Abstract
Sequencing and reversion analysis of murine hepatitis virus (MHV) temperature-sensitive (ts) viruses has identified putative ts mutations in the replicase nonstructural proteins (nsp's) of these coronaviruses. In this study, reverse transcriptase PCR sequencing of the RNA genome of an isolate of the MHV ts virus Alb ts6, referred to as Alb/ts/nsp5/V148A, identified a putative ts mutation in nsp5 (T10651C, Val148Ala), the viral 3C-like proteinase (3CLpro). The introduction of the T10651C mutation into the infectious MHV clone resulted in the recovery of a mutant virus, the nsp5/V148A virus, that demonstrated reduced growth and nsp5 proteinase activity identical to that of Alb/ts/nsp5/V148A at the nonpermissive temperature. Sequence analysis of 40°C revertants of Alb/ts/nsp5/V148A identified primary reversion to Ala148Val in nsp5, as well as two independent second-site mutations resulting in Ser133Asn and His134Tyr substitutions in nsp5. The introduction of the Ser133Asn or His134Tyr substitution into the cloned nsp5/V148A mutant virus background resulted in the recovery of viruses with increased growth fitness and the partial restoration of nsp5 activity at the nonpermissive temperature. Modeling of the nsp5 structure of Alb/ts/nsp5/V148A predicted that the Val148Ala mutation alters residue 148 interactions with residues of the substrate binding S1 subsite of the nsp5 active-site cavity. This study identifies novel residues in nsp5 that may be important for regulating substrate specificity and nsp5 proteinase activity.
Coronaviruses (CoVs) belong to a family of enveloped, positive-strand RNA viruses that cause important diseases in humans and animals. The identification of a novel human CoV as the etiological agent of severe acute respiratory syndrome (SARS) highlights the potential of this virus family to cause severe and acute disease in the human population (13, 18, 36). Additionally, the increasing identification of CoVs in bats and other animals (37), combined with the threat of CoV transspecies transmission, underscores the need to better understand the replication strategies of these viruses in order to identify improved targets for attenuation and interference with replication.
The CoV murine hepatitis virus (MHV) has been used as a model system to investigate the functions of viral proteins in replication and pathogenesis. The MHV genome is approximately 32 kb in size and contains seven genes, with the first encoding the nonstructural proteins (nsp's) and genes 2 to 7 encoding the structural and accessory proteins of the virus. The life cycle of MHV occurs exclusively in the host cell cytoplasm. Following the entry of MHV into a cell, the RNA genome is translated by host cell ribosomes from open reading frame 1 (ORF1), which consists of approximately 22 kb and encodes nsp1 to nsp16 (Fig. 1A). ORF1 is comprised of two ORFs (ORF1a and ORF1ab) that are connected by a −1 ribosomal frameshift element (7, 9, 29, 34). Translation of the ORF1a or the ORF1ab fusion polyprotein (pp1a or pp1ab, respectively) results in 495-kDa (pp1a) or 803-kDa (pp1ab) polyproteins that are processed by three virus-encoded proteinases, including two papainlike proteinase domains (PLP1 and PLP2) in nsp3 and the cysteine proteinase nsp5 (3C-like proteinase [3CLpro] or Mpro [stands for main proteinase], hereinafter referred to as nsp5 or 3CLpro), to yield intermediate and mature forms of nsp1 to nsp16 (4, 7, 16, 25, 33) (Fig. 1B).
FIG. 1.
MHV proteolytic processing of the ORF1ab polyprotein and comparison of MHV and SARS-CoV nsp5 sequences. (A) Schematic of the MHV genome. The approximately 32-kb MHV genome is shown as a line, and the locations of gene 1 (∼22 kb) and genes 2 to 7 (∼9.5 kb) are indicated. Gene 1 is composed of two ORFs (labeled arrows) connected by a −1 ribosomal frameshift element. (B) Schematic of PLP1, PLP2, and nsp5 processing of pp1a and pp1ab. Translation of ORF1a yields a pp1a of nsp1 to nsp10 or nsp11, shown on top as connected boxes with each nsp numbered. Viral proteinases (PLP1, PLP2, and nsp5) are shown as gray boxes. Bent black arrows show cleavage events mediated by the indicated proteinase. The translation of ORF1ab yields a theoretical nsp1-to-nsp16 pp1ab, shown on the bottom as connected boxes with viral proteinases shown in gray. Downstream nsp5 cleavage events are indicated by bent black arrows. (C) Sequence comparison of MHV and SARS-CoV nsp5 amino acid sequences. The MHV nsp5 sequence is shown on the top and the SARS-CoV (SCV) sequence is shown on the bottom. Residues conserved between MHV and SARS-CoV have an asterisk beneath them. Catalytic His41 and Cys145 and the conserved substrate binding S1 subsite residues Tyr161, Met162, and His163 are highlighted in gray. Arrows denote MHV Ser133, His134, and Val148.
The MHV and SARS-CoV nsp5 amino acid sequences share 50% sequence identity (Fig. 1C), and the alignment of group 1, 2, and 3 CoVs with MHV and SARS shows that they all contain the catalytic-dyad residues (His41 and C145 for MHV), as well as the conserved Tyr161-Met162-His163 motif, which comprises the substrate binding S1 subsite of the proteinase active-site cavity (Fig. 1C). MHV and SARS-CoV nsp5s cleave the ORF1ab polyprotein at 11 conserved sites following P2-Leu-Gln sequences. These processing events include the autoproteolytic liberation of nsp5 from the nascent polyprotein (Fig. 1B) (31, 33). It has been demonstrated that continuous expression and processing of the replicase polyprotein is required for ongoing and efficient RNA synthesis during MHV replication (28, 38). Additionally, it has been shown that mutation of the catalytic-dyad His41 or Cys145 residues of MHV nsp5 abolishes the proteolytic activity of the protein (32, 33).
Over the past 30 years, several panels of MHV temperature-sensitive (ts) mutant viruses were established by chemical mutagenesis and were proposed to have ts mutations in the replicase nsp's based on viral complementation studies (41). Recently, sequencing of the replicase genes of several of these previously known ts viruses identified mutations predicted to be responsible for the ts phenotypes (39), one of which has experimentally been confirmed to occur in nsp10 using reverse genetic introduction of the identified mutation into the infectious MHV clone (17). In this study, we report a novel and previously unknown mutation in the MHV nsp5 coding region of ORF1a (T10651C, Val148Ala) that is responsible for the ts phenotype of an isolate of Alb ts6, Alb/ts/nsp5/V148A. When this single-nucleotide change was engineered into the infectious MHV clone, the recovered recombinant nsp5/V148A mutant virus displayed ts replication defects associated with decreased nsp5 proteinase activity at the nonpermissive temperature. We confirmed two independent second-site reversion mutations in nsp5 that partially restored growth fitness and nsp5 proteinase activity at the nonpermissive temperature. Modeling of the nsp5 structures of MHV wild-type (WT), nsp5 ts, and nsp5 revertant viruses predicted that residue 148 of nsp5 interacts with and may help stabilize the substrate binding S1 subsite of the nsp5 active-site cavity.
MATERIALS AND METHODS
Viruses, cells, and antisera.
Recombinant WT MHV strain A59 (GenBank accession no. AY910861 [15, 40]) was used as the WT control for all experiments. An isolate of Alb ts6 from 29 June 1989, labeled Al6 R.V. 6/89 (R.V. stands for recovered virus), was kindly provided by Paul Masters (Wadsworth Center, New York State Department of Health) (41). Delayed brain tumor cells selected for high-level expression of the MHV receptor, carcinoembryonic antigen cell adhesion molecule 1 (DBT-9) (12, 23, 44), and baby hamster kidney 21 cells expressing the MHV receptor (BHK-MHVR cells) (11, 12, 44) were grown in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS) for all experiments. Medium for BHK-MHVR cells was supplemented with G418 (0.8 mg/ml) to maintain selection for cells expressing the MHV receptor. All polyclonal antisera used for biochemical experiments have previously been described. These antisera include guinea pig anti-nsp1 antiserum (GP3) (10) and rabbit anti-nsp8 antiserum (VU123) (8).
Sequence analysis of Alb/ts/nsp5/V148A.
The Alb/ts/nsp5/V148A genome was sequenced using a strategy previously described (40). The genome sequence was compared to the WT strain from which Larry Sturman originally isolated the Alb series of ts mutants (GenBank accession no. AY700211 [41]). Briefly, to obtain total intracellular RNA for sequence analysis, cells in 25-cm2 flasks were infected at 30°C using a multiplicity of infection (MOI) of ≤0.1 PFU/cell. When >10% of the cells were involved in syncytia, total intracellular RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol and used as template for reverse transcriptase PCR (RT-PCR). Reverse transcription was performed using SuperScript III RT (Invitrogen) and random hexamers at 55°C for 60 min. The Alb/ts/nsp5/V148A cDNA was then PCR amplified using Easy-A high-fidelity PCR cloning enzyme (Stratagene) or Elongase (Invitrogen) DNA polymerase and oligonucleotides specific for the MHV genome (NCBI accession no. 1NC001846). Sequencing primers were created every 600 bp, beginning with the 5′ end, from both the sense and antisense strands. These primers generated overlapping sequences spanning the cDNA at least twice. Amplicons were gel purified and directly analyzed by automated DNA sequencing. Sequences of PCR and sequencing primers are available upon request.
Reversion analysis of Alb/ts/nsp5/V148A.
Alb/ts/nsp5/V148A was grown overnight at 40°C, and at 24 h postinfection (p.i.), virus was collected from the culture supernatant. Total intracellular RNA for sequence analysis was obtained from cells in 25-cm2 flasks that were infected at an MOI of ≤0.1 PFU/cell at 40°C. When >10% of the cells were involved in syncytia, total intracellular RNA was extracted using TRIzol reagent (Invitrogen), as described above, and used as a template for RT-PCR. Reverse transcription was performed using SuperScript III RT (Invitrogen) and random hexamers at 55°C for 60 min. The genome was sequenced across the four genomic regions containing the nonsynonymous mutations identified in Alb/ts/nsp5/V148A. The nsp3 coding region was amplified using PCR with primers corresponding to nucleotides (nt) 4232 to 4248 (sense) and nt 5913 to 5929 (antisense). The nsp5 coding region was amplified using PCR with primers corresponding to nt 10157 to 10176 (sense) and nt 11782 to 11798 (antisense). The nsp7 coding region was amplified using PCR with primers corresponding to nt 11330 to 11347 (sense) and nt 13345 to 13361 (antisense). The nucleocapsid (N) coding region was amplified using PCR with primers corresponding to nt 28729 to 28746 (sense) and nt 30129 to 30145 (antisense).
Mutagenesis of MHV cDNA C fragment plasmids.
The entire MHV-A59 genome was cloned as seven cDNA fragments, A through G, each in a separate plasmid (44). The C fragment plasmid (pCR-XL-pSMART C), designed for the recovery of WT virus VUSS3 (40), was used as a template for site-directed-PCR-based mutagenesis to introduce the nsp5/Val148Ala substitution into the MHV genome. Overlapping sense and antisense primers for nsp5/V148A (Table 1) were used to generate mutant plasmids. To introduce the nsp5/Ser133Asn and nsp5/His134Tyr substitutions into the MHV genome, the C fragment plasmid was used as a template. To engineer nsp5/Ser133Asn/Val148Ala and nsp5/His134Tyr/Val148Ala substitutions into the MHV genome, the nsp5/V148A mutant plasmid was used as a template for PCR-based mutagenesis. Overlapping sense and antisense primers for nsp5/Ser133Asn and nsp5/His134Tyr (Table 1) were used to generate mutant plasmids. The sequences of all mutant fragments were verified.
TABLE 1.
Oligodeoxynucleotides used for the mutagenesis of MHV cDNA C fragment plasmids
| Primer target | Polarity | Sequencea |
|---|---|---|
| nsp5/V148A | Sense | 5′-AGT TAA AAC ATA TCC TGC AGA ACC GCA GGA TCC-3′ |
| Antisense | 5′-GGA TCC TGC GGT TCT GCA GGA TAT GTT TTA ACT-3′ | |
| nsp5/S133N | Sense | 5′-GTT ACG CTT CGT AGT AAC CAT ACC ATA AAG GGC-3′ |
| Antisense | 5′-GCC CTT TAT GGT ATG GTT ACT ACG AAG CGT AAC-3′ | |
| nsp5/H134Y | Sense | 5′-ACG CTT CGT AGT AGC TAT ACC ATA AAG GGC TCC-3′ |
| Antisense | 5′-GGA GCC CTT TAT GGT ATA GCT ACT ACG AAG CGT-3′ |
Underlined nucleotides represent nucleotides altered from the WT sequence.
Generation of MHV nsp5 mutant viruses.
Viruses containing the engineered mutations within nsp5 were produced by using the infectious cDNA assembly strategy for MHV-A59 described by Yount et al. (44) and modified by Denison et al. (15). Briefly, plasmids containing the seven cDNA cassettes of the MHV genome were digested using the appropriate restriction enzymes. Gel-purified restriction fragments were ligated together in a total reaction volume of 50 μl overnight at 16°C. Following the chloroform extraction and isopropanol precipitation of ligated DNA, full-length transcripts of MHV cDNA were generated in vitro using the mMessage mMachine T7 transcription kit (Ambion) and by following the manufacturer's protocol with modifications. Fifty-microliter reaction mixtures were supplemented with 7.5 μl of 30 mM GTP, and transcription was performed at 40.5°C for 30 min, 37°C for 60 min, 40.5°C for 30 min, 37°C for 30 min, and 40.5°C for 30 min. In parallel, transcripts encoding the MHV N protein were generated from N cDNA. To prepare for electroporation, BHK-MHVR cells were grown to subconfluence, trypsinized, and then washed twice with phosphate-buffered saline (PBS) and resuspended in PBS at a concentration of 1 × 107 cells/ml. From the resuspended cell solution, a 600-μl aliquot of cells was added to N and MHV transcripts in a 4-mm-gap electroporation cuvette. Three pulses of 850 V at 25 μF were delivered to the cuvette with a Bio-Rad Gene Pulser II electroporator. Electroporated cells were then laid over a layer of 1 × 106 uninfected DBT-9 cells in a 75-cm2 flask and incubated at 30°C and 40°C. Virus viability was determined by its cytopathic effect (syncytium formation) in the electroporated cell culture. Progeny virus in the culture medium of electroporated cells (passage 0 [P0]) was passed onto uninfected DBT-9 cells (P1 cells), and the virus released in the culture medium was designated P1 stock. For each recovered virus, RNA harvested from P1 cells was RT-PCR amplified and sequenced across the nsp5 coding region, using PCR with primers corresponding to nt 10157 to 10176 (sense) and nt 11782 to 11798 (antisense) to verify the sequence.
Viral growth analysis.
WT and nsp5 mutant viruses were analyzed for replication by using growth assays as previously described (15). Briefly, DBT-9 cells in 60-mm dishes were infected with WT or nsp5 mutant viruses in growth medium at an MOI of 0.1 PFU/cell. Following a 30-min adsorption at room temperature, the inoculum was removed, and cells were washed three times with PBS and supplied with prewarmed 10% FCS-DMEM. Cells were then incubated at either 30°C or 40°C. Samples of media were collected from 1 to 36 h p.i., and virus titers were determined by plaque assay at 30°C as described previously (28). To enhance the visualization of plaques, cells under an agar growth medium overlay were fixed with 4% formaldehyde at room temperature. The overlay was then discarded, and the fixed cell monolayer was air dried before plaques were counted.
Immunoprecipitation of radiolabeled viral proteins.
For constant-temperature experiments, confluent monolayers of DBT-9 cells in 60-mm cell culture dishes were either infected with virus at an MOI of 5 PFU/cell or mock infected with 10% FCS-DMEM at either 30°C or 40°C. At 5.5 h p.i., the medium was replaced with 10% FCS-DMEM lacking methionine and cysteine and supplemented with 5 μg/ml actinomycin D. Cells were incubated from 6 to 8 h p.i. with 100 μCi/ml of [35S]methionine-cysteine (Translabel; ICN). At 8 h p.i., cells were lysed in 500 μl of lysis buffer (150 mM sodium chloride [NaCl], 1% NP-40, 0.5% sodium deoxycholate, 50 mM Tris [pH 8.0]). Lysates were then centrifuged at 3,500 × g for 5 min to remove cell nuclei, and the supernatant was retained as the post-nuclear-removal lysate. For temperature shift experiments, confluent monolayers of DBT-9 cells in 60-mm dishes were either infected at an MOI of 5 PFU/cell or mock infected with 10% FCS-DMEM at 30°C. At 20 h p.i., the cells were shifted to 40°C. At 23.5 h p.i., the medium was replaced with 10% FCS-DMEM lacking methionine and cysteine and supplemented with 5 μg/ml actinomycin D. Cells were incubated from 24 to 26 h p.i. with 100 μCi/ml of [35S]methionine-cysteine (Translabel; ICN). At 26 h p.i., cells were lysed and post-nuclear-removal lysates were generated, as described above. Immunoprecipitations were performed in a final volume of 1 ml, using protein A-Sepharose beads (Sigma), 50 μl of radiolabeled lysate (derived from approximately 1 × 106 cells), and a 1:500 dilution of polyclonal antiserum in lysis buffer. Following an overnight incubation at 4°C, protein-bead conjugates were pelleted by centrifugation at 12,000 × g for 1 min and washed in low-salt lysis buffer (lysis buffer with 150 mM NaCl) followed by high-salt lysis buffer (lysis buffer with 1 M NaCl) and a final low-salt wash. After being rinsed, proteins were eluted by the addition of 2× lithium dodecyl sulfate buffer with 1× dichlorodiphenyltrichloroethane (NuPAGE; Invitrogen) to the beads, which were heated at 70°C for 10 min prior to electrophoresis. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 4 to 12% polyacrylamide gradient Bis-Tris gels (NuPAGE; Invitrogen) and analyzed by fluorography. The 14C-labeled high-molecular-weight standard (New England Biolabs) and full-range rainbow marker (RPN 800; Invitrogen) were used as molecular weight standards.
Modeling of MHV nsp5 structures.
The crystal structure coordinates of SARS 3CLpro (Protein Data Bank [PDB] code 2H2Z, chain A) (42) were used as a template to generate a homology model of MHV nsp5 using the online program 3D-Jigsaw in the automatic mode (5, 6, 14). The root mean square distance (RMSD) between the template and the model was determined to be 0.708 angstroms, using the molecular-modeling tool Chimera (35). The MHV nsp5 model was then used as a template for introducing the ts genotype of the nsp5/V148A virus as well as the genotypes of both of the second-site revertant mutants, the nsp5/S133N/V148A and nsp5/H134Y/V148A viruses, using the RosettaDesign Web server (http://rosettadesign.med.unc.edu/) (30). In each case, the MHV nsp5 model was analyzed using the molecular-modeling tool MacPyMOL (DeLano Scientific) to determine which amino acids were proximal to the amino acid(s) being targeted for replacement. Briefly, each amino acid to be altered was highlighted, and all other amino acids within an interaction distance of 5 angstroms were identified. Using the RosettaDesign website, the amino acid replacements were incorporated, and all amino acids within the 5-angstrom interaction distance were relaxed to allow the program to repack the side chains to an optimal energetic state. This process was repeated with each mutation and each series of mutations. Ten models were generated for each set of mutations, and the best model, based on the lowest energy score, was selected and further evaluated using MacPyMOL. In addition, an online version of the program WHAT_IF (24) was used to identify the optimal hydrogen bonding networks for the MHV nsp5 model and the nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A models.
RESULTS
Genome sequence analysis of Alb/ts/nsp5/V148A.
Previous sequence analysis of the MHV ts virus Alb ts6 identified the mutation A9494C in the nsp4 coding sequence as a candidate ts mutation (39). Because only the replicase gene was sequenced in that study, we sought to determine if additional mutations were present in genes 2 to 7 that might contribute to the ts phenotype of Alb ts6 prior to reverse genetic testing of the nsp4 mutation. We obtained an original isolate of Alb ts6, Alb/ts/nsp5/V148A. DBT-9 cells were infected with Alb/ts/nsp5/V148A at 30°C, RNA was harvested, and the complete viral genome was obtained by RT-PCR sequence analysis. The results of genome sequencing identified numerous synonymous mutations (noncoding or untranslated-region mutations) and four nonsynonymous mutations (Fig. 2). Three of the nonsynonymous mutations were located in regions of ORF1a encoding nsp3 (C5265A, Leu854Met), nsp5 (T10651C, Val148Ala), and nsp7 (C12225T, Arg83Cys). The fourth mutation was located in the N gene (G29690C, Glu8Gln). We did not detect an A9494C mutation (Asn258Thr) in the nsp4 coding sequence. These results indicate that the Alb ts6 isolate that we analyzed, Alb/ts/nsp5/V148A, is a virus different than the Alb ts6 virus described by Sawicki et al. (39).
FIG. 2.
Nonsynonymous mutations identified in Alb/ts/nsp5/V148A. (A) The first column shows the MHV protein containing the mutation. The second column shows the genome nucleotide number of the mutation. The third column shows the codon and corresponding amino acid (aa) change in the protein. The last column indicates the reversion of the nonsynonymous change following growth of the virus at 40°C. The amino acid number refers to the amino acid number within the indicated MHV protein. (B) The schematic shows MHV ORF1ab, with individual nsp's labeled as rectangular boxes 1 to 16. PLP1 and PLP2 in nsp3 are shown as stippled gray lines. Labeled arrows point to regions in the proteins where nonsynonymous changes were identified. The change in the N protein is not shown.
Identification of a novel ts mutation in MHV nsp5.
To identify the mutation(s) responsible for the ts phenotype of Alb/ts/nsp5/V148A, DBT-9 cells were infected with Alb/ts/nsp5/V148A at the nonpermissive temperature (40°C), and the virus was harvested at 24 h p.i. A plaque assay was performed at 40°C on the recovered virus, and 10 virus plaques were picked. Plaques were then expanded at 40°C in T25 flasks, and total cellular RNA was obtained. Each isolated virus was RT-PCR sequenced across the regions of the genome that contained nonsynonymous mutations in Alb/ts/nsp5/V148A. The mutations identified in nsp3, -7, and -14 and N were maintained in all 10 plaque isolates. Sequencing across nsp5 of the plaque isolates revealed either reversion to the WT sequence (C10651T, Ala148Val) or the continued presence of the T10651C mutation and an additional mutation in nsp5 at either G10606A (Ser133Asn) or C10608 (His134Tyr) (Table 2). Together, these sequencing results identified the single T10651C mutation in nsp5 as a candidate ts mutation and two additional independent mutations in nsp5 as candidate second-site revertant mutations.
TABLE 2.
Determination of nsp5 mutant virus ts phenotype
| Virus strain | Amino acid at position:
|
Titer at:a
|
EOPb | |||
|---|---|---|---|---|---|---|
| 133 | 134 | 148 | 30°C | 40°C | ||
| WT | Ser | His | Val | 9.0 × 107 | 3.0 × 108 | 3.3 |
| Alb/ts/nsp5/V148A | Ser | His | Ala | 2.8 × 108 | 8.5 × 104 | 3 × 10−4 |
| nsp5/V148A virus | Ser | His | Ala | 1.0 × 108 | 2.0 × 103 | 2 × 10−5 |
| nsp5/S133N virus | Asn | His | Val | 1.8 × 106 | 5.5 × 106 | 3.1 |
| nsp5/H134Y virus | Ser | Tyr | Val | 6.5 × 105 | 2.0 × 106 | 3.1 |
| nsp5/S133N/V148A virus | Asn | His | Ala | 2.8 × 107 | 3.8 × 107 | 1.4 |
| nsp5/H134Y/V148A virus | Ser | Tyr | Ala | 4.0 × 106 | 4.3 × 106 | 1.1 |
Values for titers indicated are in PFU/ml.
EOPs are titers at 40°C divided by titers at 30°C.
To test the phenotypes of the candidate MHV nsp5 ts and second-site revertant mutations, we used reverse genetics to engineer each mutation into the infectious MHV clone, either alone or in combination. Each virus was named on the basis of its nsp, with “nsp5” followed by a forward slash and the single-letter amino acid substitution(s), as follows: nsp5/V148A, nsp5/S133N, nsp5/H134Y, nsp5/S133N/V148A, and nsp5/H134Y/V148A. The mutant viral genomes were electroporated into cells, and cells were incubated at 30°C and monitored for the formation of virus-induced syncytia. All five mutant viruses (the nsp5/V148A, nsp5/S133N, nsp5/H134Y, nsp5/S133N/V148A, and nsp5/H134Y/V148A viruses) were recovered at 30°C, demonstrating that the mutations introduced into the MHV genome were not lethal for virus replication. Viruses harvested from electroporated cells (P0) were expanded at 30°C in T150 flasks to generate the P1 stocks that were used in all subsequent experiments. Each recovered virus was sequenced to verify that the introduced mutations were maintained in the genome and that no additional mutations were present in the nsp5 coding region.
To determine if the nsp5 mutant viruses were ts, plaque assays were performed on DBT-9 cells using WT and nsp5 mutant viruses at permissive (30°C) and nonpermissive (40°C) temperatures. Following these plaque assays, the efficiency of plating (EOP) values were calculated (Table 2). The EOP for WT virus was 3.3, confirming that there was no impairment of growth at 40°C. In contrast, the recombinant Alb/ts/nsp5/V148A virus had an EOP of 3 × 10−4, demonstrating a ts defect similar to that originally reported for Alb ts6 (41). The engineered nsp5/V148A virus had an EOP of 2 × 10−5, indicating that the single T10651C mutation was sufficient to confer the ts phenotype on the virus. The nsp5/S133N and nsp5/H134Y mutant viruses had EOPs of 3.1, very close to the EOP calculated for the WT virus. These results demonstrate that the Ser133Asn and His134Tyr mutations in nsp5 are not detrimental to virus growth and do not confer a ts phenotype on the virus. The nsp5/S133N/V148A and nsp5/H134Y/V148A mutant viruses had EOPs of 1.4 and 1.1, respectively, showing that the Ser133Asn and His134Tyr mutations were each capable of compensating for the ts defect caused by Val148Ala in this assay. These results provide experimental confirmation that the T10651C mutation in nsp5 is responsible for the ts phenotype of Alb/ts/nsp5/V148A. Additionally, the results indicate that the presence of either Ser133Asn or His134Tyr in the nsp5/V148A virus is sufficient to compensate for the ts defect at the nonpermissive temperature. Because no significant virus growth defects were observed for the nsp5/S133N or nsp5/H134Y virus, all subsequent studies were performed with WT, Alb/ts/nsp5/V148A, nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A viruses.
Growth analysis of nsp5 mutant viruses in culture.
Although the Ser133Asn and His134Tyr substitutions were individually capable of compensating for the Val148Ala ts defect as measured by EOP, the nsp5/S133N/V148A and nsp5/H134Y/V148A mutant viruses still had EOPs two- to threefold less than those of WT MHV and the nsp5/S133N and nsp5/H134Y mutant viruses. These results suggest that neither second-site mutation completely compensated for the Val148Ala ts defect. To further analyze the replication characteristics of the nsp5 mutant viruses and identify subtle replication defects, DBT-9 cells were infected with the WT, Alb/ts/nsp5/V148A, nsp5/V148A, nsp5/S133N/V148A, or nsp5/H134Y/V148A virus at an MOI of 0.01 PFU per cell at 30°C and 40°C. Samples of infected cell culture media were obtained at various times from 1 to 36 h p.i., and virus titers in each sample were determined by plaque assay at 30°C (Fig. 3).
FIG. 3.
Growth analysis of WT, ts, and revertant viruses. (A) Virus growth at 40°C. DBT-9 cells were infected with the indicated viruses at an MOI of 0.01 PFU/cell. Samples of virus supernatants were obtained at 1, 4, 8, 12, 16, 24, and 36 h p.i., and virus titers were determined by plaque assay at 30°C on DBT-9 cells. (B) Virus growth at 30°C. DBT-9 cells were infected with the indicated viruses at an MOI of 0.01 PFU/cell. Samples of virus supernatants were obtained at 1, 4, 8, 12, 16, 24, and 36 h p.i., and virus titers were determined by plaque assay at 30°C on DBT-9 cells. (V148A, nsp5/V148A virus; S133N/V148A, nsp5/S133N/V148A virus; H134Y/V148A, nsp5/H134Y/V148A virus).
Growth curves of viruses examined at 40°C mirrored the ts defect observed during the plaque reduction assays (Fig. 3A). The WT virus reached peak titers at 8 h p.i., at which time the cell monolayer was largely destroyed, and virus titers gradually declined. Growth of the Alb/ts/nsp5/V148A and nsp5/V148A ts mutant viruses was below the limits of detection of the assay until after 8 h p.i., consistent with a complete ts block in replication. After 8 h p.i., growth became exponential and reached titers of approximately 107 PFU/ml by 36 h p.i., suggesting the emergence of revertant viruses. Virus samples obtained at 24 h p.i. for Alb/ts/nsp5/V148A and the nsp5/V148A virus were sequenced across the nsp5 coding region to examine this possibility. In both cases, primary-site revertants (C10651T virus) were identified. These results confirm the ts and reversion phenotypes of Alb/ts/nsp5/V148A and the nsp5/V148A virus. The Ser133Asn and the His134Tyr mutations in the nsp5/V148A virus backbone restored the growth fitness of the ts virus at the nonpermissive temperature, but incompletely and to different extents (Fig. 3A). The nsp5/H134Y/V148A mutant virus grew with growth kinetics similar to those of the WT virus but showed a lag in growth beginning at 4 h p.i. and did not reach peak titer until 12 h p.i., at which point the cell monolayer was largely destroyed. The nsp5/S133N/V148A mutant virus reached titers 10-fold greater than those of Alb/ts/nsp5/V148A or the nsp5/V148A virus at 4 and 8 h p.i., at least 100-fold less than those of the nsp5/H134Y/V148A virus, and approximately 1,000-fold less than those of the WT virus at these same time points. Interestingly, there was exponential growth of the nsp5/S133N/V148A virus beginning between 8 and 16 h p.i., at which point the cell monolayer was destroyed. These results show that the Ser133Asn and His134Tyr mutations are each capable of compensating for the Val148Ala ts defect at the nonpermissive temperature. However, the second-site revertant viruses remain less fit than the WT virus, arguing that the Ser133Asn and His134Tyr substitutions do not completely repair the replication defect of the nsp5/V148A virus. In addition, the late change to exponential growth for both the nsp5/S133N/V148A and nsp5/H134Y/V148A viruses suggests that there remains significant pressure for additional compensating mutations in locations yet to be determined.
Growth curves of all viruses tested at 30°C had similar patterns of multiple-cycle replication following infection at a low MOI (Fig. 3B). This growth pattern manifested as exponential growth until about 4 to 8 h p.i., followed by a plateau in growth until 12 h p.i. and subsequent resumption of exponential growth. Though the growth curves had similar patterns, the times and extents of peak titer varied for each of these viruses. The WT virus grew to the highest titers of all viruses tested, achieving titers greater than 108 PFU/ml by 24 h p.i. Alb/ts/nsp5/V148A, the nsp5/V148A virus, and the nsp5/H134Y/V148A virus all grew with similar patterns of growth, showing growth reduced approximately 10-fold in comparison to that of the WT virus at up to 24 h p.i. but increasing to approximately 108 PFU/ml at 36 h p.i. Over the course of the experiment, the nsp5/S133N/V148A mutant virus grew with titers reduced 10- to 100-fold in comparison to those of the WT virus. These results indicate that the V148A substitution in nsp5 has subtle replication defects, even at the permissive temperature. Further, the compensating second-site changes S133N and H134Y that restore growth fitness to the nsp5/V148A virus at the nonpermissive temperature render the revertant viruses less fit than either the WT or the nsp5/V148A virus at the permissive temperature. Whether the ts and constitutive defects result from the same mechanism or different mechanisms remains to be determined.
Replicase protein expression and processing of nsp5 mutant viruses.
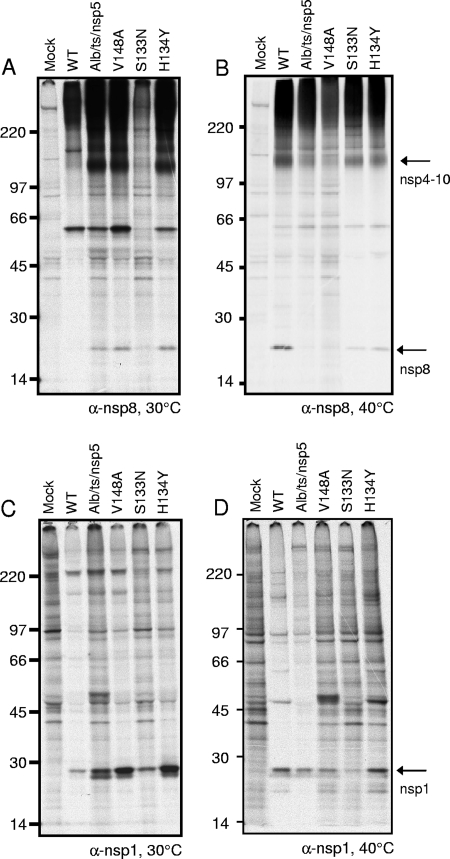
MHV nsp1 to nsp3 of the replicase polyprotein are processed by PLP1 and PLP2 in nsp3, and nsp4 to nsp16 are processed by nsp5 (3CLpro). To test the activities of nsp3 (PLP1 and PLP2) and nsp5 proteinases, DBT-9 cells were infected with the WT or nsp5 mutant virus at 30°C and radiolabeled from 6 to 9 h p.i. with [35S]Met-Cys, and cell lysates were immunoprecipitated with antibodies specific for nsp8, a viral replicase protein that is cleaved from pp1a by nsp5. At 30°C, anti-nsp8 immunoprecipitated the 22-kDa nsp8 protein from lysates of cells infected with the Alb/ts/nsp5/V148, nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A viruses but not from mock-infected cell lysates (Fig. 4A), demonstrating that nsp5 processing activity is intact at the permissive temperature for the mutant viruses. The 22-kDa nsp8 band appeared very faint in WT-infected cell lysates. This result was consistent across four individual experiments. Additionally, the 150-kDa precursor protein, nsp4 to nsp10 (22, 26, 27), was detected in all virus-infected cell lysates, confirming the translation of pp1a. Interestingly, a distinct band of unknown origin of approximately 59 kDa, seen only in the 30°C immunoprecipitations, was detected in all infected cell lysates. To test the activity of the nsp3 proteinase PLP1, antiserum specific for nsp1 was used to immunoprecipitate the 28-kDa nsp1 protein from cell lysates. Because nsp1 is cleaved from the polyprotein by PLP1, the presence of a cleavage product in the ts viruses at the nonpermissive temperature would demonstrate that the viruses were replicating and that PLP1 was active. The results showed that anti-nsp1 immunoprecipitated nsp1 from the lysates of cells infected with the WT, Alb/ts/nsp5/V148A, nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A viruses but not from mock-infected cell lysates (Fig. 4C), confirming the intact function of PLP1 in all viruses tested at 30°C. The presence of an nsp1 doublet was observed in several of the viruses, suggesting either alternative cleavage products of nsp1 or the presence of posttranslational modifications.
FIG. 4.
Activity of PLP1 and nsp5 (3CLpro) in processing replicase nsp1 and nsp8. Cytoplasmic lysates were generated from radiolabeled DBT-9 cells that were either mock infected or infected with the WT, Alb/ts/nsp5/V148A (Alb/ts/nsp5), nsp5/V148A (V148A), nsp5/S133N/V148A (S133N), or nsp5/H134Y/V148A (H134Y) virus at 30°C with labeling from 26 to 28 h p.i. (A and C) or at 30°C for 24 h followed by a shift to 40°C from 24 to 28 h p.i. with labeling from 26 to 28 h p.i. (B and D). Labeled proteins were immunoprecipitated with antiserum specific for nsp8 (A and B) or nsp1 (C and D), as indicated below each gel. Proteins were resolved by SDS-PAGE in 4 to 12% polyacrylamide gradient gels and visualized using fluorography. The numbers on the left side of the gel represent molecular mass markers (in kDa), and arrows to the right of gels point to the indicated proteins. α, anti.
It was not possible to determine the processing of the nsp5 ts viruses in cells infected at 40°C because viral protein products could not be detected. To access processing at the nonpermissive temperature, cells were infected at 30°C and then shifted to 40°C at 24 h p.i. Viral proteins were metabolically radiolabeled from 26 to 28 h p.i. with [35S]Met-Cys, and at 28 h p.i., cell lysates were harvested and immunoprecipitated with antibodies specific for nsp8 or nsp1. When lysates were immunoprecipitated with anti-nsp8, mature 22-kDa nsp8 was detected in WT-infected cell lysates following the shift to 40°C (Fig. 4B). Additionally, a 150-kDa band consistent with the known nsp4-to-nsp10 precursor was detected in the WT-infected cell lysates but not in mock-infected cell lysates. In contrast, immunoprecipitation with anti-nsp8 of lysates from cells infected with the Alb/ts/nsp5/V148A, nsp5/V148A, nsp5/S133N/V148A, or nsp5/H134Y/V148A virus detected various nsp4 to nsp10 and nsp8 band intensities following the temperature shift to 40°C. Minimal nsp4 to nsp10 and nsp8 were detected from cell lysates infected with Alb/ts/nsp5/V148A or the nsp5/V148A virus compared to the WT levels of nsp4 to nsp10 and nsp8. Cell lysates infected with the nsp5/S133N/V148A or nsp5/H134Y/V148A virus showed nsp4 to nsp10 and nsp8 at intensities less than those of WT-infected cell lysates but greater than those of Alb/ts/nsp5/V148A- or nsp5/V148A virus-infected cell lysates. As seen at 30°C, a band of approximately 59 kDa was also detected in each virus-infected cell lysate but not in the mock-infected cell lysate (Fig. 4B). It is unknown whether this band represents a cellular or viral protein. Immunoprecipitation using anti-nsp1 detected the mature 28-kDa nsp1 protein in lysates of cells infected with the WT, Alb/ts/nsp5/V148A, nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A viruses, and nsp1 was not detected in mock-infected cell lysates (Fig. 4D). The detection of nsp1 in all virus-infected lysates demonstrates that at the nonpermissive temperature, nsp's are capable of being translated from ORF1a and that the PLP1 function of nsp3 remains intact in each of the nsp5 mutant viruses following the shift to 40°C.
Modeling of MHV nsp5, nsp5/V148A, nsp5/V148A/S133N, and nsp5/V148A/H134Y.
To define the locations of the nsp5 mutations identified in this study in the MHV nsp5 protein structure, homology models of nsp5 were generated for WT MHV, using the program 3D-Jigsaw (5, 6) (Fig. 5). The mutations from nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A were added to this model by using the online program RosettaDesign (Fig. 5). The backbone of the SARS-CoV nsp5 structure (PDB code 2H2Z) (42) was then compared to the structure in the homology model of MHV nsp5 by using the MatchMaker tool under the structure comparison section of Chimera, resulting in an RMSD of 0.743 Å. This low RMSD value indicates that the structural identity between MHV nsp5 and SARS-CoV nsp5 is high. The homology models of WT MHV nsp5, nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A were then analyzed and compared.
FIG. 5.
Structural models of MHV nsp5s from WT, Alb/ts/nsp5/V148A, nsp5/S133N/V148A, and nsp5/H134Y/V148A viruses. A homology model of the MHV nsp5 protein was generated with the program 3D-Jigsaw using the coordinates of the X-ray crystal structure of SARS-CoV nsp5 (PDB accession no. 2H2Z) as a template. RosettaDesign was used to model the remaining mutations. (A) The MHV nsp5 model contains three domains shown as follows: domain I, red; domain II, blue; and domain III, gray. The substrate binding region is orange, and the catalytic dyad is green. Val148, Ser133, and His134 are shown in yellow. (B) Expanded view of the putative active-site cavity of this structural model shows that the catalytic dyad consists of His41 and Cys145, which are green. Critical substrate binding residues Tyr161, Met162, and His163 are orange. ts residues, Val148, and revertant sites Ser133 and His134 are yellow. (C to F) ts and second-site mutations in MHV nsp5. All residues are colored as for panel B, with nsp5 orientation shifted slightly to show interactions and changes associated with ts mutation and second-site revertants. Domain I and domain II colors are faded to emphasize areas of interest. (C) Substrate binding pocket of WT MHV nsp5. Predicted electron density maps, hydrophobic interactions, and side chain positions of Val148 (yellow) and Met162 (orange) proteins are indicated. (D) Substrate binding pocket of nsp5/V148A. Predicted electron density maps, hydrophobic interactions, and side chain positions of Ala148 (yellow) and Met162 (orange) proteins are indicated, depicting the loss of the predicted hydrophobic interaction between residue 148 and Met162. The model of the substrate binding pocket of the nsp5/S133N/V148A virus (E) and the nsp5/H134Y/V148A virus (F) are shown. Domain II backbone residues of the nsp5/V148A virus are pale blue, and S133N (E) and H134Y (F) substitutions are yellow. In both panels, predicted amino acid positional changes are dark blue.
Like other CoV 3C-like proteinases, MHV nsp5 is predicted to contain three domains, I to III (Fig. 5A). His41 and Cys145, which make up the catalytic dyad of the proteinase, are located in domains I and II, respectively. The active-site cavity, where substrates are bound and the catalytic dyad mediates cleavage, is formed by the interaction of domain I and domain II. The conserved Tyr161, Met162, and His163 residues are found in domain II in the active-site cavity and comprise the substrate binding S1 subsite (Fig. 5B) (1). The nsp5/Val148 residue is conserved between MHV and SARS-CoV and, based on the MHV nsp5 homology model, is packed 3.91 Å from Met162 (Fig. 5C). This close proximity of Val148 to Met162 predicts that hydrophobic interactions occur between the side chains of the two amino acids. Modeling of the Ala148 mutation encoded by Alb/ts/nsp5/V148A and the nsp5/V148A virus does not predict major disruptions in protein structure or organization (Fig. 5D). Rather, Val148Ala is predicted to alter or limit the hydrophobic interactions of residue 148 with Met162, resulting in additional rotameric positions of Met162. Overall, the model suggests that Val148 stabilizes the position of Met162 in the S1 subsite of the active-site cavity.
The Ser133 and His134 residues are not predicted to be found within or adjacent to the active-site cavity of nsp5 but rather are predicted to be located in domain II on the opposing side of the Tyr161-Met162-His163 motif from Val148 (Fig. 5B, E, and F). Superimposition of the Ser133Asn and His134Tyr substitutions onto the nsp5/V148A model predicts that Ser133Asn and His134Tyr do not interact directly or indirectly with the catalytic-dyad His41/Cys145 residues, the substrate binding Tyr161-Met162-His163 residues, or residue Val148Ala. In addition, there was no predicted propagation of structural changes into the active-site cavity of the second-site revertant viruses. Modeling of the His134Tyr mutation in nsp5/V148A predicts only subtle alteration at residues Leu130, Ala131, Ser132, His172, and Tyr182 (Fig. 5F). Interestingly, His172 has been identified as a crucial residue required for the stabilization of the interaction between the S1 subsite binding residue His163 and the Gln residue in position 1 of the substrate (43). In contrast, modeling of the Ser133Asn mutation predicted no interactions or changes proximal to the active-site cavity (Fig. 5E). Instead, modeling predicted side chain modifications of Ala131, Val195, Gly196, and Ala197, which are directed away from the active-site cavity. Virus replication and protein processing of the nsp5/S133N/V148A virus were more impaired than those of the nsp5/H134Y/V148A virus, suggesting that modifications induced by Ser133Asn compensate the nsp5/Val148Ala defect less efficiently than His134Tyr. The impact of these mutations on substrate selection or substrate binding remains to be determined. However, the direct juxtaposition of the residues selected for changes (Ser133 and His134 in MHV) argues for conserved roles in the structure and function of nsp5.
DISCUSSION
In this study, we identified a novel mutation (T10651C) that results in a Val148Ala substitution in nsp5, the 3C-like proteinase of MHV, that is responsible for the ts phenotype of Alb/ts/nsp5/V148A, an original isolate of the MHV ts Alb ts6 mutant virus. Additionally, we identified two second-site reversion mutations in nsp5 that partially compensate for the replication and protein-processing defects that manifest at the nonpermissive temperature. A recent published report on Alb ts6 identified a mutation in nsp4 (A9494C, Asn258Thr) as the Alb ts6 ts mutation (39). Interestingly, there was no overlap between the nsp4 sequence results reported by Sawicki et al. (39) for Alb ts6 and the results of the complete genome sequencing of Alb/ts/nsp5/V148A reported in this paper. We therefore conclude that the previously reported Alb ts6 virus is distinct from Alb/ts/nsp5/V148A characterized in this study. The sequencing and reverse genetic studies presented in this paper argue that the complete genome sequencing and recapitulation of ts mutations by use of reverse genetic strategies will be critical for future studies of ts mutant viruses.
Met162 and the S1 subsite of the MHV nsp5 active-site cavity.
The CoV nsp5 protein is a highly conserved cysteine proteinase that is essential for virus polyprotein processing and replication. The 3C-like proteinase activity of nsp5 was first experimentally confirmed in our laboratory (33) and, since the SARS outbreak in 2003, has been a highly active area of research and a major target for antiviral development (19, 21). The results of this study provide new insights into the dynamic structure-function relationships that occur during nsp5 processing of replicase polyprotein substrates and identify novel residues that may be critical for the proper function or regulation of nsp5 activity. In particular, Met162 is known to be part of the Tyr161-Met162-His163 (Y-M-H) motif that is conserved in the substrate binding S1 subsite of the nsp5 active-site cavity. Previously, Met162 was described as the “X” residue of a “Y-X-H” S1 subsite motif analogous to the “G-X-H” S1 site of other viral 3C and 3C-like proteinases, with the His163 residue concluded to be critical for binding to the P1-Gln residue of the substrate and the buried Tyr161 thought to be critical for the stable tautomeric positioning of His163 (Fig. 5B) (1, 45). When WT MHV nsp5 was modeled, the Val148 residue was found to be directly juxtaposed to the Met162 residue. This same positioning of the Val residue proximal to the Tyr161-Met162-His163 S1 subsite is reflected in the crystal structures of transmissible gastroenteritis virus and SARS-CoV (2, 3). Modeling of the Val148Ala mutation of Alb/ts/nsp5/V148A predicts the loss of hydrophobic interactions between residue 148 and Met162, allowing for greater rotameric flexibility of the Met162 side chain. Overall, these results suggest that the nsp5/Met162 MHV and its highly conserved CoV nsp5 orthologs play an important role in the S1 subsite that was not previously predicted by structural studies.
The rapid emergence of Alb/ts/nsp5/V148A revertant viruses at 40°C demonstrates selection for a functional nsp5 molecule. Identification and confirmation of two independent second-site revertant mutations (Ser133Asn and His134Tyr in nsp5) indicate that the partial restoration of nsp5 function can occur in more than one manner and that the adjacent Ser133 and His134 residues may play a role in the stability or function of nsp5. Because Val148Ala is predicted to allow increased rotameric flexibility of the Met162 side chain, which may destabilize the active-site cavity of nsp5, it is reasonable to hypothesize that the compensating mutations function to restabilize the active-site cavity. The His134Tyr mutation is predicted to subtly shift the positions of the His172 and Tyr182 residues that face Tyr161 and His163 (Fig. 5B). Orthologs of His172 and Tyr182 proteins are highly conserved across different CoV groups and are proposed to participate in forming the base of the S1 subsite, which helps to maintain the stability of His163 and interactions with the substrate. The Ser133Asn substitution in nsp5 did not appear to modify any residues that directly or indirectly interact with the active-site cavity or the S1 subsite. In fact, the predicted changes are in amino acid side chains that are directed away from the active site. One of these affected residues, Arg131, is conserved across all CoV groups and thus might affect nsp5's function in an unknown fashion. It is also conceivable that at higher temperatures, the Ser133Asn substitution may propagate changes through His134 in a manner analogous to that predicted for His134Tyr. Interestingly, this revertant virus was less competent in virus growth at 30°C and 40°C than either the WT or the nsp5/H134Y/V148 virus, suggesting that Ser133Asn is a less-than-optimal “second-choice” reversion. These possibilities remain to be tested by genetic and biochemical studies.
Impact of nsp5/V148A on the design of small-molecule inhibitors of nsp5 for SARS-CoV.
There is current research to define and test inhibitors of SARS-CoV nsp5 3C-like proteinases (19-21). Recently, a cluster of serine residues (Ser139, Ser144, and Ser147) was identified near the active-site pocket of SARS-CoV nsp5 as being susceptible to targeting by compounds containing boronic acid (3). Both Ser139 and Ser147 are conserved in the nsp5s of all CoVs, further defining this region of nsp5 as an attractive target for drug development. Interestingly, the Val148Ala mutation in nsp5 of Alb/ts/nsp5/V148A is located adjacent to Ser147. Our results indicate that CoV nsp5 proteins may possess an arsenal of adaptive mechanisms to respond to adverse conditions. The same type of subtle modifications that allowed rapid reversion from a nonfunctional ts nsp5/V148A mutant virus might provide the capacity for the virus to exclude protein, peptide, or chemical substrate analog inhibitors of nsp5 proteinases while retaining the capacity to process viral polyprotein substrates. It will be critical and highly informative to test all proposed inhibitors for resistant mutant viruses and to completely sequence the nsp5 coding region and possibly the entire genomes of revertant viruses that arise during tests of nsp5 inhibitors.
Acknowledgments
We express our appreciation to Lance Eckerle, Mark Gadlage, and Michelle Becker for their critical advice and reviews of the manuscript.
This work was supported by Public Health Service award T32 CA009385 (J.S.S.) from the National Institutes of Health, Public Health Service award R01 AI50083 (M.R.D.) from the National Institute of Allergy and Infectious Diseases, and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service award CA68485 for the Vanderbilt University DNA Sequencing Shared Resource.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 213213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 3001763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Bacha, U., J. Barrila, A. Velazquez-Campoy, S. A. Leavitt, and E. Freire. 2004. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry 434906-4912. [DOI] [PubMed] [Google Scholar]
- 4.Baker, S. C., K. Yokomori, S. Dong, R. Carlisle, A. E. Gorbalenya, E. V. Koonin, and M. M. C. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 676056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates, P. A., L. A. Kelley, R. M. MacCallum, and M. J. Sternberg. 2001. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 45(Suppl. 5)39-46. [DOI] [PubMed] [Google Scholar]
- 6.Bates, P. A., and M. J. Sternberg. 1999. Model building by comparison at CASP3: using expert knowledge and computer automation. Proteins 37(Suppl. 3)47-54. [DOI] [PubMed] [Google Scholar]
- 7.Bonilla, P. J., A. E. Gorbalenya, and S. R. Weiss. 1994. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology 198736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 28521-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bredenbeek, P. J., C. J. Pachuk, A. F. H. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. M. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 181825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockway, S. M., X. T. Lu, T. R. Peters, T. S. Dermody, and M. R. Denison. 2004. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 7811551-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, W., and R. S. Baric. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 703947-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, W., V. J. Madden, C. J. Bagnell, and R. S. Baric. 1997. Host-derived intracellular immunization against mouse hepatitis virus infection. Virology 228318-332. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, V. C., S. K. Lau, P. C. Woo, and K. Y. Yuen. 2007. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 20660-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Contreras-Moreira, B., and P. A. Bates. 2002. Domain fishing: a first step in protein comparative modelling. Bioinformatics 181141-1142. [DOI] [PubMed] [Google Scholar]
- 15.Denison, M. R., B. Yount, S. M. Brockway, R. L. Graham, A. C. Sims, X. Lu, and R. S. Baric. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 785957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denison, M. R., P. W. Zoltick, S. A. Hughes, B. Giangreco, A. L. Olson, S. Perlman, J. L. Leibowitz, and S. R. Weiss. 1992. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson, E. F., A. C. Sims, R. L. Graham, M. R. Denison, and R. S. Baric. 2007. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 816356-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 3481967-1976. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh, A. K., K. Xi, V. Grum-Tokars, X. Xu, K. Ratia, W. Fu, K. V. Houser, S. C. Baker, M. E. Johnson, and A. D. Mesecar. 2007. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 175876-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh, A. K., K. Xi, K. Ratia, B. D. Santarsiero, W. Fu, B. H. Harcourt, P. A. Rota, S. C. Baker, M. E. Johnson, and A. D. Mesecar. 2005. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 486767-6771. [DOI] [PubMed] [Google Scholar]
- 21.Goetz, D. H., Y. Choe, E. Hansell, Y. T. Chen, M. McDowell, C. B. Jonsson, W. R. Roush, J. McKerrow, and C. S. Craik. 2007. Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry 468744-8752. [DOI] [PubMed] [Google Scholar]
- 22.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 763697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano, N., K. Fujiwara, and M. Matumoto. 1976. Mouse hepatitis virus (MHV-2). Plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20219-225. [PubMed] [Google Scholar]
- 24.Hooft, R. W., C. Sander, and G. Vriend. 1996. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins 26363-376. [DOI] [PubMed] [Google Scholar]
- 25.Kanjanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 747911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanjanahaluethai, A., and S. C. Baker. 2001. Processing of the replicase of murine coronavirus: papain-like proteinase 2 (PLP2) acts to generate p150 and p44. Adv. Exp. Med. Biol. 494267-273. [DOI] [PubMed] [Google Scholar]
- 27.Kanjanahaluethai, A., D. Jukneliene, and S. C. Baker. 2003. Identification of the murine coronavirus MP1 cleavage site recognized by papain-like proteinase 2. J. Virol. 777376-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. C., R. A. Spence, P. F. Currier, X. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 2081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, H.-J., C.-K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. LaMonica, J. Tuler, A. Bagdzhadhzyan, and M. M. C. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Y., and B. Kuhlman. 2006. RosettaDesign server for protein design. Nucleic Acids Res. 34W235-W238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, X., Y. Lu, and M. R. Denison. 1996. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology 222375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, Y., and M. R. Denison. 1997. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology 230335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, Y., X. Lu, and M. R. Denison. 1995. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 693554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pachuk, C. J., P. J. Bredenbeek, P. W. Zoltick, W. J. Spaan, and S. R. Weiss. 1989. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology 171141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 36.Poon, L. L., K. H. Chan, O. K. Wong, T. K. Cheung, I. Ng, B. Zheng, W. H. Seto, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2004. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin. Chem. 5067-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon, L. L. M., D. K. W. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. C. Leung, S. K. P. Lau, P. C. Y. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 792001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawicki, S. G., and D. L. Sawicki. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 793391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturman, L. S., C. Eastwood, M. F. Frana, C. Duchala, F. Baker, C. S. Ricard, S. G. Sawicki, and K. V. Holmes. 1987. Temperature-sensitive mutants of MHV-A59. Adv. Exp. Med. Biol. 218159-168. [DOI] [PubMed] [Google Scholar]
- 42.Xue, X., H. Yang, W. Shen, Q. Zhao, J. Li, K. Yang, C. Chen, Y. Jin, M. Bartlam, and Z. Rao. 2007. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 366965-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, H., M. Yang, Y. Ding, Y. Liu, Z. Lou, Z. Zhou, L. Sun, L. Mo, S. Ye, H. Pang, G. F. Gao, K. Anand, M. Bartlam, R. Hilgenfeld, and Z. Rao. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 10013190-13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 7611065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 28757-94. [DOI] [PMC free article] [PubMed] [Google Scholar]