required for the mutant external scaffolding protein to enter the morphogenetic pathway. Identical results were obtained for wild-type φX174-infected cells expressing the G61V and G61P external scaffolding proteins.
FIG. 3.
Analysis of early assembly intermediates synthesized in wild-type-infected cells. For these experiments, each gradient was prepared as described in Materials and Methods and separated into approximately 50 80-μl fractions. Every other fraction was analyzed by SDS-15% PAGE. In the figure, only the relevant fractions are depicted, i.e., fractions 16 to 30 from the bottom of the tube. The 12S* particle is located in fractions 18 to 20, and the 9S* particle is seen in fractions 26 to 28. The third lane from the left contains a molecular weight marker.
The accumulation of the 9S* particle may indicate that the mutant D protein, in concert with the wild-type protein, is removing assembly intermediates from the pathway. To investigate the possibility of aggregation, supernatant and pellet fractions from wild-type-infected cells expressing either a wild-type D or mutant G61D gene were analyzed by SDS-PAGE. No significant differences, vis-à-vis the identity of the expressed gene, were observed in either fraction. Moreover, 5 to 20% sucrose gradients were run to detect the possible comigration of major spike protein G pentamers (6S) and D protein. Again, no significant differences were observed.
Isolation of resistant mutants.
To select for resistant mutants, wild-type φX174 was plated on cells expressing the G61V, G61K, and G61D proteins. Selections were initially conducted with a wild-type φX174 stock. The mutants recovered in that selection are listed in Table 3. To investigate whether additional resistance mutations could be recovered, 10 single plaques were isolated from the initial wild-type stock. Each plaque was plated for a resistant mutant on cells expressing the G61V and G61D proteins. Four additional mutations—mh3dr(F)R214H, mh3dr(F)R233H, mh3dr(F)F408L, and mh3dr(B)A87S—were recovered. The other isolates were identical to previously recovered mutants. The entire genomes of two resistant mutants and the parental strain were sequenced, and only one substitution was found in both cases, indicating that the identified changes are most likely necessary and sufficient to confer the resistance phenotype. The identified nucleotide changes reside in either the major coat or the internal scaffolding genes (Table 3). The affected amino acids in the internal scaffolding protein are located in the primary region of coat internal scaffolding protein contacts. This region, as well as the substitutions at amino acids 212, 214, 227, 233, and 408, would most likely be located under and surrounding the D3 subunit (see Fig. 1E and F and the Discussion).
TABLE 3.
Plating efficiency of wild-type and mh3dr mutant strains of φX174
| Straina | Plating efficiencyb of protein with a substitution for G61 expressed in vivo
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gc | V | D | K | P | S | A | T | |
| Wild type | 1.0 | 10−2 | <10−4 | <10−4 | <10−4 | 1.0 | 1.0 | 1.0 |
| mh3dr(F)S426L mutant | 1.0 | 1.0 | 10−4 | 0.3 | 0.7 | 0.4 | 0.4 | 0.6 |
| mh3dr(F)M212I mutant | 1.0 | 0.8 | 10−4 | 0.3 | 0.5 | 0.5 | 0.5 | 0.5 |
| mh3dr(F)M212V mutant | 1.0 | 0.7 | 0.3 | <10−4 | 0.3 | 0.3 | 0.3 | 0.3 |
| mh3dr(F)S227P mutant | 1.0 | 1.0 | 10−3 | 0.5 | 0.8 | 0.9 | 0.7 | 1.0 |
| mh3dr(B)P102L mutant | 1.0 | 1.0 | 1.0 | <10−5 | 10−2 | 10−2 | 10−2 | 10−2 |
| mh3dr(B)A87S mutant | 1.0 | 10−2 | 1.0 | <10−4 | 0.8 | 0.7 | 0.2 | 0.5 |
Mutant names reflect conferred amino acid substitutions, for example, mh3dr(F)S426L represents “mutant α-helix 3 D protein resistant in coat protein F, serine 426→leucine.” In a second selection, four additional substitutions were identified: mh3dr(F)R214H(isolated on cells expressing the G61V protein), mh3dr(F)R233H (isolated on the cells expressing the G61V protein), mh3dr(B)R106H(isolated on cells expressing the G61P protein), and mh3dr(F)S408L (isolated on cells expressing the G61K protein).
The plating efficiency for wild-type φX174 is the titer on a cell line expressing a mutant external scaffolding protein/titer on cell line expressing wild-type external scaffolding protein. Values for a cell line on which a mutant was isolated are indicated in boldface.
That is, the wild-type protein.
The results of plating assays (Table 3) indicate that the resistance mutations do not exhibit strict allele specificity. However, no mutation confers resistance to both G61D and G61K proteins. Each mutation is active against only one of these proteins, indicating that electrostatic interactions are influencing resistance. Thus, despite the internal location of the internal scaffolding protein and the buried nature of some of the coat protein substitutions, the outer surface of the capsid or 9S* particle is most likely altered. These alterations do not appear to be a function of the charge of the substituted amino acid side chain. For example, there are two neutral substitutions for M212 that confer different resistance phenotypes vis-à-vis the G61D and G61K proteins. In regard to the structure of α-helix 3 of protein D, almost all of the resistance mutations are active against the G61P protein, which would affect this helix in a different manner than the other substitutions. Although the other substitutions would theoretically inhibit the helix from forming a fully kinked conformation, the proline substitution would most likely break the helix or lock it in a bent position. Together, these observations suggest a general resistance mechanism.
DISCUSSION
In the φX174 procapsid atomic structure α-helix 3 of the external scaffolding protein D is found in one of two conformations: straight or kinked 30° at glycine residue 61. The kinked conformations occur in subunits D1 and D3 (Fig. 1C). This kink appears to be necessary for proper D-D, D-coat, and D-spike protein interactions within the non-quasi-equivalent external scaffolding protein lattice (6, 8). The two α-helix 3 conformations are also present in the structure of assembly naive D-protein dimer (18), suggesting that this conformational variance is critical at the very onset of assembly. Moreover, there appears to be stringent evolutionary pressure to retain this glycine residue. It is conserved in all known Microviridae external scaffolding proteins, of which there are more than 40, and always overlaps with the gene E start codon (19).
To determine whether amino acid substitutions could be tolerated at this site, mutant genes with G61 substitutions were cloned and assayed for the ability to inhibit wild-type progeny production. The results from plating and burst experiments indicate that all mutant proteins inhibit morphogenesis, with severity being a function of the substituted amino acid size. Larger side chains (V, D, and K) substituted at G61 result in very low burst size and efficiency-of-plating values of <10−4. Glycine 61 was also substituted with the rotationally restrictive amino acid proline, which resulted in similar plating efficiencies. In contrast, smaller side chains (S, A, and T) result in only a moderate decreases in phage production and plaque size.
The assembly intermediates made in wild-type-infected cells expressing the strongly inhibitory G61D, G61V, and G61P external scaffolding proteins were analyzed. In these experiments, both the mutant and the wild-type D proteins were present. Under these conditions, 9S* particles, containing the coat F and internal scaffolding B proteins, accumulate. However, in the complete absence of any D protein, assembly proceeds through the formation of the 12S* particle, which is a later assembly intermediate. 12S* particles also accumulate in nullD mutant-infected cells expressing inhibitory proteins. Thus, wild-type subunits must be present for the mutant subunits to interfere with morphogenesis, suggesting that heterodimers are the inhibitory species. These heterodimers may directly prevent 12S* particle formation or channel 12S* particles into an off-pathway reaction, the product of which dissociates into early assembly intermediates in vivo or during sample preparation.
Mutants resistant to the G61V, G61D, and G61K external scaffolding proteins were isolated via direct genetic selections and occurred at a frequency of approximately 10−6. The amino acid substitutions are located in the coat and internal scaffolding proteins. The affected amino acids in the internal scaffolding protein are located in the primary region of coat-internal scaffolding protein contacts, which resides under the D3 subunit (Fig. 1A and E). The depiction of the coat and internal scaffolding proteins in Fig. 1 are from the atomic structure of the closed procapsid, which most likely represents an off-pathway product. During crystallization, the coat protein underwent maturation to its virion conformation. In the cryo-electron microscopy reconstruction, which most likely represents the native species, there are large pores at the threefold axes of symmetry (1). To form these 30-Å pores, the two α-helices that transverse the twofold axis of symmetry would be shifted upward, moving amino acids 212, 214, and 227 toward the center of the asymmetric unit.
Several genetic analyses designed to investigate different φX174 assembly phenomena have been conducted (2-5, 9, 15, 24, 25). In these studies second-site mutations were most frequently found in the coat protein, often clustering to discreet regions. The vast majority of these substitutions have been located in the protein's insertion loops, as opposed to the β-barrel core, suggesting that more flexible regions mediate dynamic assembly processes. While most analyses have identified unique residues, the resistance mutations at amino acids 212, 214, and 227 were previously isolated for their ability to utilize chimeric external scaffolding proteins with foreign first α-helices (24, 25). The other mutants in that class mapped throughout the large threefold related coat protein α-helix and were not isolated as resistance mutations. Since both phenomena involve external scaffolding-coat protein dynamics, it is not surprising that some substitutions have pleiotropic effects. The other resistance mutations isolated here are novel, especially those in the internal scaffolding protein. Previous analyses have not yielded second-site mutations in this protein. Although the structure of the satellite virus P4 external scaffolding Sid protein forms a different lattice than the φX174 D protein, there are some structural and genetic commonalities. Sid subunits, such as the φX174 D protein, appear to use dimers to construct the external lattice (8). Vis-à-vis the P2 helper virus, Sid protein can be regarded as P2 assembly inhibitor. P2 mutations resistant to the actions of the Sid protein define a specific region of the P2 coat protein that can exclude the external scaffolding protein (7, 17, 22), much like the φX174 mutations described here may exclude inhibitory D proteins from the D3 positions.
The resistance mutations isolated here do not exhibit strict allele specificity and are active against the G61P protein, which would affect the structure of α-helix 3 in a different manner than the other substitutions. Although the other substitutions would theoretically inhibit the helix from forming the fully kinked conformation, the proline substitution would lock the helix in a bent position or break it altogether. Thus, it is most likely that the mechanism of resistance involves excluding a heterodimer from the D3D4 position. At present, it is not known whether heterodimers directly prevent 12S* particle formation or channel 12S* particles into an off-pathway reaction, the product of which dissociates into early assembly intermediates. In the former model, the heterodimers would have the ability to prematurely interact with 9S* particles. The resistance mutations would prevent this premature association. In the latter model, exclusion would prevent the off-pathway reaction. Analyzing the D-protein stoichiometry of “resistant” procapsids and attempts to stabilize possible off-pathway products should elucidate more fully the molecular mechanisms of inhibition and resistance.
Acknowledgments
We thank Min Chen for technical assistance and expertise.
This research was supported by National Science Foundation grant MCB 054297 to B.A.F., an American Society for Microbiology Undergraduate and NSF Graduate Research Fellowship to J.E.C., the Undergraduate Biological Research Program at the University of Arizona, and the McNair Achievement Program.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Bernal, R. A., S. Hafenstein, N. H. Olson, V. D. Bowman, P. R. Chipman, T. S. Baker, B. A. Fane, and M. G. Rossmann. 2003. Structural studies of bacteriophage alpha3 assembly. J. Mol. Biol. 32511-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burch, A. D., and B. A. Fane. 2000. Foreign and chimeric external scaffolding proteins as inhibitors of Microviridae morphogenesis. J. Virol. 749347-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burch, A. D., and B. A. Fane. 2003. Genetic analyses of putative conformation switching and cross-species inhibitory domains in Microviridae external scaffolding proteins. Virology 31064-71. [DOI] [PubMed] [Google Scholar]
- 4.Burch, A. D., J. Ta, and B. A. Fane. 1999. Cross-functional analysis of the Microviridae internal scaffolding protein. J. Mol. Biol. 28695-104. [DOI] [PubMed] [Google Scholar]
- 5.Chen, M., A. Uchiyama, and B. A. Fane. 2007. Eliminating the requirement of an essential gene product in an already very small virus: scaffolding protein B-free φX174, B-free. J. Mol. Biol. 373308-314. [DOI] [PubMed] [Google Scholar]
- 6.Dokland, T., R. A. Bernal, A. Burch, S. Pletnev, B. A. Fane, and M. G. Rossmann. 1999. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus φX174. J. Mol. Biol. 288595-608. [DOI] [PubMed] [Google Scholar]
- 7.Dokland, T., B. H. Lindqvist, and S. D. Fuller. 1992. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2-P4 bacteriophage system. EMBO J. 11839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dokland, T., R. McKenna, L. L. Ilag, B. R. Bowman, N. L. Incardona, B. A. Fane, and M. G. Rossmann. 1997. Structure of a viral procapsid with molecular scaffolding. Nature 389308-313. [DOI] [PubMed] [Google Scholar]
- 9.Ekechukwu, M. C., D. J. Oberste, and B. A. Fane. 1995. Host and φX174 mutations affecting the morphogenesis or stabilization of the 50S complex, a single-stranded DNA synthesizing intermediate. Genetics 1401167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fane, B. A., K. L. Brentlinger, A. D. Burch, S. L. Hafenstein, E. Moore, C. R. Novak, and A. Uchiyama. φX174, in press. In R. Calendar (ed.), The bacteriophages. Oxford Press, London, England.
- 11.Fane, B. A., and M. Hayashi. 1991. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage φX174. Genetics 128663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fane, B. A., S. Head, and M. Hayashi. 1992. Functional relationship between the J. proteins of bacteriophages φX174 and G4 during phage morphogenesis. J. Bacteriol. 1742717-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fane, B. A., and P. E. Prevelige, Jr. 2003. Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem. 64259-299. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein, R., J. Lengyel, G. Pruss, K. Barrett, R. Calendar, and E. Six. 1974. Head size determination and the morphogenesis of satellite phage P4. Curr. Top. Microbiol. Immunol. 68:59-75. [DOI] [PubMed] [Google Scholar]
- 15.Hafenstein, S., and B. A. Fane. 2002. phi X174 genome-capsid interactions influence the biophysical properties of the virion: evidence for a scaffolding-like function for the genome during the final stages of morphogenesis. J. Virol. 765350-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi, M., A. Aoyama, D. L. Richardson, and N. M. Hayashi. 1988. Biology of the bacteriophage φX174, p. 1-71. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Press, Inc., New York, NY. [Google Scholar]
- 17.Kim, K. J., M. G. Sunshine, B. H. Lindqvist, and E. W. Six. 2001. Capsid size determination in the P2-P4 bacteriophage system: suppression of sir mutations in P2's capsid gene N by supersid mutations in P4's external scaffold gene sid. Virology 28349-58. [DOI] [PubMed] [Google Scholar]
- 18.Morais, M. C., M. Fisher, S. Kanamaru, L. Przybyla, J. Burgner, B. A. Fane, and M. G. Rossmann. 2004. Conformational switching by the scaffolding protein D directs the assembly of bacteriophage φX174. Mol. Cell 15991-997. [DOI] [PubMed] [Google Scholar]
- 19.Rokyta, D. R., C. L. Burch, S. B. Caudle, and H. A. Wichman. 2006. Horizontal gene transfer and the evolution of microvirid coliphage genomes. J. Bacteriol. 1881134-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roof, W. D., H. Q. Fang, K. D. Young, J. Sun, and R. Young. 1997. Mutational analysis of slyD, an Escherichia coli gene encoding a protein of the FKBP immunophilin family. Mol. Microbiol. 251031-1046. [DOI] [PubMed] [Google Scholar]
- 21.Shore, D., G. Deho, J. Tsipis, and R. Goldstein. 1978. Determination of capsid size by satellite bacteriophage P4. Proc. Natl. Acad. Sci. USA 75400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Six, E. W., M. G. Sunshine, J. Williams, E. Haggard-Ljungquist, and B. H. Lindqvist. 1991. Morphopoietic switch mutations of bacteriophage P2. Virology 18234-46. [DOI] [PubMed] [Google Scholar]
- 23.Tonegawa, S., and M. Hayashi. 1970. Intermediates in the assembly of φX174. J. Mol. Biol. 48219-242. [DOI] [PubMed] [Google Scholar]
- 24.Uchiyama, A., M. Chen, and B. A. Fane. 2007. Characterization and function of putative substrate specificity domain in microvirus external scaffolding proteins. J. Virol. 818587-8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchiyama, A., and B. A. Fane. 2005. Identification of an interacting coat-external scaffolding protein domain required for both the initiation of φX174 procapsid morphogenesis and the completion of DNA packaging. J. Virol. 796751-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, S., P. Palasingam, R. H. Nokling, B. H. Lindqvist, and T. Dokland. 2000. In vitro assembly of bacteriophage P4 procapsids from purified capsid and scaffolding proteins. Virology 275133-144. [DOI] [PubMed] [Google Scholar]