Abstract
Foamy viruses (FVs) are ancient retroviruses that are ubiquitous in nonhuman primates (NHPs). While FVs share many features with pathogenic retroviruses, such as human immunodeficiency virus, FV infections of their primate hosts have no apparent pathological consequences. Paradoxically, FV infections of many cell types in vitro are rapidly cytopathic. Previous work has shown that low levels of proviral DNA are found in most tissues of naturally infected rhesus macaques, but these proviruses are primarily latent. In contrast, viral RNA, indicative of viral replication, is restricted to tissues of the oral mucosa, where it is abundant. Here, we perform in situ hybridization on tissues from rhesus macaques naturally infected with simian FV (SFV). We show that superficial differentiated epithelial cells of the oral mucosa, many of which appear to be shedding from the tissue, are the major cell type in which SFV replicates. Thus, the innocuous nature of SFV infection can be explained by replication that is limited to differentiated superficial cells that are short-lived and shed into saliva. This finding can also explain the highly efficient transmission of FVs among NHPs.
Foamy viruses (FVs) are complex retroviruses that are widespread in nonhuman primates (NHPs), felines, equines, and bovines. Most biological and molecular biological analyses have been done with NHP FVs, primarily using an isolate called prototype FV, which is of chimpanzee origin but was isolated from a human tumor in culture (9). Simian FVs (SFVs) from NHPs can be zoonotically transmitted to humans, primarily through bites or other wounds. Among people who are at high risk due to close contact with NHPs, the infection rate is about 2 to 3% (22, 26). However, no horizontal transmission has been documented between infected humans. Infectious SFV has been infrequently isolated from some human peripheral blood mononuclear cells (PBMCs) and oral swabs (3, 8). While both zoonotically infected humans and naturally infected NHPs acquire lifelong infections, in all cases examined, infections appeared to be nonpathogenic (reviewed in reference 14). The lack of evident pathogenicity in infected NHPs or humans is in marked contrast to the ability of FVs to induce rapid cytopathicity in a variety of tissue culture cell types derived from humans and other species. Why this viral infection fails to cause disease or pathology in its hosts despite its cytopathic outcome in vitro is unknown.
We previously showed that there are high levels of FV RNA in oropharyngeal tissues from naturally infected rhesus macaques and that oropharyngeal swabs have the highest levels of viral RNA, up to 4.7 × 104 FV RNA copies per cell equivalent (18). These results support the idea that virus is shed into saliva, although the salivary glands themselves do not have detectable viral RNA. We also found that low levels of proviral DNA are detectable in most tissues and in PBMCs, as previously reported (7). However, since these tissues do not have detectable levels of FV RNA, they are primarily latently infected. In tissue culture systems, once viral mRNAs and proteins are produced, the infected cells rapidly die (15). This suggests that FV replication in vivo might be limited to expendable cell types whose death would not lead to obvious pathology. In order to determine the cells that are permissive for FV replication in vivo, we developed an in situ hybridization (ISH) assay for FV RNA and used immunohistochemistry for cell-type-specific markers to further identify these permissive cells.
MATERIALS AND METHODS
Animals and tissues.
Rhesus macaques (Macaca mulatta) of Indian origin between the ages of 2 and 21 years, of either sex, were bred and housed at the Oregon National Primate Research Center (ONPRC). All studies were conducted in accordance with the standards of the ONPRC's Animal Use and Care Committee and the approved protocols in the Guide for the Care and Use of Laboratory Animals (1). Animals were screened for FV by the presence of FV RNA in oral swabs, as previously described (18). Tissues obtained from animals at necropsy, including the buccal and pharyngeal epithelium, tongue, and tonsil, were placed either in 10% neutral buffered formalin for ISH and immunohistochemistry or in RNAlater (Ambion) for RNA isolation and quantitative reverse transcription (RT)-PCR for FV gag RNA, as previously described (18). Our ISH analyses used sections from tongues obtained from three FV-positive (FV+) animals, the pharyngeal epithelium and tonsil from an additional FV+ animal, and a control tongue from an FV-negative (FV−) animal.
Probe generation.
Sense and antisense regions of FV gag were generated to use as RNA probes for ISH. First, a 679-base region of FV gag (bases 1253 to 1932) of FV isolate 5, GenBank accession number DQ120934, was PCR amplified from cDNA prepared by oligo(dT) priming of RNA extracted from FV isolate 5-infected TF cells, a rhesus macaque fibroblast cell line (13), using primers FV gag1316_F1 (CTGGACAAGCTGTAGTTACTGCTAT) and FVgagR1 (GTTCCCTTGATTTCCGCTTCCAGAG).
The PCR-amplified products were cloned by standard methods into the pCR4-TOPO vector (Invitrogen), downstream of the T7 RNA polymerase priming site. Clones with the FV gag regions in both sense and antisense orientations were verified by both sequencing and restriction digestion. Each construct was linearized with PmeI (New England Biolabs), as confirmed by gel analysis, and used for in vitro transcription according to the manufacturer's instructions for the Riboprobe system (Promega).
ISH for FV RNA.
Formalin-fixed, paraffin-embedded tissues from three FV+ macaques and an FV− macaque were cut into 4-μm sections, placed on positively charged slides (Fisher), and baked at 56°C for 30 min. ISH similar to that previously described was performed (4). Details of the hybridization procedures are available upon request. Each tissue sample was hybridized with the sense and antisense FV gag probes in at least three independent ISH assays, with two time points per assay.
Immunohistochemistry for cell-type-specific and cell proliferation markers.
Slides with serial sections of tissues were deparaffinized as described for the ISH slides and rehydrated in Dako wash buffer. CD45 and cytokeratin slides were steamed for 40 min in preheated target retrieval solution (Dako), pH 6, in a steamer and cooled for 20 min. Slides were rinsed three times in wash buffer, and all subsequent staining steps were performed at room temperature using Dako autostainer. Endogenous peroxide activity was blocked using 3% H2O2 for 8 min, followed by protein blocking by incubation in 15% swine serum and 5% human serum (or in 5% mouse serum for Ki67 staining) in Tris-buffered saline containing 1% bovine serum albumin for 10 min. All antibodies were incubated on the tissue for 30 min and then washed with wash buffer. CD45 leukocyte common antigen (Dako) was used at a concentration of 4.7 μg/ml, cytokeratin 8.13 (Sigma-Aldrich) was used at a dilution of 1:20, and Ki67 RM-9106 (Lab Vision) was used at a concentration of 0.5 μg/ml. The antibodies were detected by Envision Plus horseradish peroxidase mouse-specific polymer (Dako) for 30 min. Staining for all slides was visualized with 3,3′-diaminobenzidine (Dako) for 7 min, and the sections were counterstained with hematoxylin (Dako) for 2 min. Concentration-matched isotype control slides were run for each tissue sample (Jackson ImmunoResearch Laboratories). Images were taken with a Nikon E800 microscope equipped with a CoolSNAP color video camera (Photometrics), using Metamorph software, version 5.0 (Universal Imaging Corporation).
RESULTS
First, we performed ISH with a rhesus macaque fibroblast cell line (13) uninfected or infected with FV in vitro. We could detect high levels of FV RNA in the infected but not uninfected cells, and the signal was specific for the antisense gag probe (Fig. 1). We then screened oropharyngeal tissues for FV RNA levels by using a quantitative RT-PCR (18). Tissues in which we could detect the highest levels of viral RNA (>100 copies of gag per cell equivalent, by RT-PCR) were used for these studies. For our ISH analyses, we utilized oropharyngeal tissues, including the tongue, pharyngeal epithelium, and tonsil. Our previous work showed that, in a tissue culture model, transcription of viral RNA from the long terminal repeat, including the gag gene, was consistently associated with viral replication (15). Therefore, we assumed that the presence of viral gag RNA is a marker of viral replication and hence denotes permissive cells.
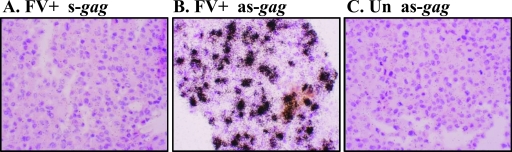
FIG. 1.
FV replication in uninfected or FV-infected macaque fibroblasts measured by ISH. Dark silver grains indicate an FV RNA-specific signal in rhesus macaque immortalized TF fibroblast cells either infected with FV (FV+) (A and B) or uninfected (Un) (C). (A) Sense gag probe (s-gag); (B and C) antisense gag probe (as-gag). Cells were counterstained with hematoxylin and eosin.
Oropharyngeal tissues are classified either as keratinized mucosa, having an outer cornified epithelial layer, such as the (dorsal) tongue, or as nonkeratinized mucosa, such as the pharyngeal epithelium and tonsil. The oral mucosal epithelium is organized on an underlying basement membrane and is composed of basal cells, from which layers of more-differentiated, squamous epithelial cells originate. Below the epithelial basement membrane lies mesenchymal or connective tissue, comprised mostly of fibroblasts, leukocyte-derived cells, and other cells, including endothelial cells, in a collagen-rich matrix (17).
An examination of oropharyngeal tissue sections from rhesus monkeys naturally infected with FV revealed that FV replication was concentrated in discrete foci and localized to the outer epithelium, specifically to the superficial epithelium. We consistently observed a patchy distribution, with two to eight foci of FV RNA+ cells identified within each of the oropharyngeal tissue sections we examined. For example, the pharyngeal epithelium from an FV+ animal (Fig. 2A) shows eight foci of replication. At higher magnifications, FV RNA+ cells were seen exclusively in the superficial epithelium (Fig. 2B). FV RNA consistently localized to the differentiated cells and was not evident in basal cells or cells of the intermediate layers. No positive signal was detected in the same region by using the gag sense probe (Fig. 2C). Many FV RNA+ cells appeared to be in the process of desquamating from the tissues, seen clearly in the pharyngeal epithelium (Fig. 3). The underlying mesenchyme and other sites within the oropharyngeal tissues were carefully examined, but no signal indicative of FV RNA was observed. In the keratinized epithelium of the tongue, FV RNA+ cells also consistently localized to the superficial differentiated epithelium but were excluded from the keratinized region (demarcated by dark pink staining in Fig. 4A and B). Many of the infected cells appear to be desquamating from the tissues (Fig. 4A). The same types of permissive cells were found for tongues from two additional FV+ animals, but no specific signal was observed using the gag antisense probe with tongue tissue from an FV− animal (data not shown). FV replication in the tonsil also demonstrated this pattern of localization and distribution (data not shown). Thus, in all tissues examined, the cell types supporting viral replication localized to the superficial epithelium.
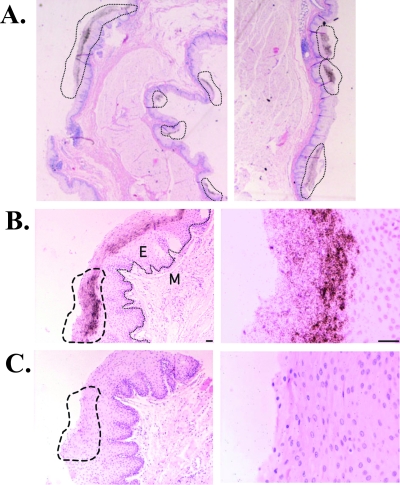
FIG. 2.
FV RNA expression localizes to the superficial epithelium. Dark silver grains overlying cells indicate an FV RNA-specific signal in the rhesus macaque pharyngeal epithelium. FV RNA was detected by ISH with a 35S-labeled FV RNA probe (679-nucleotide fragment of gag in antisense orientation) (A and B) or a control sense probe (C), using tissues that were cut into 4-μm sections and counterstained with hematoxylin and eosin. Shown are bright-field micrographs of FV RNA+ regions at ×20 (A), ×100 (left) and ×400 (right) (B and C) magnifications. Dashed lines indicate FV RNA+ regions or the same region, using the sense probe (C). A line indicates the basement membrane in panel B (left). M, mesenchyme; E, epithelium. Cells were counterstained with hematoxylin and eosin. Scale bar, 50 μm.
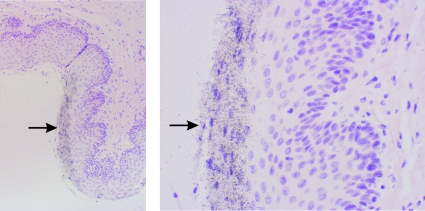
FIG. 3.
FV replication in the pharyngeal epithelium localizes to cells sloughing from the tissue. Sections were treated as described in the legend to Fig. 1. An FV RNA+ section of the epithelium is shown at ×100 (left) and ×400 (right) magnifications. Cells were counterstained with hematoxylin and eosin. A region where infected cells are desquamating from the tissue is denoted by arrows.
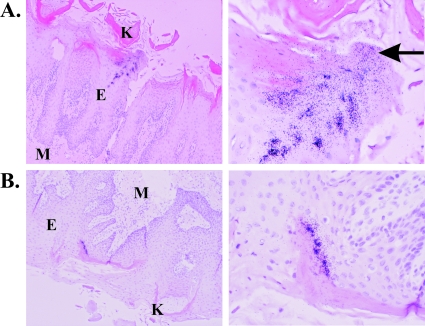
FIG. 4.
FV replication in the superficial epithelium of the tongue localizes to the superficial epithelium. Sections were treated as described in the legend to Fig. 1. Two FV+ regions from an infected tongue are shown. Cells were counterstained with hematoxylin and eosin, and the keratinized regions are demarcated by the dense pink staining. Images are shown at ×100 (left) and ×400 (right) magnifications. M, mesenchyme; E, epithelium; K, keratinized layer. A region where infected cells are sloughing from the tissue is denoted by an arrow.
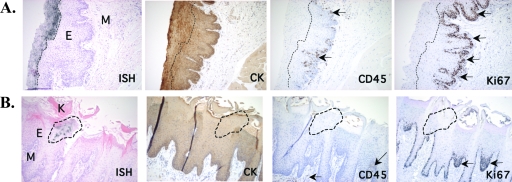
To determine the specific cell type in which FV replicates, we used antibodies specific to epithelium (cytokeratin)- and leukocyte (CD45)-derived cells, as well as an antibody to Ki67, a cell proliferation antigen (21), and performed immunohistochemistry on tissue sections adjacent to those used for ISH (Fig. 5). Cytokeratin expression was pronounced and colocalized with FV RNA+ regions in the pharyngeal epithelium (Fig. 5A) and the tongue (Fig. 5B). Cells expressing CD45 were present in the epithelium but at deeper sites, along the basement membrane, and did not colocalize with FV RNA+ regions (Fig. 5A and B). Thus, the majority of FV-permissive cells are epithelially derived.
FIG. 5.
FV replication localizes to suprabasal epithelial cells. Immunohistochemistry for cytokeratin (CK), CD45, or Ki67 was performed with tissue sections contiguous to those used for ISH. Bright-field micrographs show FV RNA+ regions of the rhesus macaque pharyngeal epithelium at ×100 magnification. Brown deposits indicate CK-, CD45-, or Ki67-specific staining. Arrows point to CD45+ or Ki67+ cells. Dashed lines indicate FV RNA+ regions. M, mesenchyme; E, epithelium; K, keratinized layer. (A) Section of the pharyngeal epithelium of an SFV+ animal. (B) Section of the tongue of an SFV+ animal.
In the mucosal epithelium, basal cells are normally the only dividing cells. In both the pharyngeal epithelium (Fig. 5A) and tongue (Fig. 5B), cells expressing the cell proliferation marker Ki67 were confined mostly to the basal epithelial cells along the basement membrane, and only a few Ki67+ cells were scattered throughout the superficial epithelium. Thus, the majority of FV RNA+ cells did not colocalize with cells expressing Ki67, and therefore, epithelium-derived, nonproliferating cells were confirmed to be the permissive cell type. Together, these observations establish that, in vivo, FV replicates in nonproliferating superficial epithelial cells of the oral mucosal epithelium.
DISCUSSION
FVs are the most ancient RNA viruses, having coevolved with their primate hosts for over 30 million years (23). This long-term virus-host coevolution may have resulted in the establishment of a viral replicative niche that is innocuous to its host while also providing a facile means of efficient viral transmission to new hosts. In a previous study, SFV expression was examined in four SFV+ African green monkeys. Only one oral cavity was found to be SFV+, and the site of SFV expression in the oral mucosal tissue of this animal was examined by ISH with env, tas, and pol probes (7). Those authors found a few SFV+ cells along the stroma-epithelium border of an oral mucosal section. In contrast, we found that all SFV+ macaques have detectable levels of RNA in a number of different oral tissues (18), and in the current study, we detected RNA+ cells in a variety of oral tissues from a number of different SFV+ macaques, but only at the superficial epithelium. Our findings show that FV replication is limited to cells at a very late stage of epithelial cell differentiation, when the cells are about to be shed from the tissue. Within the oral mucosal epithelium, the complete differentiation process, from a basal epithelial cell to a terminally differentiated shed epithelial cell, takes about 7 days (17). The most superficial terminally differentiated cells in the epithelial cell layer turn over in about 3 hours (6). Thus, the limitation of viral replication to a relatively expendable, superficial cell type could account for the innocuous character of FV infection in vivo. FV-infected cells may be shed from the tissue before virus can spread to adjacent susceptible cells, limiting replication to a focus, as we have observed. Perhaps the evolved kinetics of viral infection in vivo simply allows no time for adjacent differentiated epithelial cells to become permissively infected. The contrasting in vitro situation, in which FV infection spreads rapidly and is cytopathic to the entire culture, may be an artifact of culture systems that do not represent the differentiated cell types or cell turnover rates within mucosal epithelial tissues.
The pattern of FV replication appears similar to that observed for some other viruses, most notably human papillomavirus (19) and Kaposi's sarcoma-associated herpesvirus (10). Human papillomavirus and Kaposi's sarcoma-associated herpesvirus establish latent infections in basal epithelial cells of the mucosa, and viral replication is then activated as epithelial cells differentiate. Infectious virions are produced only in terminally differentiated cells and released when these superficial cells are sloughed. Although we do not have direct evidence of infection of basal cells, our data are consistent with such a model for FV. Numerous in vitro studies have shown that cell division is required for the integration and replication of a variety of culture cells by both wild-type FV and FV-based vectors (2, 24). We do not know whether initial viral infection is of dividing cells, but our data are consistent with a model in which FV integrates into dividing cells and expression is induced only when the cells differentiate.
Robust viral replication in sloughed-off cells of the oral cavity can explain the efficient transmission of FV, which is postulated to occur via saliva through biting or other methods of introducing saliva at skin lesions, such as grooming (reviewed in reference 14), and which results in infection rates approaching 100% in some NHP populations (11). A high incidence of infection, salivary mode of transmission, and replication in the oropharyngeal tissues are also found in herpesviruses, such as Epstein-Barr virus (EBV). In EBV-associated hairy leukoplakia lesions, EBV-infected CD14+ monocytes are first infected and these cells migrate into the epithelium, where virus is spread to oral epithelial cells (25). As in FV infections, productive EBV infection occurs only in the terminally differentiated layers, highly analogous to the results presented here for FV. In the case of FV, several studies indicate that PBMCs are latently infected well before viral replication can be detected in the oral mucosa (5; J. Yee and N. Lerche, personal communication). Thus, it is possible that migratory cells, such as macrophages and other leukocytes, are initially infected and that such cells eventually traffic to the oropharyngeal tissues, where they localize to the mesenchyme-epithelium cell border and may spread the latent infection to less differentiated epithelial cells.
Like FVs, herpesviruses (12) and papillomaviruses (20), with similar replication patterns, are also ancient viruses. Differentiation-specific viral activation within differentiated cells of the oral mucosa or the skin arose independently in these diverse virus families. This mechanism of viral replication promotes efficient virus transmission via shed cells while limiting viral replication to a superficial site and thus minimizing host tissue damage. Indeed, in the majority of infections, none of these viruses leads to pathogenic sequelae.
At the molecular level, it is not known what limits FV replication to cells of the oral mucosal epithelium. Previous work has shown that in vitro, FV is inhibited in latently infected cells by a transcriptional block that can be alleviated by protein kinase C inducers or by the addition of high levels of the viral transcriptional transactivator protein Tas (15, 16). Thus, it may be that replication in vivo is limited to cells that contain a specific transcription factor(s) or lack a transcriptional inhibitor(s). Further work is needed to molecularly characterize limitations to FV infection in latently infected cells both in vitro and in vivo.
FVs are genetically stable in vivo, in stark contrast to other retroviruses such as human immunodeficiency virus (HIV) (23). Several reasons have been suggested. The first is that FV infections are primarily latent. However, it is known that there is a high level of replication in oral mucosal tissues (18). The second is that FV reverse transcriptase might be less error prone than that of HIV. However, at least in vitro, the overall error rate for FV reverse transcriptase is at least equal to that for HIV reverse transcriptase (4). The limited focal replication of FV in vivo can help explain this paradox. Our data suggest that each viral replication is an ongoing process, with each initial infection limited to just a few rounds of further infection. The daughter viruses are then sloughed into saliva, and new initiating infections occur from the parental infecting virus. In this model, there are not enough rounds of replication from any initial infection to drive diversity.
It remains unknown why there is no human-specific FV. Possible explanations lie in a lack of human behaviors required for efficient transmission, such as biting and grooming. Alternatively, there may be a biological restriction to replication in the oral cavity of humans, rendering it nonpermissive for viral replication. Although virus has been isolated from the oral cavity of one infected human (3), it is not known whether the virus was recovered from latently or permissively infected cells. The data presented here, which define the cells that support FV replication, will aid in the determination of whether the oral cavities of humans are permissive for viral replication as in natural hosts.
We have shown that the niche of in vivo FV replication in natural primate hosts is the differentiated superficial epithelial cell within the oropharyngeal tissues, a short-lived reservoir. We propose that replication in this cell type accounts for the nonpathogenic character of FV infections.
Acknowledgments
We thank Michael Emerman, Beverly Dale-Crunk, and Martha Jackson for critical reading of the manuscript, Jeff Vieira and Carl Chatzky for helpful discussions, and J. Yee and N. Lerche for sharing unpublished results. We also thank Anne Lewis for helpful discussions and for collecting rhesus macaque tissues, Julie Randolph-Haebecker, Sue Knoblaugh, and Min Spencer for technical assistance, and Al Legasse and Shannon Planer for animal care.
This project was financially supported by NIH ROI CA 81297 and funds from FHCRC to M.L.L., NIH training grant CA09229 to S.M.M., ONPRC core grant P51 RR00163 and RO1-AI054292 to L.J.P., and grants U42-RR016025 and U24-RR018107 to M.K.A.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.AVMA Panel on Euthanasia. 2001. 2000 Report of the AVMA Panel on Euthanasia. J. Am. Vet. Med. Assoc. 218669-696. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz, P. D., R. A. Weiss, and M. O. McClure. 1995. Cell cycle dependence of foamy retrovirus infection. J. Virol. 697295-7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boneva, R. S., W. M. Switzer, T. J. Spira, V. B. Bhullar, V. Shanmugam, M. E. Cong, L. Lam, W. Heneine, T. M. Folks, and L. E. Chapman. 2007. Clinical and virological characterization of persistent human infection with simian foamy viruses. AIDS Res. Hum. Retrovir. 231330-1337. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, P. L., C. R. Stenbak, D. Hoberman, M. L. Linial, and S. H. Hughes. 2007. In vitro fidelity of the prototype primate foamy virus (PFV) RT compared to HIV-1 RT. Virology 367253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, J. I., H. W. Merks, J. Fournier, R. S. Boneva, and P. A. Sandstrom. 2007. Characterization of blood-borne transmission of simian foamy virus. Transfusion 47162-170. [DOI] [PubMed] [Google Scholar]
- 6.Dawes, C. 2003. Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch. Oral Biol. 48329-336. [DOI] [PubMed] [Google Scholar]
- 7.Falcone, V., J. Leupold, J. Clotten, E. Urbanyi, O. Herchenröder, W. Spatz, B. Volk, N. Bölm, A. Toniolo, D. Neumann-Haefelin, and M. Schweizer. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 2577-14. [DOI] [PubMed] [Google Scholar]
- 8.Heneine, W., W. M. Switzer, P. Sandstrom, J. Brown, S. Vedapuri, C. A. Schable, A. S. Khan, N. W. Lerche, M. Schweizer, D. Neumann-Haefelin, L. E. Chapman, and T. M. Folks. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4403-407. [DOI] [PubMed] [Google Scholar]
- 9.Herchenroder, O., R. Turek, D. Neumann-Haefelin, A. Rethwilm, and J. Schneider. 1995. Infectious proviral clones of chimpanzee foamy virus (SFVcpz) generated by long PCR reveal close functional relatedness to human foamy virus. Virology 214685-689. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, A. S., N. Maronian, and J. Vieira. 2005. Activation of Kaposi's sarcoma-associated herpesvirus lytic gene expression during epithelial differentiation. J. Virol. 7913769-13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones-Engel, L., K. A. Steinkraus, S. M. Murray, G. A. Engel, R. Grant, N. Aggimarangsee, B. P. Y.-H. Lee, C. May, M. A. Schillaci, C. Somgird, T. Sutthipat, L. Vojtech, J.-Y. Zhao, and M. L. Linial. 2007. Sensitive assays for simian foamy viruses reveal a high prevalence of infection in commensal, free-ranging, Asian monkeys. J. Virol. 817330-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlin, S., E. S. Mocarski, and G. A. Schachtel. 1994. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J. Virol. 681886-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchoff, V., S. Wong, J. S. St, and G. S. Pari. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147321-333. [DOI] [PubMed] [Google Scholar]
- 14.Linial, M. L. 2007. Foamy viruses, p. 2245-2263. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia, PA.
- 15.Meiering, C. D., and M. L. Linial. 2002. Reactivation of a complex retrovirus is controlled by a molecular switch and is inhibited by a viral protein. Proc. Natl. Acad. Sci. USA 9915130-15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meiering, C. D., C. Rubio, C. May, and M. L. Linial. 2001. Cell-type-specific regulation of the two foamy virus promoters. J. Virol. 756547-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, J., S. J. Squier, and S. J. Gerson. 1984. The structure and function of oral mucosa. Pergamon Press, Oxford, United Kingdom.
- 18.Murray, S. M., L. J. Picker, M. K. Axthelm, and M. L. Linial. 2006. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J. Virol. 80663-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakahara, T., A. Nishimura, M. Tanaka, T. Ueno, A. Ishimoto, and H. Sakai. 2002. Modulation of the cell division cycle by human papillomavirus type 18 E4. J. Virol. 7610914-10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong, C. K., S. Y. Chan, M. S. Campo, K. Fujinaga, P. Mavromara-Nazos, V. Labropoulou, H. Pfister, S. K. Tay, J. ter Meulen, and L. L. Villa. 1993. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J. Virol. 676424-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholzen, T., and J. Gerdes. 2000. The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 182311-322. [DOI] [PubMed] [Google Scholar]
- 22.Switzer, W. M., V. Bhullar, V. Shanmugam, M. E. Cong, B. Parekh, N. W. Lerche, J. L. Yee, J. J. Ely, R. Boneva, L. E. Chapman, T. M. Folks, and W. Heneine. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 782780-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Switzer, W. M., M. Salemi, V. Shanmugam, F. Gao, M. E. Cong, C. Kuiken, V. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, Z. Tooze, F. Villinger, E. C. Holmes, and W. Heneine. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434376-380. [DOI] [PubMed] [Google Scholar]
- 24.Trobridge, G., and D. W. Russell. 2004. Cell cycle requirements for transduction by foamy virus vectors compared to those of oncovirus and lentivirus vectors. J. Virol. 782327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tugizov, S., R. Herrera, P. Veluppillai, J. Greenspan, D. Greenspan, and J. M. Palefsky. 2007. Epstein-Barr virus (EBV)-infected monocytes facilitate dissemination of EBV within the oral mucosal epithelium. J. Virol. 815484-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfe, N. D., W. M. Switzer, J. K. Carr, V. B. Bhullar, V. Shanmugam, U. Tamoufe, A. T. Prosser, J. N. Torimiro, A. Wright, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. Heneine. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363932-937. [DOI] [PubMed] [Google Scholar]