Abstract
Herpesviruses specify a ubiquitin-specific protease activity located within their largest tegument protein. Although its biological role is still largely unclear, mutation within the active site abolished deubiquitinating (DUB) activity and decreased virus replication in vitro and in vivo. To further elucidate the role of DUB activity for herpesvirus replication, the conserved active-site cysteine at amino acid position 26 within pUL36 of Pseudorabies virus (PrV) (Suid herpesvirus 1), a neurotropic alphaherpesvirus, was mutated to serine. Whereas one-step growth kinetics of the resulting mutant virus PrV-UL36(C26S) were moderately reduced, plaque size was decreased to 62% of that of the wild-type virus. Ultrastructural analysis revealed large accumulations of unenveloped nucleocapsids in the cytoplasm, but incorporation of the tegument protein pUL37 was not abolished. After intranasal infection with PrV-UL36(C26S) mice showed survival times two times longer than those of mice infected with wild-type or rescued virus. Thus, the DUB activity is important for PrV replication in vitro and for neuroinvasion in mice.
Herpesviruses possess a large double-stranded DNA genome, comprising more than 70 open reading frames (ORFs) encoding viral proteins, of which at least 30 are incorporated into the mature virus particle. The most complex virion component is the tegument, which, in the alphaherpesviruses, contains more than 15 different viral proteins. Operationally, the tegument has been divided into an “inner,” capsid-associated part and an “outer,” envelope-juxtaposed portion (reviewed in references 42 and 43). The innermost part contains the largest herpesvirus tegument protein, which, in the prototypic alphaherpesvirus herpes simplex virus type 1 (HSV-1), is the product of the UL36 gene. Homologs of this protein, for clarity henceforth named pUL36, are present in all members of the Herpesviridae which have been analyzed in this respect to date.
Previous studies revealed two conserved domains within the N termini of pUL36 homologs. One is required for interaction with pUL37, another conserved tegument protein (4, 22, 33, 44, 50), whereas the second specifies a ubiquitin-specific cysteine protease activity (20, 24, 27, 47, 48, 51). This activity is not required for herpesvirus replication, as has been shown for human cytomegalovirus (HCMV) (51) and Marek's disease virus (MDV) (24). Nevertheless, abrogation of deubiquitinating activity resulted in impairment of replication in vitro (51) and in vivo (24).
In pseudorabies virus (PrV), pUL36 is the only truly essential tegument protein (14), and its deletion completely blocks viral replication. Interestingly, recent data demonstrated that about one-third of PrV pUL36 located in the C-terminal half can be deleted without drastic impairment of viral replication. In contrast, the extreme C terminus of pUL36 is essential (6, 35), probably due to its association with the capsid-associated pUL25 (10). Moreover, deletion of an N-terminal domain of about 200 amino acids comprising the deubiquitinating module (7, 35) resulted in impairment, but not abrogation, of viral replication, indicating that the deubiquitinating activity is not essential for PrV replication. However, a more-detailed mutagenesis of specific amino acids involved in deubiquitination has not yet been performed.
The herpesvirus replication cycle is a well-organized procedure and relies on numerous enzyme activities as well as protein-protein interactions (reviewed in reference 43). In the last decade it has become clear that various cellular pathways, e.g., cell cycle control, signal transduction, protein trafficking, or immune response, are modulated by covalent attachment of ubiquitin or ubiquitin-like polypeptides, leading to proteasomal degradation, activation/inactivation of intrinsic enzyme activity, or translocation to specific cellular compartments depending on the nature of the modification (17, 23, 28). Furthermore, deubiquitinating enzymes have been identified by chemical or bioinformatical tools in nearly all kingdoms of life (39, 45, 46), corroborating a key role for the ubiquitin modification machinery. In consequence, many intracellular pathogens hijack the ubiquitin pathways, and several interactions of viral and bacterial proteins with the host cell ubiquitin machinery have been elucidated (reviewed in references 16, 36, and 38). For example, the HSV-1 immediate-early protein ICP0 has been shown to interact with the cellular ubiquitin-specific protease USP7/HAUSP, linking herpesvirus replication to the p53 pathway (12). It also contains two different regions possessing E3-ubiquitin ligase activity that mediate binding to cellular proteins (8, 49). Furthermore, in addition to the conserved cysteine protease module located in the N terminus of pUL36 (27, 39, 47) several other viral proteins with deubiquitinase activity have been identified: the adenovirus proteinase adenain (1) and the papain-like protease of severe acute respiratory syndrome-associated coronavirus, PLpro (37), indicating a widespread mechanism of linking viral replication to the host cell machinery.
The catalytic residues of the cysteine protease are highly conserved throughout all herpesvirus subfamilies (48), and deubiquitinating activity of pUL36 has been shown in vitro for HSV-1 (27), HCMV (51) and murine cytomegalovirus (47), Epstein-Barr virus (47), and MDV (24). Despite this conservation, the biological relevance of pUL36-dependent deubiquitination for herpesvirus biology remains unclear. An HCMV mutant carrying a single-amino-acid exchange in the active-site cysteine residue mutated to isoleucine showed slower production of infectious virus but no obvious differences in ultrastructural analysis compared to wild-type virus (51). Replication in vitro of MDV carrying a cysteine-alanine exchange was also only slightly reduced, and lytic replication in the natural host, chicken, was not affected. Interestingly, MDV oncogenicity was reduced significantly (24).
To analyze the role of pUL36-dependent deubiquitination in PrV infection in detail, we mutated the active-site cysteine at amino acid position 26 to serine with the aim of abolishing deubiquitinating activity but not interfering with the overall structure of the protein. The resulting mutant virus as well as a rescuant was tested for replication in cell culture and neuronal spread in our standard animal model.
MATERIALS AND METHODS
Viruses and cells.
PrV strain Kaplan (PrV-Ka [26]) was used as the parental wild-type strain. Viruses were propagated in rabbit kidney (RK13) or porcine kidney (PSEK) cells. PrV-ΔUL36F, which lacks almost the complete UL36 coding region, was propagated on RK13-UL36 cells (14).
Plasmid constructs and isolation of virus recombinants.
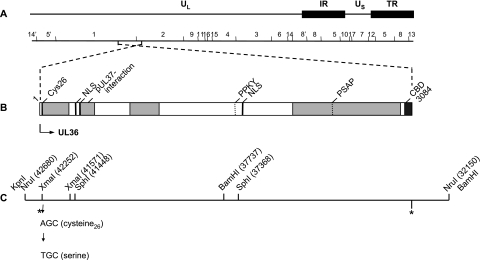
For site-directed mutagenesis of the active-site cysteine (C26), plasmid pUC-UL36, containing a 10.5-kb genomic NruI fragment comprising the complete ORF of UL36 (14) (Fig. 1C), was used for PCR. With primers sb_UL36CFOR (5′-CACACCCGGGTCGGGCGTCTCGAGCC-3′; nucleotides [nt] 42237 to 42257 [GenBank accession no. BK001744]; the mutated nucleotide is shown in bold, and the XmaI site is shown in italics) and sb_UL36CREV (5′-CACACGACGGCGAGGACGGGGATGGC-3′; nt 41551 to 41570 [GenBank accession no. BK001744]), an approximately 700-bp fragment was amplified. After digestion with XmaI (nt 41571 and 42252), the resulting 681-bp fragment was cloned into XmaI-cleaved pUC19 (NEB) resulting in pUC-XmaI(C26S). Correct amplification and mutagenesis were verified by sequencing. This modified XmaI fragment was used to substitute for the original fragment in the cloned 1.2-kb KpnI/SphI fragment (Fig. 1C). After insertion of a 4-kb SphI/SphI fragment (nt 41448 to 37368), the 4.9-kb KpnI/BamHI(C26S) fragment (nt 42680 to 37737) was cloned into appropriately cleaved pcDNA3. The full-length UL36 ORF was reconstituted after insertion of a 5.5-kb BamHI fragment comprising the C-terminal half of UL36 to yield pcDNA-UL36(C26S) (Fig. 1C). Correct reconstitution of the UL36 ORF was confirmed by restriction enzyme analyses.
FIG. 1.
Schematic overview of PrV pUL36 and construction of PrV-UL36(C26S). (A) Diagram of the PrV genome divided into unique long (UL) and unique short (US) regions by internal (IR) and terminal (TR) repeats. Locations of BamHI restriction sites are also shown. (B) Confirmed and putative functional domains in PrV pUL36. Cys26, active-site cysteine for deubiquitinating activity in PrV; pUL37 binding site; CBD, capsid binding domain (10, 35); NLS, nuclear localization signals; late domain motifs (PPKY and PSAP). Light gray boxes indicate nonessential regions. (C) UL36 gene region with relevant restriction enzyme cleavage sites and corresponding nucleotide numbers (GenBank accession no. BK001744) and adjacent restriction sites provided by the cloning vectors pUC19 and pcDNA3. Single-nucleotide mutation resulted in cysteine-serine exchange. Locations of the UL36 start and stop codons are indicated by asterisks.
Virus recombinant containing the point mutation C26S was isolated after cotransfection of pcDNA-UL36(C26S) with PrV-ΔUL36F DNA (14) into RK13 cells. Viral DNA of several progeny plaque isolates was tested for correct UL36 insertion (data not shown). To confirm that the point mutation was present, the relevant part of UL36 was amplified by PCR using primers UL362FOR and UL36rev (7) and sequenced. Since the viral DNA of all plaque isolates showed the same correct sequence, one isolate was chosen, named PrV-UL36(C26S), and further characterized in this study.
To verify that the defects in replication of PrV-UL36(C26S) were caused by the single-amino-acid exchange, DNA of PrV-UL36(C26S) was cotransfected with plasmid pUC-KpnI/SphI containing the corresponding wild-type UL36 sequence. DNA of progeny plaques was analyzed by sequencing of the PCR products as described above. One plaque isolate which specified the wild-type sequence was chosen randomly, named PrV-UL36(C26S)R, and included in the experiments.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
To verify incorporation of pUL36(C26S) into virus particles, virions of PrV-UL36(C26S) as well as of PrV-Ka and PrV-UL36(C26S)R were purified as described previously (7). After separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransfer, membranes were incubated with a monospecific rabbit antiserum directed against a glutathione S-transferase-pUL36 fusion protein containing amino acids 1371 to 2158 (dilution, 1:100,000), and bound antibody was detected as described previously (7). Parallel blots were probed with monospecific antisera against pUL37 (1:100,000) (32) and the major capsid protein, pUL19 (1:300,000) (31).
In vitro growth studies.
For determination and analysis of one-step growth kinetics and plaque size, RK13 cells were infected with PrV-Ka, PrV-UL36(C26S), and PrV-UL36(C26S)R and processed as described previously (7). Supernatants and cell lysates were combined and titrated.
Electron microscopy.
For analysis of ultrathin sections, RK13 cells were infected with PrV-Ka, PrV-UL36(C26S), and PrV-UL36(C26S)R at a multiplicity of infection of 1 and incubated for 14 h at 37°C. Fixation and embedding were done as described previously (18). Ultrathin sections were counterstained with uranyl acetate and lead salts and examined with a Tecnai 12 electron microscope (Philips, Eindhoven, The Netherlands).
In vivo studies.
To investigate the influence of the point mutation on neuroinvasion of PrV, a previously described mouse infection model was used (30). Mean survival times and transneuronal spread were analyzed after intranasal infection of 10 6- to 8-week-old CD-1 mice with 104 PFU of PrV-UL36(C26S) and PrV-Ka in 10 μl (5 μl per nostril), respectively. Four animals were inoculated with PrV-UL36(C26S)R. Animals were observed three times a day for clinical signs, and one mouse of each group was euthanized and necropsied every 24 h after inoculation to follow the kinetics of viral spread by histopathology and immunohistochemistry as described previously (21, 30).
RESULTS
Isolation of PrV-UL36(C26S) and revertant.
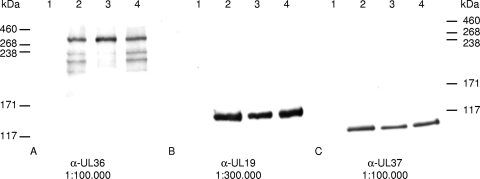
To investigate the functional role of the deubiquitinating activity for PrV replication, we altered the predicted active-site cysteine residue in PrV pUL36 to serine by mutation of a single nucleotide at position 42239(T→A), to abolish thioether linkage to lysine residues of bound ubiquitin due to lack of sulfur in serine but to maintain an amino acid structure highly similar to that of cysteine in order not to alter the overall structure of the protein. In addition we restored the wild-type amino acid sequence after cotransfection of PrV-UL36(C26S) with the pUC-KpnI/SphI fragment to test for effects of possible second-site mutations. Correct mutagenesis and reversion of the mutation were verified by sequencing and restriction enzyme analyses of the constructed plasmids as well as of the generated virus recombinants PrV-UL36(C26S) and PrV-UL36(C26S)R (data not shown). Protein expression and incorporation of the mutated pUL36 were demonstrated by indirect immunofluorescence (data not shown) and Western blot analysis of purified virions showing that pUL36(C26S) is incorporated and correctly expressed in the full-length form (Fig. 2A). Since mutagenesis resulted only in the exchange of one amino acid, no differences in molecular masses between pUL36 and pUL36(C26S) were detected (Fig. 2A). The minor bands prominent in PrV-Ka and PrV-UL36(C26S)R, which varied between preparations, may be degradation products of pUL36 which, depending on the preparation, were sometimes also observed in PrV-UL36(C26S). Parallel blots probed with antisera against the major capsid protein pUL19 (Fig. 2B), as well as the inner tegument protein and interaction partner of pUL36, pUL37 (Fig. 2C), showed no drastic differences between the different viruses.
FIG. 2.
Western blot analysis of purified virions. Purified virions of PrV-Ka (lanes 2), PrV-UL36(C26S) (lanes 3), and PrV-UL36(C26S)R (lanes 4) as well as noninfected RK13 cells (lanes 1) were separated on a 6% polyacrylamide gel and probed with antisera against pUL36 (A), pUL19 (B), and pUL37 (C). Locations of molecular mass marker proteins (high-molecular-weight markers; Invitrogen) are indicated on the left (A and B) or right (C), respectively.
In vitro replication studies.
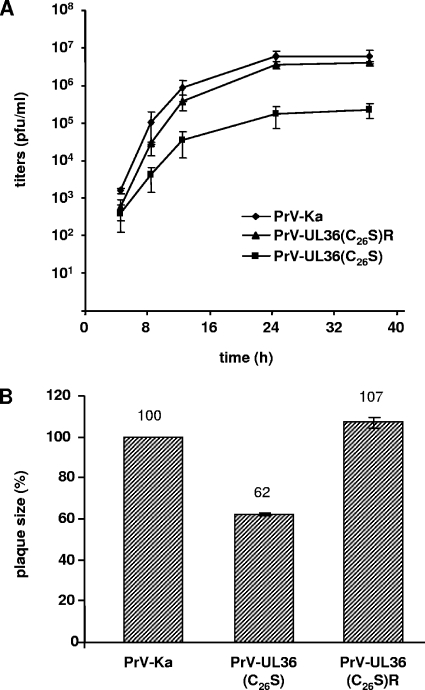
Infectious progeny could be isolated on noncomplementing RK13 cells, a finding which confirms previous results with PrV mutants lacking about 200 amino acids encompassing the complete deubiquitinating module (7, 35). However, corresponding one-step growth kinetics (Fig. 3A) revealed 20- to 30-fold-reduced final titers for PrV-UL36(C26S) (2.3 × 105) compared to those of PrV-Ka (6.1 × 106), while the rescue mutant PrV-UL36(C26S)R (4 × 106) replicated with kinetics similar to those of PrV-Ka. Plaque diameters were only 62% for PrV-UL36(C26S) compared to wild-type PrV-Ka, set as 100%. This reduction was fully corrected (107%) in PrV-UL36(C26S)R (Fig. 3B).
FIG. 3.
Replication properties. (A) One-step growth kinetics. Replication of PrV-Ka, PrV-UL36(C26S), and PrV-UL36(C26S)R was assessed after infection of RK13 cells. Average titers and standard deviations of three independent experiments are shown. (B) Determination of plaque sizes. Plaques of PrV-Ka, PrV-UL36(C26S), and PrV-UL36(C26S)R on RK13 cells were measured, and mean plaque diameters of 30 plaques, determined in at least three independent experiments, were calculated as percentages of plaque size by PrV-Ka. Standard deviations are indicated.
Ultrastructural analyses.
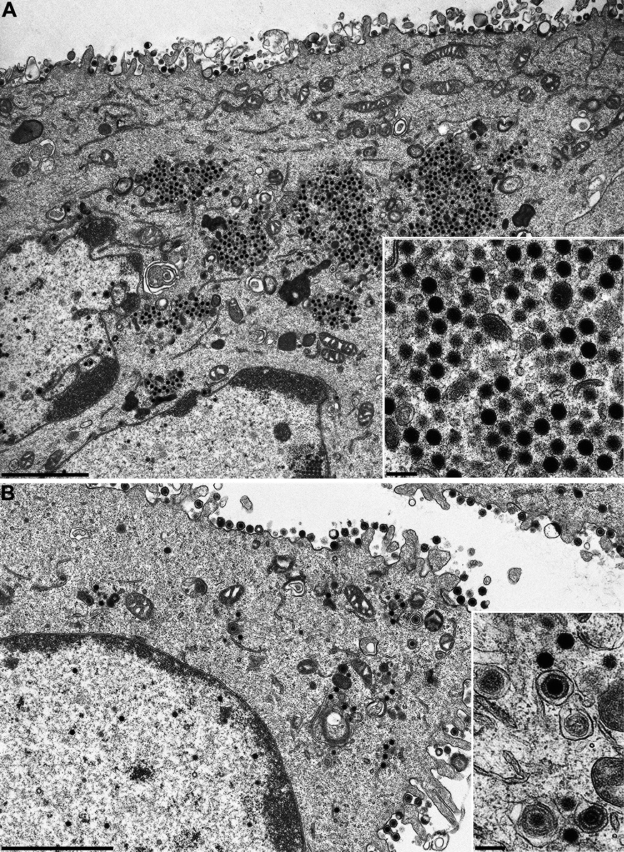
To study virus replication in cell culture in more detail, RK13 cells were analyzed by electron microscopy 14 h after infection with PrV-Ka, PrV-UL36(C26S) (Fig. 4A), and PrV-UL36(C26S)R (Fig. 4B). Correlating with the results obtained from one-step growth kinetics, normal virion morphogenesis including secondary envelopment (Fig. 4B, inset) was observed in PrV-UL36(C26S)R-infected cells with numerous extracellular virus particles lining the plasma membrane (Fig. 4B). In contrast, large accumulations of nonenveloped nucleocapsids were found in the cytoplasm of PrV-UL36(C26S)-infected cells (Fig. 4A, inset), indicating that abrogation of deubiquitinating activity interferes with cytoplasmic stages of PrV maturation in RK13 cells and impairs secondary envelopment late in infection.
FIG. 4.
Ultrastructural analysis of PrV-UL36(C26S)- and PrV-UL36(C26S)R-infected cells. RK13 cells were infected at a multiplicity of infection of 1 and analyzed 14 h after infection. (A) Overview of a PrV-UL36(C26S)-infected cell and higher magnification of cytoplasmic accumulations (inset). (B) Overview of a PrV-UL36(C26S)R-infected cell and higher magnification of intracytoplasmic capsids and secondary envelopment (inset). Bars, 2.4 μm (A and B) and 250 nm (A and B, insets).
Animal infection and immunohistopathology.
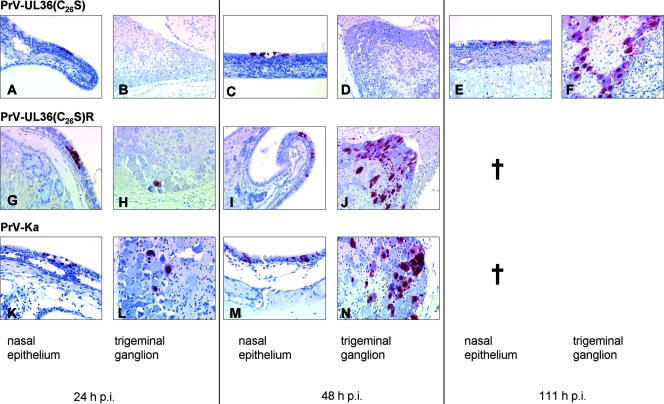
Ubiquitin plays a key role in various cellular and immunological pathways, and many pathogens hijack the host ubiquitin machinery. To assay whether abrogation of the catalytic activity of the deubiquitinating protease in PrV pUL36 has any effects on PrV replication in vivo, we used our small-animal infection model (29, 30). After bilateral intranasal infection of mice with PrV-Ka or PrV-UL36(C26S)R, the first clinical signs were observed at day 2 postinfection (p.i.). Mice became anorectic and depressed, cowered in a hunched position, or showed extensive scratching of the facial skin, causing dermal erosions or ulcerations with hemorrhage. All infected mice died at an average time of 50 h p.i and 53 h p.i., respectively (Table 1). Histopathology revealed focal degeneration (swelling and hypereosinophilia) and necrosis of nasal mucosa with subadjacent lymphocytic infiltration. No inflammatory changes were seen in ganglia (trigeminal ganglia, pterygopalatine ganglia, and optic ganglia) and in the brain. Immunohistochemistry for the pUL19 major capsid protein was positive within groups of degenerated ciliated epithelial cells in the respiratory nasal mucosa at 24 h p.i (Fig. 5G and K). In addition, few infected neurons were detectable in the trigeminal ganglia at 24 h p.i. (Fig. 5H and L). At 48 h p.i. the number of infected cells in the ganglion increased (Fig. 5J and N). At this time, immunostaining in the pterygopalatine and optic ganglia was strongly positive (data not shown). In the brain, the spinal trigeminal nucleus (Sp5) was also infected (Table 1).
TABLE 1.
Kinetics of neuroinvasion in the murine trigeminal pathway
| Virus | Mean time to death (h) | Clinical symptom(s) at day p.i.a:
|
Immunohistochemistry result (no. of days p.i.)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Nasal cavity | Neurons
|
||||
| First order | Second order | Cortical | |||||||||
| PrV-Ka | 50.3 | 0 | +++ | † | 1 | 1 | 2 | ||||
| PrV-UL36 (C26S)R | 53.4 | 0 | +++ | † | 1 | 1 | 2 | ||||
| PrV-UL36(C26S) | 111.7 | 0 | + | ++ | +++ | † | 1 | 3 | 4 | 5 | |
Clinical symptoms: 0, clinically normal mice; +, slight depression, hunched position, and ruffled hair coat; ++, apathy, anorexia, moderate dyspnea, and slight facial pruritus; +++, severe attacks of excitation, self-mutilation, skin erosions, and heavy dyspnea; †, animal moribund or dead.
For immunohistochemistry, values are numbers of days after infection at which first pUL19 detection occurred at the different levels of the trigeminal pathway. Nasal cavity, respiratory mucosal epithelium; first-order neurons, trigeminal ganglion; second-order neurons, spinal trigeminal nucleus (Sp5); cortical neurons, ectorhinal cortex.
FIG. 5.
Immunohistochemistry. Detection of pUL19 (PrV major capsid protein) in the nasal mucosa and in the trigeminal ganglion of mice infected with PrV-UL36(C26S), PrV-UL36(C26S)R, and PrV-Ka. †, all animals dead.
After infection with PrV-UL36(C26S), a considerably prolonged mean time to death of 111 h was observed. However, time to death of individual mice varied rather widely between 96 and 120 h p.i.. First clinical signs occurred at 48 h p.i., when animals started to have slightly reduced activity. At day 3 p.i. mice were anorectic and cowered in a hunched position. At day 4 p.i. all mice showed excitation and were hyperactive with attacks of scratching the facial skin, which finally led to severe dermal lesions. Immunohistochemistry revealed a delay in the transsynaptic and transneuronal spread. Whereas immunostaining of the nasal respiratory epithelium showed positive cells already at 24 h p.i. (Fig. 5A), a result which is comparable to those for PrV-Ka infections (Fig. 5K), PrV antigen in neurons was not detectable until 72 h p.i. in only a small number of neurons in the trigeminal ganglion (Table 1). The number of infected ganglion cells increased at 96 h p.i. At 111 h p.i. the amount of immunostaining in the trigeminal ganglion was comparable to the extent of infection with PrV-Ka at 48 h p.i. (Fig. 5F). At this time several second- and third-order neurons were also infected (Table 1).
DISCUSSION
To investigate whether specific alteration of the predicted catalytic-site residue within the cysteine protease module encoded in the N terminus of pUL36 has any effects on virus replication in vitro and in vivo, we engineered a PrV mutant carrying a single amino acid substitution resulting in a change of cysteine at position 26 to serine. We report here that the mutation reduced PrV replication in RK13 cells, resulting in an impairment in secondary envelopment, and strongly delayed neuroinvasion in a murine infection model.
To date, only a few studies of viruses carrying specific alterations in the cysteine protease module of the herpesvirus large tegument protein pUL36 have been published (24, 51). Although we have not tested for the deubiquitinating activity in PrV pUL36 in a biochemical assay, the high degree of conservation of the catalytic triad module as well as demonstration of protease activity and its distortion by single point mutations in pUL36 homologs of all three herpesvirus subfamilies implies functional conservation of the deubiquitination activity (20, 24, 27, 47, 48, 51).
A mutant of HCMV carrying an isoleucine residue instead of the catalytic cysteine (C24I) in the pUL36 homolog has been shown to be replication competent but developed cytopathic effects more slowly in human foreskin fibroblasts than did the wild type (51). Unlike our data showing large cytoplasmic accumulations of unenveloped nucleocapsids in PrV-UL36(C26S)-infected cells, the HCMV mutant displayed no obvious defects in ultrastructural analyses. Thus, our mutant is the only one described so far in which mutation of the conserved deubiquitination motif has a clear effect on virion morphogenesis.
Mutations in several proteins have been shown to result in accumulation of viral capsids in the cytosol. These include pUL37 (32), pUL47 (34), and pUL48 (15). pUL37 forms a complex with pUL36 (33), and it may be hypothesized that the pUL36 mutation impairs complex formation with pUL37. However, our data do not support this, since pUL37 appears present in purified virions in amounts similar to those found in wild-type or rescued virus. Moreover, the accumulation of capsids in the cytosol observed in PrV-UL36(C26S)-infected cells did not show the hexagonal symmetry observed in the absence of pUL37 (32). Thus, the defect in morphogenesis should not be due to indirect impairment of pUL37 binding.
In MDV replacement of the active-site cysteine by alanine (C98A) only slightly diminished viral replication in cell culture and affected plaque formation in a cell-type-dependent manner (24). Furthermore, lytic replication in vC98A-infected chickens was similar to that of wild-type virus infection but the formation of T-cell lymphomas was drastically impaired, indicating an important role for the deubiquitinating activity in in vivo T-cell transformation.
In our mouse infection model (30) we investigated the influence of the C26S mutation on PrV neuroinvasion and transneuronal spread. Mice infected with PrV-UL36(C26S) showed a mean survival time two times longer than that of mice infected with wild-type or rescued virus. However, typical clinical symptoms like pruritus and hyperesthesia occurred 24 h before death, demonstrating that this mutant is still neuropathogenic. Kinetic analysis showed that the delay in death was associated with a delay in neuroinvasion. Infection of neurons of the trigeminal circuit was first observed 72 h after infection, which is ca. 48 h later than in mice infected with wild-type or rescued virus. By 111 h p.i. the infection of the trigeminal ganglion, as judged from the number of immunostained neurons, was comparable to that in mice infected with wild-type or rescued virus at 48 h p.i. Interestingly, due to the longer survival times of the infected mice, PrV-UL36(C26S) was able to reach cortical neurons (third neuronal level), which were not infected by wild-type or rescued virus.
This impairment in neuroinvasion parallels the replication deficiency in cell culture, although it is not clear whether the latter is the sole reason for the delay in neuronal infection. However, the observed defect in morphogenesis can also explain the reduction in egressing axonal capsids at late times after infection with a PrV mutant lacking the whole deubiquitinating module (35).
Interestingly, in previous studies using a PrV mutant lacking the complete deubiquitinating module (PrV-UL36Δ22-248 [7]) we observed a reduction in viral titers which was similar to that of PrV-UL36(C26S), which parallels data from work by others (35). However, plaque sizes of PrV-UL36Δ22-248 were reduced only to 82% of original size and mean time to death in mice increased to ca. 73 h. In contrast, PrV-UL36(C26S) exhibited plaque sizes which were reduced to 62% of the original size and mean time to death increased to ca. 111 h. In line with these observations, ultrastructural analyses did not show a specific defect in PrV-UL36Δ22-248-infected cells compared to wild-type virus, whereas accumulations of cytoplasmic capsids were observed in cells infected by PrV-UL36(C26S). These differences could not be attributed to a differential effect of the mutation on pUL37 incorporation (Fig. 2). Thus, the more-specific mutation resulted in a stronger phenotype than did the ca.-200-amino-acid deletions encompassing the whole deubiquitinating module (7, 35).
The more drastic phenotype of PrV-UL36(C26S) could have been induced by fortuitous secondary mutations. However, rescue of the defect in PrV-UL36(C26S)R by the wild-type KpnI/SphI fragment demonstrated that mutations outside this fragment played no role. Moreover, we sequenced the whole 1.2-kbp KpnI/SphI fragment in PrV-UL36(C26S) and found only the engineered mutation. Thus, the observed phenotype is clearly due to the C26S exchange. One possible explanation is that the specific exchange of the catalytic cysteine blocks enzymatic activity but, presumably, not substrate binding, resulting in a blocked state in which substrate is bound to the catalytic center. This could either sterically hinder other important interactions of pUL36 (although interaction with pUL37 still occurs) or distort pUL36 structure to a larger extent than deletion of the whole module could.
Ubiquitination has been shown to be involved in targeting of proteins toward the ESCRT machinery, which is required for maturation of infectious virions in retro-, rhabdo-, and filoviruses (reviewed in reference 5). Recently published studies showed that dominant-negative variants of ESCRT proteins may also impair maturation of HSV-1 (9, 11). In contrast, depletion of ESCRT proteins increased HCMV assembly (13). The modules which interact with ESCRT proteins have been termed “late domains” since mutation of these conserved sequence motifs resulted in a block of retrovirus infection late in the replication cycle (reviewed in reference 5).
Based on sequence analysis, several putative late domain motifs have been found within the PrV pUL36 protein. Deletion of the PSAP motif in the carboxy-terminal half of PrV pUL36 affected virus replication in vitro only marginally, whereas mean time to death of infected mice was significantly increased (7). However, so far it is unclear whether PrV uses the ESCRT machinery for its replication.
Since pUL36 remains bound to the incoming capsid during its transit through the cytosol to the nuclear pore (18, 19, 40), it represents a prime candidate for a viral protein interacting with either the cellular motor system or the nuclear pore or both. Indeed, a temperature-sensitive mutant of HSV-1 pUL36 exhibits a defect in early stages of infection (2, 3). Recently, cleavage of pUL36 has been suggested to be required for the release of the viral genome into the nucleus (25). Furthermore, Wang et al. suggested a role for pUL36 deubiquitinating activity in directing capsids after cell entry to the nuclear pore complex (51), and several reports linked ubiquitin-related SUMO proteases to the nuclear pore complex (reviewed in reference 41). However, we did not observe a defect in early stages of infection of cells with PrV-UL36(C26S), indicating that deubiquitinating activity is not required for transport of incoming capsids, docking at the nuclear pore, and genome release.
Taken together, we report here that a PrV mutant carrying a specific alteration of the active-site cysteine residue within the deubiquitinating module in the essential PrV tegument protein pUL36 exhibits a defect in virion morphogenesis accompanied by an impairment of neuroinvasion in a mouse infection model. Further studies need to address the identity of cellular or viral substrates of the cysteine protease in pUL36 to further elucidate its biological role in herpesvirus replication.
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG Me 854/8-2; Me 854/9-1).
We thank D. Werner and P. Meyer for expert technical assistance and M. Jörn for photographic help.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 766323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J. Virol. 46371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 761043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 34455-63. [DOI] [PubMed] [Google Scholar]
- 6.Böttcher, S., B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C Mettenleiter. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 809910-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher, S., H. Granzow, C. Maresch, B. Möhl, B. G. Klupp, and T. C. Mettenleiter. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 8113403-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calistri, A., P. Sette, C. Salata, E. Cancellotti, C. Forghieri, A. Comin, H. Göttlinger, G. Campadelli-Fiume, G. Palù, and C. Parolin. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 8111468-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coller, K. E., J. I. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 8111790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 817380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 161519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraile-Ramos, A., A. Pelchen-Matthews, C. Risco, M. T. Rejas, V. C. Emery, A. F. Hassan-Walker, M. Esteban, and M. Marsh. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell. Microbiol. 92955-2967. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 7811879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., H. Granzow, B. G. Klupp, A. Karger, K. Michael, C. Maresch, R. Klopfleisch, and T. C. Mettenleiter. 2007. Relevance of the interaction between alphaherpesvirus UL3.5 and UL48 proteins for virion maturation and neuroinvasion. J. Virol. 819307-9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, G., and H. Luo. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 845-14. [DOI] [PubMed] [Google Scholar]
- 17.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82373-428. [DOI] [PubMed] [Google Scholar]
- 18.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 793200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 712072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gredmark, S., C. Schlieker, V. Quesada, E. Spooner, and H. L. Ploegh. 2007. A functional ubiquitin-specific protease embedded in the large tegument protein (ORF64) of murine gammaherpesvirus 68 is active during the course of infection. J. Virol. 8110300-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harkness, J. E., and J. E. Wagner. 1995. The biology and medicine of rabbits and rodents. Williams & Wilkins, Baltimore, MD.
- 22.Harmon, M.-E., and W. Gibson. 1996. High molecular weight virion protein of human cytomegalovirus forms complex with product of adjacent open reading frame, abstr. W35-4, p.144. Proc. Am. Soc. Virol.
- 23.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 24.Jarosinski, K., L. Kattenhorn, B. Kaufer, H. Ploegh, and N. Osterrieder. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 10420025-20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for the release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 823311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7394-407. [DOI] [PubMed] [Google Scholar]
- 27.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19547-557. [DOI] [PubMed] [Google Scholar]
- 28.Kirkin, V., and I. Dikic. 2007. Role of ubiquitin- and Ubl-binding proteins in cell signalling. Curr. Opin. Cell Biol. 19199-205. [DOI] [PubMed] [Google Scholar]
- 29.Klopfleisch, R., B. G. Klupp, W. Fuchs, M. Kopp, J. P. Teifke, and T. C. Mettenleiter. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J. Virol. 805571-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klopfleisch, R., J. P. Teifke, W. Fuchs, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 782956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 7410063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 758927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 763065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 768820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 8012086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindner, H. A. 2007. Deubiquitination in virus infection. Virology 362245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Ménard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 7915199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loureiro, J., and H. L. Ploegh. 2006. Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv. Immunol. 92225-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love, K. R., A. Catic, C. Schlieker, and H. L. Ploegh. 2007. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat. Chem. Biol. 3697-705. [DOI] [PubMed] [Google Scholar]
- 40.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 1025832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28612-618. [DOI] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106167-180. [DOI] [PubMed] [Google Scholar]
- 43.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113163-169. [DOI] [PubMed] [Google Scholar]
- 44.Mijatov, B., A. L. Cunningham, and R. J. Diefenbach. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 36826-31. [DOI] [PubMed] [Google Scholar]
- 45.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123773-786. [DOI] [PubMed] [Google Scholar]
- 46.Rawlings, N. D., F. R. Morton, C. Y. Kok, J. Kong, and A. J. Barrett. 2008. MEROPS: the peptidase database. Nucleic Acids Res. 36320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 7915582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlieker, C., W. A. Weihofen, E. Frijns, L. M. Kattenhorn, R. Gaudet, and H. L. Ploegh. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 9677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 988815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 799566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 806003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]