Abstract
Papillomavirus E6 proteins are adapters that change the function of cellular regulatory proteins. The bovine papillomavirus type 1 E6 (BE6) binds to LXXLL peptide sequences termed LD motifs (consensus sequence LDXLLXXL) on the cellular protein paxillin that is a substrate of Src and focal adhesion kinases. Anchorage-independent transformation induced by BE6 required both paxillin and BE6-binding LD motifs on paxillin but was independent of the major tyrosine phosphorylation sites of paxillin. The essential role of paxillin in transformation by BE6 highlights the role of paxillin in the transduction of cellular signals that result in anchorage-independent cell proliferation.
Anogenital type human papillomavirus (HPV) E6 oncoproteins and E6 from bovine papillomavirus type 1 (BE6) interact with cellular proteins by binding to an eight-amino-acid peptide displayed on the target protein (XLXXLLXX, abbreviated LXXLL here) (3, 7, 10, 30); this interaction is required for cellular transformation by BE6 (1, 30). While anogenital HPV E6 proteins bind to a LXXLL motif on the ubiquitin ligase E6AP, BE6 binds to LXXLL motifs found on the cellular adapter protein paxillin, while HPV-16 E6 binds paxillin poorly (26, 30).
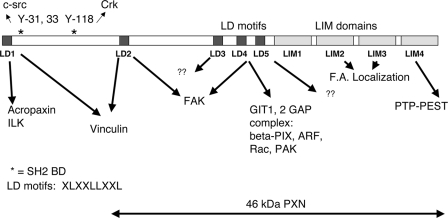
Paxillin was first identified as a hyperphosphorylated protein in Src transformed cells. It localizes to focal adhesions and associates with focal adhesion proteins implicated in the regulation of cell attachment, spreading, and migration, including focal adhesion kinase (FAK), GIT1, PAK, Src, and Crk (reviewed in reference 6 and illustrated in Fig. 1). Paxillin tyrosine phosphorylation sites bind to the SH2-containing proteins Src (at Y31) and CRKL (at Y118). Paxillin also contains five copies of a peptide motif termed LD1 through LD5 (consensus LXXLLXXL) that serve as binding sites for cellular proteins; LD1 interacts with acropaxin and ILK (16, 17), LD2 interacts with FAK and vinculin, LD4 interacts with GIT1 and FAK (29), and the LD3 and LD5 interaction partners remain uncharacterized, although LD5 is functionally required to support FAK tyrosine phosphorylation in embryonic stem (ES) cells (32). LD motifs also bind the BE6 oncoprotein (26, 30). The carboxy terminus of paxillin contains four LIM domains: LIM2 and -3 localize paxillin to focal adhesions (5, 32), and LIM1, -2, and -3 support the tyrosine phosphorylation of FAK in ES cells (32). LIM4 interacts with the tyrosine phosphatase PTP-PEST (8, 21).
FIG. 1.
Paxillin domains. Diagram of paxillin mutants indicating the location of five LD motifs (peptide motifs with the consensus sequence LXXLLXXL) and four zinc-finger LIM domains. Y31, 33, and Y118 are tyrosine phosphorylation sites. Binding partners for the LD motifs, tyrosine phosphorylation sites, and functions ascribed to the LIM domains are from Brown and Turner (6). “F.A. localization” refers to focal adhesion localization, a function ascribed to LIM domains 2 and 3 (6).
Paxillin regulates focal adhesion turnover, since cells from which paxillin has been deleted or that express paxillin mutants have delayed focal adhesion turnover and cell migration (33, 34), and paxillin has been shown to influence the activity of proteins, including the Rho family of small GTP-binding proteins that are involved in regulating actin dynamics (reviewed in references 19 and 35). HIC-5 is a paxillin family member that is highly similar to paxillin but differs in its binding to FAK and GIT proteins and is differentially expressed in differentiated tissues and in response to morphogenic signaling (14, 25, 28). HIC-5 is expressed in paxillin-null mouse embryonic fibroblasts (MEFs) (12, 31).
Although the interaction of paxillin with BE6 correlates with transformation by BE6 and transforming BE6 mutants (9), it is unclear whether paxillin is essential for BE6 transformation. In the present study, we demonstrate through the use of paxillin-null mouse fibroblasts that both paxillin and BE6-binding sites on paxillin are required for the transformation by BE6.
MATERIALS AND METHODS
Cells and tissue culture.
Murine C127 and paxillin-null MEFs were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum; assays for anchorage-independent cell growth were performed as previously described (1).
Plasmids.
Glutathione S-transferase (GST) and maltose-binding protein (MBP) fusions to BE6 and paxillin and paxillin mutants have been previously described (30). MBP and GST fusions were expressed in Escherichia coli strain BL21(DE3)-RIL and purified onto amylose-Sepharose or glutathione-Sepharose beads as described previously (1).
Immunoprecipitations and Western blots.
A total of 5 × 107 cells per lane were lysed in NP-40 lysis buffer on ice (150 mM NaCl, 50 mM Tris [pH 7.5], 50 mM NaF, 5 mM sodium pyrophosphate, 1% NP-40, 0.01% phenylmethylsulfonyl fluoride, 1 mM sodium vanadate, 1 μg of leupeptin-aprotinin/ml), clarified by centrifugation, equalized for protein content, immunoprecipitated with Flag (Sigma) or paxillin clone 349 monoclonal antibody (BD biosciences), and analyzed by Western blotting for BE6 and paxillin. To ascertain BE6 expression levels in transduced MEFs, 107 retrovirally transduced murine fibroblasts were lysed in 0.5 ml of 1% sodium dodecyl sulfate (SDS), diluted 1:10 with NP-40 lysis buffer, and immunoprecipitated with 5 μl of rabbit anti-BE6 antisera. Immunoprecipitates were analyzed by Western blot and probed with rabbit anti-BE6 antibody.
RESULTS
BE6 association with paxillin was found through a yeast two-hybrid screen and by association with transiently overexpressed and epitope-tagged BE6 (26, 30). Thus, the reported association could represent an overexpression artifact. To determine whether paxillin associates with native BE6 in stable expression conditions, paxillin-null MEFs (12) were stably transduced by native BE6 alone, Flag-tagged paxillin alone, or BE6 together with Flag-paxillin. Precipitation of BE6 by Flag antibodies was only observed if coexpressed with Flag-paxillin (Fig. 2A). In order to determine whether BE6 was associated with endogenous paxillin under stable expression conditions in transformed cells, murine C127 cells or C127 cells transformed by BE6 were analyzed by immunoprecipitation of endogenous paxillin and immunoblot analysis of BE6. BE6 was immunoprecipitated by antibodies to paxillin but not isotype control (EE) antibodies, was only observed in BE6 transduced cells, and was independent of cell attachment (Fig. 2B).
FIG. 2.
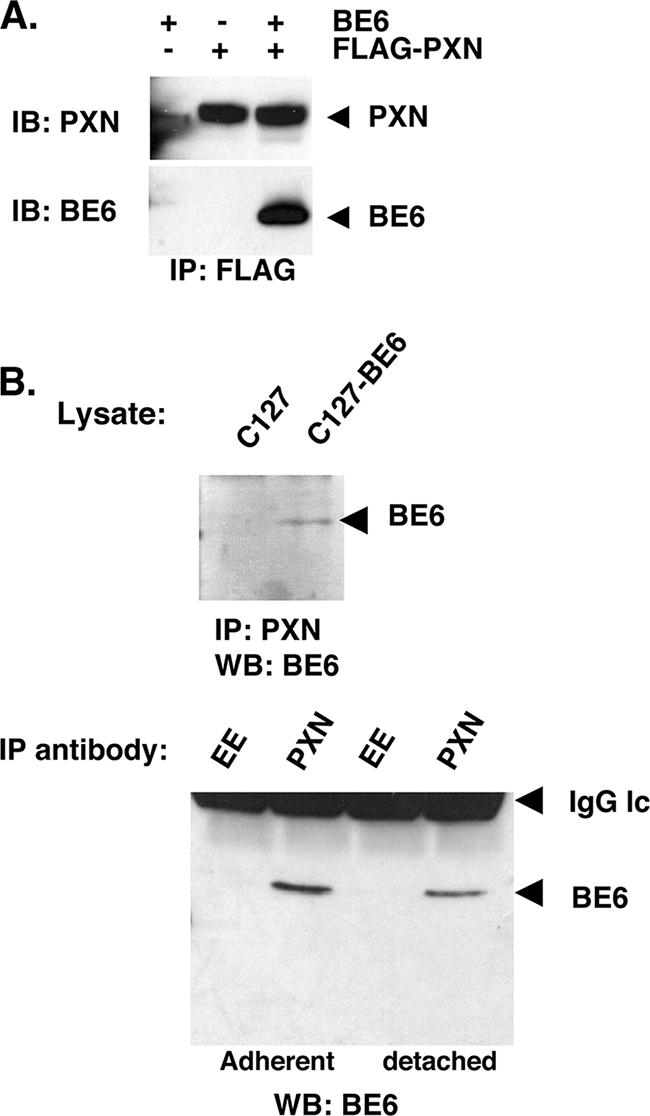
Association of BE6 with paxillin in MEFs. (A) Paxillin-null MEFs (12) were stably transduced with Flag-paxillin and or BE6. BE6 cannot be directly detected in lysates by Western blotting. (B) C127 cells transduced with either vector or wild-type BE6 were immunoprecipitated at 4°C for 1 h with either paxillin monoclonal antibody 349 (BD Biosciences) or isotype control EE monoclonal antibody as indicated. SDS-eluted samples from washed beads were Western blotted as described in Materials and Methods. Detached cells were incubated in complete medium at 37°C for 1 h prior to lysis.
In order to determine the LD motifs of paxillin or HIC-5 that might bind BE6, each paxillin LD motif was expressed as a GST fusion; BE6 bound to LD1, -2, and -4, whereas LD3 and LD5 interacted poorly (Fig. 3A). In contrast to paxillin, BE6 only bound the LD1 motif of HIC-5, since deletion of only HIC-5 LD1 eliminated the interaction of HIC-5 containing the remaining LD motifs with BE6 (Fig. 3B); the LD motifs of paxillin and HIC-5 are similar but not identical (4, 25).
FIG. 3.
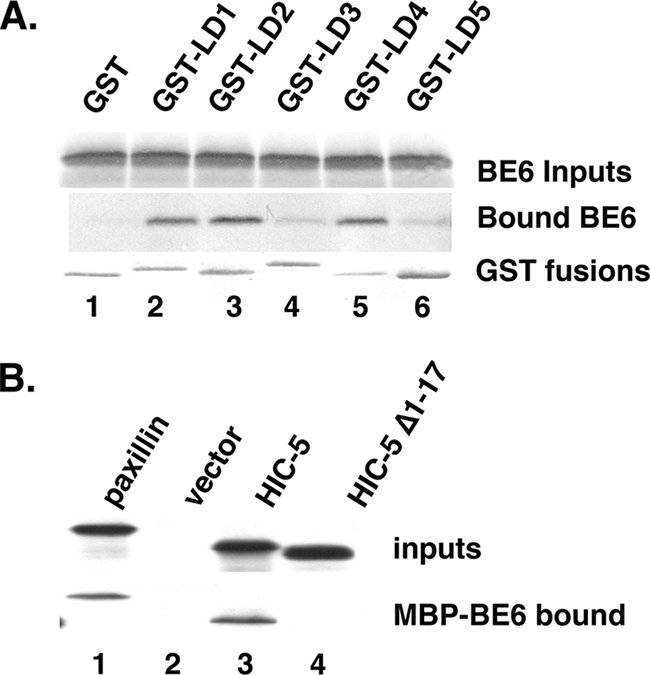
Mapping the BE6 interaction motifs of paxillin and HIC-5. (A) In vitro binding of in vitro-translated BE6 with GST fusions to LD motifs were performed as previously described (9, 30). (B) Association of HIC-5 LD1 with BE6. In vitro-translated HIC-5 and HIC-5 deleted of the first 17 residues that contain LD1 were tested for in vitro association with bacterially expressed MBP-BE6 as described previously (30).
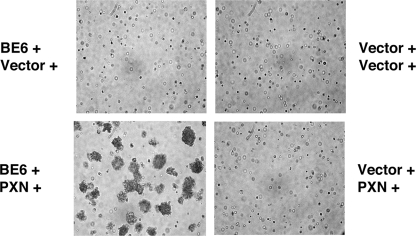
To determine whether paxillin is required for transformation by BE6, drug-selected cells were suspended in agarose and photographed for the formation of anchorage independent colonies. Only cells transduced with both BE6 and paxillin could form anchorage-independent colonies (Fig. 4). Similar, although less-efficient, transformation results were obtained with paxillin-null T17 cells (31; data not shown). Although the paxillin-null MEFs express HIC-5, reexpression of paxillin was required for colony formation.
FIG. 4.
Requirement of paxillin for transformation by BE6. The indicated retrovirally transduced paxillin−/− MEFs (paxillin with puromycin and BE6 with hygromycin selection or empty vectors as indicated) were photographed for anchorage-independent colony formation 10 days after seeding in agarose as described previously (2).
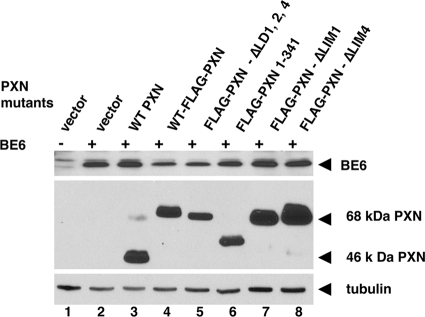
Paxillin mutants were constructed to analyze their ability to support BE6 transformation. Paxillin mutants were constructed that deleted all of the BE6 binding motifs (PXN-ΔLD1,2,4), all of the LIM domains (PXN 1-341), the amino-terminal tyrosine phosphorylation sites at amino acids 31 and 118 (PXN-46 kDa), only LIM domain 4 (required for association with PTP-PEST that is implicated in the activation of Src family members), or only LIM domain 1 (required to support the tyrosine phosphorylation of FAK in ES cells (32) (Fig. 1). Each of these mutants was expressed together with BE6 when transduced in paxillin-null MEFs (Fig. 5). The 46-kDa paxillin molecule has been proposed to be produced by either internal translation initiation or by proteolytic cleavage of full-length paxillin (22, 27); the 46-kDa form is deleted of LD1 and the major tyrosine phosphorylation sites at amino acids 31, 33, 40, and 118 that are binding sites for c-src and Crk family proteins, as well as serine phosphorylation sites implicated in the regulation of migration in response to HGF (13). In paxillin−/− MEFs, the great majority of reexpressed wild-type paxillin is processed from the full-length 68-kDa form to the cleaved 46-kDa form; interestingly, an amino-terminal Flag epitope blocks this cleavage from occurring (Fig. 5). The processing of paxillin to the 46-kDa form is independent of BE6 expression (data not shown). Although Flag-paxillin was less efficient at supporting BE6 transformation than native paxillin, Flag-paxillin ΔLD1,2,4 did not induce any anchorage-independent colony formation alone or in combination with BE6 (Table 1). The 46-kDa paxillin fully supported BE6 transformation (Table 1).
FIG. 5.
Expression of BE6 and reexpression of paxillin and paxillin mutants in paxillin−/− MEFs. The indicated paxillin mutants were stably retrovirally transduced with or without BE6 (indicated by “+” and “-” signs). Paxillin and tubulin expression was detected in lysates by Western blotting with rabbit anti-paxillin and mouse anti-tubulin, respectively. BE6 was detected by immunoprecipitation from 107 SDS-lysed cells, followed by Western blotting as described in Materials and Methods.
TABLE 1.
Mutational analysis of paxillin in transformation of MEFs by BE6
| Paxillin moleculea | Anchorage-independent colony formationb:
|
|
|---|---|---|
| Without BE6 | With BE6 | |
| Wild-type PXN | - | ++++ |
| Flag-PXN | - | ++ |
| Flag-46 kDa-PXN | - | ++++ |
| Flag-PXN-ΔLD1,2,4 | - | - |
| Flag-PXN 1-341 | - | - |
| Flag-PXN ΔLIM1 | - | - |
| Flag-PXN-ΔLIM4 | - | - |
The paxillin (PXN) mutants previously described (32).
Colony formation in agarose was determined four times as shown in Fig. 3, and average results are presented. Scale: ++++, colony formation >50% that observed by wild-type paxillin; ++, colony formation 25 to 50% that of WT paxillin in three separate assays; −, no colony formation.
Paxillin is required to support the tyrosine phosphorylation of FAK in ES cells (32). In that study, LIM domains 1 to 3 were each required, whereas LIM4 is dispensable. Paxillin molecules deleted of either all of the LIM domains (paxillin 1-341) or containing in-frame deletions of LIM domains 1 and 4 were not able to support anchorage-independent growth induced by BE6 (Table 1). Each of the paxillin mutants shown in Table 1 was expressed, as was BE6, in these cells (Fig. 5).
DISCUSSION
Normal keratinocytes require both soluble growth factors and contact with an extracellular-matrix-coated surface for cell proliferation. In the papillomavirus life cycle, virus-infected cells in the spinous layer that no longer contact matrix aberrantly reenter the cell cycle in order to amplify viral DNA from the low copies found in basal keratinocyte to high copies for the production of virus. In HPVs, this is accomplished through the interaction of the E7 oncoprotein with members of the retinoblastoma family of proteins. Unlike other papillomaviruses, papovaviruses, or adenoviruses, a strong direct interaction between BPV-1 oncoproteins and retinoblastoma family members has not yet been demonstrated. Therefore, it is likely that BPV-1 oncoproteins manipulate signal transduction in order to modify retinoblastoma family proteins and allow for S-phase reentry in spinous epithelial cells that have lost contact with the matrix-rich basement membrane.
We have used the ability of BE6 to induce anchorage-independent cell proliferation as an initial model to study the role of BE6 in this process. The mechanism by which BE6 interaction with paxillin allows for anchorage-independent cell growth is as yet unclear, but a critical analysis of the differences between paxillin (which supports BE6 transformation) and HIC-5 (which is expressed in MEFs but does not support transformation) should be informative.
The 46-kDa paxillin is deleted of major tyrosine phosphorylation sites and Crk-family binding sites at the amino terminus of paxillin (one of the differences between paxillin and HIC-5 [25]), indicating that these binding sites are dispensable for BE6 transformation. Although the BE6-binding LD motifs 2 and 4 of paxillin are required, it is possible, but not proven, that BE6 transforms through competitive interactions between cellular proteins and BE6 for these particular LD motifs. It is significant that paxillin deleted of the BE6 binding motifs does not induce transformation in the absence of BE6 (paxillin ΔLD1,2,4; Table 1), arguing against a simple competitive binding model. Alternatively, it may be that BE6 interaction with paxillin results in a gain of function for the complex that results in transformation, so it is possible that BE6 transformation requires both paxillin and an additional interaction partner(s). Indeed, the interaction of BE6 with paxillin is not sufficient for transformation since BE6 transforms murine C127 cells to anchorage independence but does not readily transform NIH 3T3 cells, despite the expression of paxillin in both cell types (20; S. Vande Pol, unpublished observations). Although it seems less likely, it is also possible that BE6 transformation does not require direct interaction with paxillin at all, but that paxillin could be an essential scaffolding molecule required to propagate a signal that originates from an interaction between BE6 and a different (nonpaxillin) cellular factor.
Attachment-independent signaling and cell cycle progression are a hallmark of malignant cells. It was first observed that primary cell cultures require attachment for cell proliferation, whereas virally transformed cells do not (15, 24), and that attachment-dependent but immortalized cells arrest in G1 upon suspension (18). A close correlation exists between attachment-independent cell proliferation and tumorigenicity in nude mice (11, 23). The association of viral oncoproteins with their cellular targets has provided critical clues for the identification of cellular proteins that regulate signal transduction and the cell cycle. The essential role of paxillin in transformation by BE6 should focus attention upon paxillin and its role in facilitating anchorage-independent cell proliferation, a hallmark of tumorigenicity.
Acknowledgments
This study was supported by NIH grants CA-69292 and CA-120352 to S.V.P.
We thank Elliot Androphy, Jason Chen, and Janet Cross for critical reading of the manuscript.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Bohl, J., K. Das, B. Dasgupta, and S. B. Vande Pol. 2000. Competitive binding to a charged leucine motif represses transformation by a papillomavirus E6 oncoprotein. Virology 271163-170. [DOI] [PubMed] [Google Scholar]
- 2.Bohl, J., B. Hull, and S. B. Vande Pol. 2001. Cooperative transformation and coexpression of bovine papillomavirus type 1 E5 and E7 proteins. J. Virol. 75513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brimer, N., C. Lyons, and S. B. Vande Pol. 2007. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 358303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M. C., M. S. Curtis, and C. E. Turner. 1998. Paxillin LD motifs may define a new family of protein recognition domains. Nat. Struct. Biol. 5677-678. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. C., J. A. Perotta, and C. E. Turner. 1996. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 1351109-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. C., and C. E. Turner. 2004. Paxillin: adapting to change. Physiol. Rev. 841315-1339. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. J., Y. Hong, E. Rustamzadeh, J. D. Baleja, and E. J. Androphy. 1998. Identification of an alpha helical motif sufficient for association with papillomavirus E6. J. Biol. Chem. 27313537-13544. [DOI] [PubMed] [Google Scholar]
- 8.Cote, J. F., C. E. Turner, and M. L. Tremblay. 1999. Intact LIM 3 and LIM 4 domains of paxillin are required for the association to a novel polyproline region (Pro 2) of protein-tyrosine phosphatase-PEST. J. Biol. Chem. 27420550-20560. [DOI] [PubMed] [Google Scholar]
- 9.Das, K., J. Bohl, and S. B. Vande Pol. 2000. Identification of a second transforming region in bovine papillomavirus type 1 E6 and the role of E6 interaction with paxillin, E6BP, and E6AP. J. Virol. 74812-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elston, R. C., S. Napthine, and J. Doorbar. 1998. The identification of a conserved binding motif within human papillomavirus type 16 E6 binding peptides, E6AP and E6BP. J. Gen. Virol. 79371-374. [DOI] [PubMed] [Google Scholar]
- 11.Freedman, V. H., and S. I. Shin. 1974. Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell 3355-359. [DOI] [PubMed] [Google Scholar]
- 12.Hagel, M., E. L. George, A. Kim, R. Tamimi, S. L. Opitz, C. E. Turner, A. Imamoto, and S. M. Thomas. 2002. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell. Biol. 22901-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishibe, S., D. Joly, Z. X. Liu, and L. G. Cantley. 2004. Paxillin serves as an ERK-regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol. Cell 16257-267. [DOI] [PubMed] [Google Scholar]
- 14.Matsuya, M., H. Sasaki, H. Aoto, T. Mitaka, K. Nagura, T. Ohba, M. Ishino, S. Takahashi, R. Suzuki, and T. Sasaki. 1998. Cell adhesion kinase beta forms a complex with a new member, Hic-5, of proteins localized at focal adhesions. J. Biol. Chem. 2731003-1014. [DOI] [PubMed] [Google Scholar]
- 15.McPherson, I., and L. Montagnier. 1964. Agar suspension culture for the selective assay of cells transformed by polyoma virus. Virology 23291-294. [DOI] [PubMed] [Google Scholar]
- 16.Nikolopoulos, S. N., and C. E. Turner. 2000. Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J. Cell Biol. 1511435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolopoulos, S. N., and C. E. Turner. 2001. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J. Biol. Chem. 55. [DOI] [PubMed] [Google Scholar]
- 18.Otsuka, H., and M. J. Moskowitz. 1975. Arrest of 3T3 cells in G1 phase in suspension culture. J. Cell Physiol. 87213-220. [DOI] [PubMed] [Google Scholar]
- 19.Raftopoulou, M., and A. Hall. 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 26523-32. [DOI] [PubMed] [Google Scholar]
- 20.Schiller, J. T., W. C. Vass, and D. R. Lowy. 1984. Identification of a second transforming region in bovine papillomavirus. Proc. Natl. Acad. Sci. USA 817880-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen, Y., G. Schneider, J. F. Cloutier, A. Veillette, and M. D. Schaller. 1998. Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin. J. Biol. Chem. 2736474-6481. [DOI] [PubMed] [Google Scholar]
- 22.Shim, S. R., S. Kook, J. I. Kim, and W. K. Song. 2001. Degradation of focal adhesion proteins paxillin and p130cas by caspases or calpains in apoptotic rat-1 and L929 cells. Biochem. Biophys. Res. Commun. 286601-608. [DOI] [PubMed] [Google Scholar]
- 23.Shin, S., V. H. Freedman, R. Risser, and R. Pollack. 1975. Tumorigenicity of virus transformed cells in nude mice is correlated specifically with anchorage-independent growth in vitro. Proc. Natl. Acad. Sci. USA 724435-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoker, M., C. O'Neill, S. Berryman, and V. Waxman. 1968. Anchorage and growth regulation in normal and virus-transformed cells. Int. J. Cancer 3683-693. [DOI] [PubMed] [Google Scholar]
- 25.Thomas, S. M., M. Hagel, and C. E. Turner. 1999. Characterization of a focal adhesion protein, Hic-5, that shares extensive homology with paxillin. J. Cell Sci. 112181-190. [DOI] [PubMed] [Google Scholar]
- 26.Tong, X., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 944412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tumbarello, D. A., M. C. Brown, S. E. Hetey, and C. E. Turner. 2005. Regulation of paxillin family members during epithelial-mesenchymal transformation: a putative role for paxillin delta. J. Cell Sci. 1184849-4863. [DOI] [PubMed] [Google Scholar]
- 28.Tumbarello, D. A., and C. E. Turner. 2007. Hic-5 contributes to epithelial-mesenchymal transformation through a RhoA/ROCK-dependent pathway. J. Cell Physiol. 211736-747. [DOI] [PubMed] [Google Scholar]
- 29.Turner, C. E., M. C. Brown, J. A. Perrotta, M. C. Riedy, S. N. Nikolopoulos, A. R. McDonald, S. Bagrodia, S. Thomas, and P. S. Leventhal. 1999. Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: a role in cytoskeletal remodeling. J. Cell Biol. 145851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vande Pol, S. B., M. C. Brown, and C. E. Turner. 1998. Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene 1643-52. [DOI] [PubMed] [Google Scholar]
- 31.Wade, R., J. Bohl, and S. Vande Pol. 2002. Paxillin null embryonic stem cells are impaired in cell spreading and tyrosine phosphorylation of focal adhesion kinase. Oncogene 2196-107. [DOI] [PubMed] [Google Scholar]
- 32.Wade, R., and S. Vande Pol. 2006. Minimal features of paxillin that are required for the tyrosine phosphorylation of focal adhesion kinase. Biochem. J. 393565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb, D. J., C. M. Brown, and A. F. Horwitz. 2003. Illuminating adhesion complexes in migrating cells: moving toward a bright future. Curr. Opin. Cell Biol. 15614-620. [DOI] [PubMed] [Google Scholar]
- 34.Webb, D. J., K. Donais, L. A. Whitmore, S. M. Thomas, C. E. Turner, J. T. Parsons, and A. F. Horwitz. 2004. FAK-Src signaling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6154-161. [DOI] [PubMed] [Google Scholar]
- 35.Wehrle-Haller, B., and B. A. Imhof. 2003. Actin, microtubules and focal adhesion dynamics during cell migration. Int. J. Biochem. Cell. Biol. 3539-50. [DOI] [PubMed] [Google Scholar]