Abstract
Poliovirus (PV) 2A protease (2APro) cleaves eukaryotic initiation factors 4GI and 4GII (eIF4GI and eIF4GII) within virus-infected cells, effectively halting cap-dependent mRNA translation. PV mRNA, which does not possess a 5′ cap, is translated via cap-independent mechanisms within viral protease-modified messenger ribonucleoprotein (mRNP) complexes. In this study, we determined that 2APro activity was required for viral polysome formation and stability. 2APro cleaved eIF4GI and eIF4GII as PV polysomes assembled. A 2ACys109Ser (2APro with a Cys109Ser mutation) protease active site mutation that prevented cleavage of eIF4G coordinately inhibited the de novo formation of viral polysomes, the stability of viral polysomes, and the stability of PV mRNA within polysomes. 2ACys109Ser-associated defects in PV mRNA and polysome stability correlated with defects in PV mRNA translation. 3CPro activity was not required for viral polysome formation or stability. 2APro-mediated cleavage of eIF4G along with poly(rC) binding protein binding to the 5′ terminus of uncapped PV mRNA appear to be concerted mechanisms that allow PV mRNA to form mRNP complexes that evade cellular mRNA degradation machinery.
Poliovirus (PV) (genus Enterovirus, family Picornaviridae) has a single-stranded, positive-sense RNA genome that functions as an mRNA. The 5′-terminal VPg (viral protein of the genome) is removed by a cellular enzyme before PV mRNA engages ribosomes (1, 2). Uncapped PV mRNA has a long 5′ nontranslated region (5′ NTR) containing a cloverleaf RNA structure and an internal ribosome entry site (IRES), an open reading frame (ORF) 6,627 nucleotides long, a 3′ NTR, and a poly(A) tail (19, 33, 39, 44). Typically, host cell mRNA degradation machinery degrades uncapped mRNAs (32, 38). Poly(rC) binding proteins (PCBPs) interact with the PV 5′-terminal cloverleaf RNA as viral polysomes form, protecting the nascent viral mRNA from 5′ exonuclease (23, 31).
Two viral proteases encoded within the viral ORF, 2A protease (2APro) and 3C protease (3CPro), are responsible for the proteolytic processing of the viral polyprotein as well as cleavage of specific host proteins (13, 25, 26). 2APro is a cysteine protease with amino acid residues His 20, Asp 38, and Cys 109 comprising the catalytic core (47). The primary viral polyprotein cleavage mediated by 2APro liberates the P1 capsid polyprotein precursor from the P23 nonstructural polyprotein precursor (18). Cleavage of P1 from the viral polyprotein precursor occurs cotranslationally, precluding synthesis of P123 polyproteins corresponding to the entire ORF. 2APro also cleaves viral protein 3CD and larger polyproteins containing 3CD, although the biological significance of this cleavage within 3CD is unclear because it is not required for viral replication (18). 3CPro and 3CDPro mediate a catalytic cascade responsible for PV polyprotein processing at the VP2-VP3, VP3-VP1, 2A-2B, 2B-2C, 2C-3A, 3A-3B, 3B-3C, and 3C-3D junctions (17). Variable kinetics of cleavage at each of these sites within various polyprotein intermediates leads to the characteristic pattern of PV proteins.
In addition to their roles in viral polyprotein processing, PV proteases cleave specific cellular proteins. 2APro cleavage of eukaryotic initiation factors 4GI and 4GII (eIF4GI and eIF4GII) inhibits cap-dependent translation of host mRNAs early during the course of PV infection without inhibiting cap-independent translation of PV mRNAs (15). The COOH-terminal portion of eIF4G (eIF4GCOOH) interacts with the PV IRES to mediate the initiation of translation (35). The NH-terminal portion of eIF4G (eIF4GNH), with binding domains mediating interactions with the cap binding protein eIF4E and poly(A) binding protein (PABP), is not required for IRES-mediated mRNA translation (35). PABP is cleaved by 2APro, by 3CPro, and by cellular caspases (21, 27, 29); however, the biological significance of PABP cleavage during PV infections is unclear because cleavage is typically slow and incomplete relative to the kinetics of PV RNA translation and replication.
PV 2APro increases the stability, translation, and replication of PV mRNA (22), yet the mechanisms responsible for these phenotypes are undefined. In this study, we examined de novo viral polysome formation and stability. 2APro activity was required for viral mRNA and polysome stability. We discuss how 2APro-mediated cleavage of eIF4G underlies a concerted strategy to both inhibit host gene expression and coordinately allow for uncapped PV mRNA to form polysomes uncoupled from host mRNA degradation machinery.
MATERIALS AND METHODS
PV mRNAs.
cDNA clones containing subgenomic replicons (RNA2 and DJB13) of PV RNA (Mahoney type 1) were kindly provided by James B. Flanegan, University of Florida, Gainesville. The plasmid encoding RNA2, a subgenomic replicon of PV type 1, contained an in-frame deletion of nucleotides 1175 to 2956, removing VP2 and VP3 of the capsid genes (10). Site-directed mutagenesis (QuikChange mutagenesis kit; Stratagene, La Jolla, CA) was utilized to introduce single amino acid substitutions within the 2APro and 3CPro genes. The oligonucleotides used to introduce a Cys109Ser mutation into 2APro (2ACys109Ser) were 5′-CGCATCTCCAGGGGATTCCGGAGGCATACTCAG-3′ and 5′-CTGAGTATGCCTCCGGAATCCCCTGGAGATGCG-3′. The oligonucleotides used to introduce a His39Glu mutation into 3CPro (3CHis39Glu) were 5′-GCTATTTTACCAACCGAAGCTTCACCTGGTG-3′ and 5′-CACCAGGTGAAGCTTCGGTTGGTAAAATAGC-3′. Both oligonucleotide sets were used individually in the construction of PV RNA2 2APro(−) and PV RNA2 3CPro(−) (PV RNA2 encoding mutant viral proteases that expressed viral proteins with altered polyprotein processing) and sequentially in the construction of PV RNA2 2APro(−)3CPro(−). The plasmid encoding DJB13 contained a deletion in the 5′ NTR of PV type 1 encompassing nucleotides 41 to 70 of the cloverleaf RNA structure.
PV mRNAs were transcribed from MluI-linearized cDNA templates by T7 transcription (Epicenter, Madison, WI). [α-32P]CTP was included to make radiolabeled RNA when necessary. When indicated, PV mRNAs were capped with unmethylated GTP caps using the ScriptCap m7G capping system (Epicenter, Madison, WI) according to the manufacturer's instructions. RNA was precipitated with 2.5 M ammonium acetate, washed with ethanol, and quantified by UV absorption at 260 nm.
Cell-free translation.
Cytoplasmic HeLa cell extracts were incubated with PV mRNAs as previously described (5, 6, 30). Reaction mixtures (20 to 300 μl) contained 50% (by volume) S10, 20% (by volume) initiation factors (IFs), 10% (by volume) 10× nucleotide reaction mix (10 mM ATP, 2.5 mM GTP, 2.5 mM CTP, 600 mM KCH3CO2, 300 mM creatine phosphate, 4 mg/ml creatine kinase, and 155 mM HEPES-KOH [pH 7.4]), 2 mM guanidine hydrochloride, and T7 transcripts of viral RNA at 50 μg/ml. Reaction mixtures were incubated at 34°C for the indicated periods of time. [35S]methionine (Amersham, Piscataway, NJ) was included in reaction mixtures to radiolabel proteins for kinetics studies of protein synthesis. [35S]methionine pulse-chase labeling was performed by adding puromycin at 200 μg/ml to initiate the chase period. Samples (1 μl) of reaction mixtures containing [35S]methionine were precipitated with 5% trichloroacetic acid, filtered on nitrocellulose filters, and quantified by scintillation counting to determine the magnitude of protein synthesis. Samples (4 μl) of reaction mixtures containing [35S]methionine were also solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (2% SDS [Sigma, St. Louis, MO], 62.5 mM Tris-HCl [pH 6.8], 0.5% 2-mercaptoethanol, 0.1% bromophenol blue, 20% glycerol), heated at 100°C for 5 min, and separated by gel electrophoresis in 9 to 18% SDS-polyacrylamide gels. [35S]methionine-labeled proteins were detected by phosphorimaging.
PV mRNA stability.
PV mRNA stability was assayed by incubating 32P-labeled PV mRNAs in HeLa cell-free translation reaction mixtures as previously described (31). Portions of the reaction mixtures were removed at the times indicated and solubilized in 0.5% SDS buffer (0.5% SDS, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl). A portion of each sample was precipitated in 7% trichloroacetic acid and 50 mM sodium pyrophosphate and filtered onto nitrocellulose filters, and radiolabeled RNA retained on the filters was quantified by scintillation counting. Another portion of each sample was extracted with phenol-chloroform, precipitated with ethanol, and fractionated by electrophoresis in 1% agarose Tris-borate-EDTA (TBE). RNA within the gels was stained with ethidium bromide and detected with UV light. Radiolabeled RNAs within gels were detected by phosphorimaging.
Polysomes.
Sucrose gradients were prepared, and samples were treated using minor modifications to previously published protocols (16, 36, 43). Sucrose gradients were prepared in 14- by 89-mm polypropylene tubes (Seton, Los Gatos, CA) by layering 3 ml of 0.5 M sucrose on top of 8 ml of 1.5 M sucrose (0.5 or 1.5 M sucrose, 0.3 M NaCl, 10 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 100 μg cycloheximide per ml, and 100 μg heparin per ml). Tubes were capped with parafilm and laid on their sides at room temperature for 1 h to facilitate gradient formation.
Cell-free translation reaction mixtures (300 μl) were diluted with an equal volume of homogenization buffer solution (0.1 M Tris-HCl [pH 7.6], 0.1 M NaCl, 40 mM MgCl2, 0.5 M sucrose, 300 μg cycloheximide per ml, and 100 μg heparin per ml) to stabilize polysomes and 60 μl (1/10 volume) of a 10× nonionic detergent solution (5% Triton X-100, 121 mM sodium deoxycholate). Samples were layered on top of the gradients and subjected to ultracentrifugation at 36,000 rpm (∼160,000 × g) for 3 h at 4°C using an SW41 rotor (Beckman, Fullerton, CA). Sucrose gradients were fractionated using a BR-184 tube piercer and a syringe pump (Brandel, Gaithersburg, MD) and a UA-6 UV detector (ISCO, Lincoln, NE). Fluorinert FC-40 (ISCO) was pumped into the bottom of sucrose gradients (pump speed of 3.0, ∼1 ml per min). The gradient was pushed up through the tube and carried through the UV detector (sensitivity of 0.5, noise filter set at 1.5, peak separator off, and chart speed of 150). Digital data (100 samples per second) were collected during fractionation using a DI-158U USB data acquisition device (DATAQ Instruments, Akron, OH) connecting the UV detector to a personal computer. The DI-158U interface received output through the 1-V recorder output of the UA-6 UV detector into the positive port of analog channel 1 of the DI-158U device. The digital data were processed using WINDAQ/HS and WINDAQ/XL software (DATAQ Instruments), and UV absorption at 254 nm was plotted versus mobility (from top to bottom or fraction number of sucrose gradients) using Microsoft Excel.
Twenty 0.5-ml fractions were collected from each gradient. When 32P-labeled viral RNA was used, it was detected by Cerenkov counting on total fractions from the sucrose gradient. Phenol-chloroform extraction and ethanol precipitation were used to extract total RNA from the fractions of the sucrose gradients. RNA was separated by electrophoresis in 1% agarose TBE gels, stained with ethidium bromide, and detected with UV light or by phosphorimaging for 32P-labeled viral RNA.
Western blots.
Proteins fractionated by SDS-PAGE were transferred to Hybond-P membranes (Amersham, Piscataway, NJ) using a constant current of 1.6 A for 1.5 h in transfer buffer (25 mM Tris-HCl, 0.2 M glycine, 0.05% SDS, and 20% methanol). Membranes were soaked for 1 h in blocking buffer (20 mM Tris-HCl [pH 7.6], 14 mM NaCl, 0.05% Tween 20, and 5.0% powdered dry milk) and then gently rocked overnight at 4°C with 1:1,000 dilutions of eIF4GNH rabbit polyclonal antibody (AbCAM, Cambridge, MA), eIF4GCOOH mouse monoclonal antibody (BD Biosciences, San Jose, CA), and eIF4GII mouse monoclonal antibody (Novus Biologicals, Littleton, CO) diluted in blocking buffer. Membranes were then washed three times for 15 min each time with blocking buffer. Membranes were soaked with secondary antibodies (1:10,000 goat anti-mouse antibody conjugated to horseradish peroxidase (Sigma, St. Louis, MO) or 1:25,000 donkey anti-rabbit antibody conjugated to horseradish peroxidase [Jackson ImmunoResearch, West Grove, PA]) diluted in blocking buffer for 1 h with gentle rocking at room temperature. Each blot was then washed three times for 15 min each time with blocking buffer and developed using Lightning Plus chemiluminescence (Perkin Elmer, Waltham, MA). Blots were exposed to blue-light autoradiography film (ISC BioExpress, Kaysville, UT) for 5 or 20 min and developed.
RESULTS
Translation of PV mRNAs with mutations in the viral proteases.
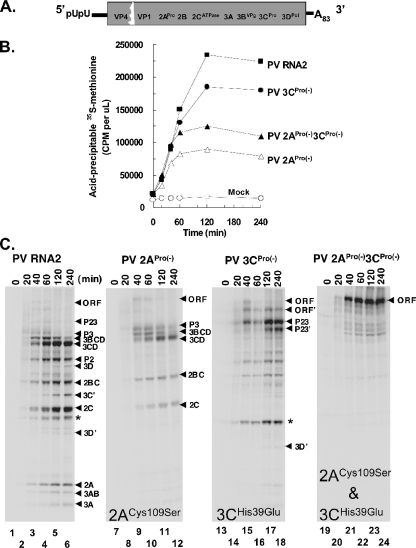
PV RNA2 was the wild-type control mRNA for the experiments in this study. PV RNA2 is a viral mRNA and replicon that functions efficiently both in vivo (10) and in cell-free translation-replication reaction mixtures (28). PV RNA2 contains an in-frame deletion in the portion of the ORF encoding VP2 and VP3 capsid proteins (Fig. 1A). 2ACys109Ser and 3CHis38Glu mutations that prevent 2A and 3C protease activity (3, 47) were engineered individually and together into PV RNA2 (Fig. 1A).
FIG. 1.
2ACys109Ser mutation diminished translation of PV mRNAs. (A) Diagram of PV RNA2, a wild-type PV mRNA with an in-frame capsid protein deletion (10, 28). (B) Kinetics and magnitude of PV mRNA translation in cell-free reaction mixtures as measured by the incorporation of [35S]methionine (in cpm per μl) into acid-precipitable material after 0, 20, 40, 60, 120 and 240 min of incubation. Reaction mixtures containing 50 μg/ml PV RNA2 mRNA, PV 2APro(−) mRNA, PV 3CPro(−) mRNA, PV 2APro(−)3CPro(−) mRNA, or no mRNA (Mock) are indicated. (C) PV polyprotein processing revealed by SDS-PAGE and phosphorimaging. [35S]methionine-labeled proteins after 0, 20, 40, 60, 120, and 240 min of incubation in reaction mixtures containing PV RNA2 mRNA, PV 2APro(−) mRNA, PV 3CPro(−) mRNA, or PV 2APro(−)3CPro(−) mRNA. The migration positions of specific viral proteins are indicated by the black arrowheads to the right of the gels. The migration position of the capsid protein fragment is indicated by an asterisk.
PV mRNAs were programmed into cell-free translation reactions containing [35S]methionine to determine the effects of protease mutations on viral mRNA translation and polyprotein processing. PV mRNA translation was monitored by the incorporation of [35S]methionine into acid-precipitable proteins (Fig. 1B) and by SDS-PAGE (Fig. 1C). Acid-precipitable [35S]methionine accumulated for up to 2 h in reaction mixtures containing PV RNA2 (Fig. 1B, PV RNA2). Mock reactions without viral mRNA synthesized no acid-precipitable proteins (Fig. 1B, Mock). PV mRNAs encoding mutant viral proteases synthesized less acid-precipitable material than wild-type PV mRNA, especially the two mRNAs encoding mutant 2APro (Fig. 1B). Defective translation of PV mRNAs with 2APro mutations was most profound between 1 and 2 h of translation (Fig. 1B).
[35S]methionine was incorporated into PV RNA2 proteins as expected (Fig. 1C, lanes 1 to 6). Mature PV proteins were not detectable during the first 20 min of incubation (Fig. 1C, PV RNA2, lane 2). Mature viral proteins were detected by 40 min of translation and accumulated over time (Fig. 1C, lanes 3 to 6). PV mRNAs encoding mutant viral proteases expressed viral proteins with altered polyprotein processing as expected (Fig. 1C, lanes 7 to 12 for PV 2APro(−), lanes 13 to 18 for PV 3CPro(−), and lanes 19 to 24 for PV 2APro(−)3CPro(−)). A smear of nascent nearly full-length viral polyproteins was detectable during the first 20 min of PV 2APro(−)3CPro(−) mRNA translation (Fig. 1C, lane 20), and unprocessed viral polyprotein corresponding to the entire ORF accumulated in the reaction mixture containing PV 2APro(−)3CPro(−) mRNA (Fig. 1C, lanes 21 to 24). These results indicated that the 2ACys109Ser and 3CHis38Glu mutations effectively prevented viral protease-mediated polyprotein processing, that ribosomes required approximately 20 min to traverse the ORF of the viral mRNA, and that 2APro activity was especially important for ongoing PV protein accumulation after ribosomes had traversed the viral ORF.
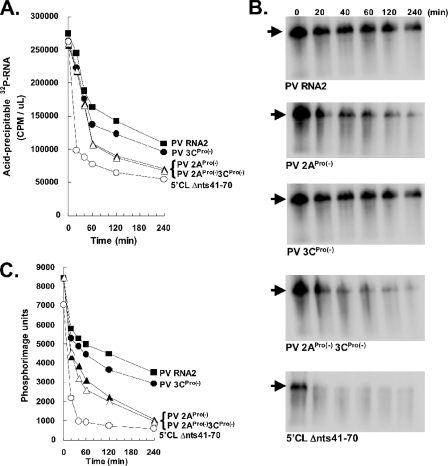
2ACys109Ser mutation decreased PV mRNA stability.
Because defects in PV mRNA stability could explain the reduced amounts of viral mRNA translation associated with 2APro mutation (22), we examined the kinetics and magnitude of PV mRNA stability within the cell-free translation-replication reaction mixtures. 32P-labeled PV mRNAs were programmed into cell-free translation reactions as in Fig. 1, and the amounts of acid-precipitable PV mRNA were determined over time (Fig. 2A), the integrity of PV mRNA within the reactions was examined by agarose gel electrophoresis (Fig. 2B), and the amount of intact PV mRNA within the gel was plotted versus the time of incubation (Fig. 2C). Wild-type PV mRNA was the most stable over time (Fig. 2, PV RNA2). After rapidly declining for 20 to 40 min, 20 to 30% of the PV mRNA remained stable for several hours (Fig. 2, PV RNA2). In contrast, PV mRNA with a 5′ cloverleaf mutation was completely degraded within 20 to 40 min of incubation (Fig. 2, 5′ CL Δnts41-70). Mutations in the 5′ cloverleaf that disrupt the binding of PCBP render PV mRNA extremely susceptible to 5′ exonuclease (31). PV mRNAs with protease mutations were less stable than wild-type PV mRNA but more stable than PV mRNA with a 5′ cloverleaf mutation (Fig. 2). While PV mRNAs expressing mutant 2APro were stable as ribosomes initially loaded (0 to 20 min), they became increasingly less stable than wild-type mRNAs after 40 to 60 min of incubation.
FIG. 2.
2ACys109Ser mutation decreased PV mRNA stability. Cell-free translation reactions were programmed with 50 μg/ml of 32P-labeled PV RNA2, PV 2APro(−), PV 3CPro(−), PV 2APro(−)3CPro(−), or 5′CL Δ41-70 mRNA. (A) Acid-precipitable 32P-labeled RNA (32P-RNA) (in cpm per μl) was measured after 0, 20, 40, 60, 120, and 240 min of incubation. (B) 32P-labeled PV mRNAs from the reaction mixtures in panel A were fractionated by agarose gel electrophoresis and detected by phosphorimaging. (C) Amounts of intact PV mRNAs within agarose gels quantified by phosphorimaging and plotted versus time of incubation in the reaction mixtures.
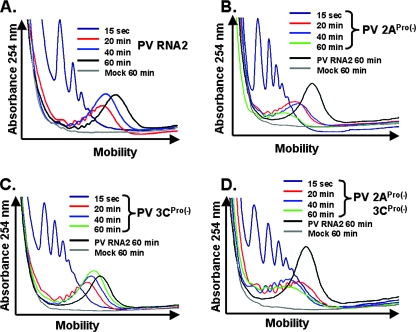
FIG. 5.
2ACys109Ser mutation inhibited polysome formation and stability. Cell-free translation reaction mixtures containing 50 μg/ml PV RNA2 mRNA, PV 2APro(−) mRNA, PV 3CPro(−) mRNA, PV 2APro(−)3CPro(−) mRNA, or no mRNA (Mock) were incubated for 15 seconds (0.25 min) or 20, 40, and 60 min as indicated. Products from the reactions were fractionated by sucrose gradient centrifugation, and polysomes were detected by UV absorbance during sucrose gradient fractionation as described in Materials and Methods. The UV trace from the portion of the gradients with polysomes was cropped and magnified for clarity.
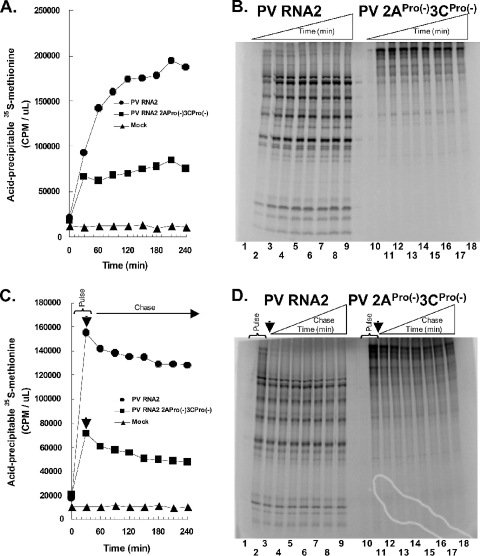
2ACys109Ser and 3CHis39Glu mutations did not affect PV protein stability.
The amounts of radiolabeled viral proteins accumulating in Fig. 1 reflect not only protein synthesis but also protein degradation. Therefore, the diminished accumulation of PV proteins associated with the 2ACys109Ser and 3CHis39Glu mutations in Fig. 1 could be due, in part, to decreased stability of PV precursor proteins relative to the stability of viral proteins naturally processed from their precursors. To examine this possibility, we compared [35S]methionine continuous radiolabeling (Fig. 3A and B) and [35S]methionine pulse-chase experiments (Fig. 3C and D) to determine whether PV precursor proteins associated with protease mutations were less stable than viral proteins naturally processed from their precursors. Continuous labeling conditions demonstrated that polyprotein corresponding to the poliovirus ORF stopped accumulating after 30 min of incubation in reaction mixtures containing PV mRNA encoding 2ACys109Ser and 3CHis39Glu mutations (Fig. 3A and B, PV 2APro(−)3CPro(−)), whereas the amounts of [35S]methionine in wild-type PV proteins increased for up to 120 min (Fig. 3A and B, PV mRNA). In the pulse-chase experiment, puromycin was added after 30 min of incubation to stop ongoing protein synthesis, precluding additional incorporation of [35S]methionine into PV proteins (Fig. 3C and D). PV proteins radiolabeled with [35S]methionine during the initial 30-min pulse were relatively stable during the following 3.5-h chase period (Fig. 3C and D). The amounts of acid-precipitable [35S]methionine decreased modestly during the 3.5-h chase period (Fig. 3C). Furthermore, the PV polyprotein expressed from PV mRNA encoding 2ACys109Ser and 3CHis39Glu mutations was not degraded any more rapidly than mature viral proteins (Fig. 3C and D). Thus, the reduced incorporation of [35S]methionine associated with the translation of PV mRNA encoding 2ACys109Ser and 3CHis39Glu mutations was due predominantly, perhaps even exclusively, to the decreased PV mRNA stability demonstrated in Fig. 2.
FIG. 3.
2ACys109Ser and 3CHis39Glu mutations did not affect PV protein stability. Cell-free translation reactions containing [35S]methionine were programmed with 50 μg/ml of PV RNA2 mRNA, PV 2APro(−)3CPro(−) mRNA, or no mRNA (Mock) as indicated. (A and B) Continuous [35S]methionine labeling. [35S]methionine incorporation (in cpm per μl) into proteins was measured after 0, 30, 60, 90, 120, 150, 180, 210, and 240 min of incubation [indicated by the height of the triangle labeled Time (min)] by quantifying the amounts of acid-precipitable radiolabel (A) and by SDS-PAGE followed by phosphorimaging (B). (C and D) [35S]methionine pulse-chase. Puromycin (200 μg per ml) was added to reaction mixtures containing [35S]methionine after 30 min of incubation (arrow indicates time of puromycin addition) to prevent ongoing protein synthesis. [35S]methionine incorporation into proteins was measured after 0, 30, 60, 90, 120, 150, 180, 210, and 240 min of incubation [indicated by the height of the triangle labeled Chase Time (min) in panel D] by quantifying the amounts of acid-precipitable radiolabel (C) and by SDS-PAGE followed by phosphorimaging (D).
PV 2ACys109Ser mRNA degradation correlated with the kinetics of 2APro-mediated cleavage of eIF4G.
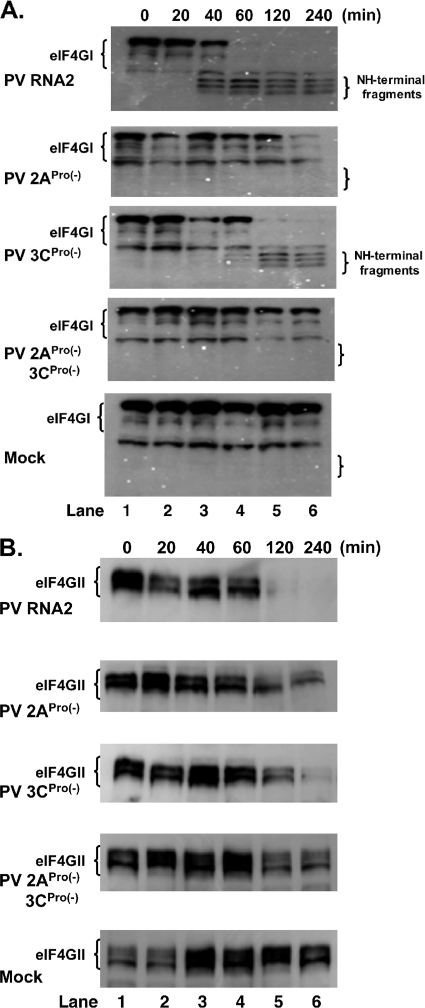
The integrity of eIF4GI and eIF4II during PV mRNA translation was determined by Western blotting (Fig. 4). The integrity of eIF4GI was unaltered over 4 h of incubation in mock reactions (Fig. 4A, Mock). In contrast, eIF4GI was cleaved rapidly between 20 and 60 min of incubation in reaction mixtures containing PV mRNA (Fig. 4A, PV RNA2). A number of eIF4GI isoforms with variable-length NH termini are present in HeLa cells (7, 8). NH-terminal fragments corresponding to the three most abundant isoforms were most prominent after cleavage by 2APro (Fig. 4A, NH-terminal fragments). eIF4GI was not cleaved in reaction mixtures containing PV 2APro(−) or PV 2APro(−)3CPro(−) mRNAs (Fig. 4A). The lack of eIF4GI cleavage between 20 and 60 min of incubation in reaction mixtures containing PV 2APro(−) and PV 2APro(−)3CPro(−) mRNAs corresponded with the impaired translation of these mRNAs after 20 to 60 min of incubation (compare Fig. 1B with Fig. 4A) and with the defect in PV mRNA stability first evident between 20 and 60 min of incubation (Fig. 2).
FIG. 4.
2ACys109Ser mutation prevented eIF4GI and eIF4GII cleavage during PV mRNA translation. Cell-free translation reaction mixtures containing 50 μg/ml PV RNA2 mRNA, PV 2APro(−) mRNA, PV 3CPro(−) mRNA, PV 2APro(−)3CPro(−) mRNA, or no mRNA (Mock) were incubated for 0, 20, 40, 60, 120, and 240 min as indicated. (A) Proteins from the reaction mixtures were fractionated by SDS-PAGE and probed by Western blotting with eIF4GI polyclonal antibody that recognizes an epitope within the NH-terminal portion of the protein. The migration positions of intact eIF4GI and NH-terminal fragments are indicated to the left of the gels. (B) Proteins from the reaction mixtures were fractionated by SDS-PAGE and probed by Western blotting with eIF4GII monoclonal antibody. The migration positions of intact eIF4GII proteins are indicated to the left of the gels.
eIF4GII, which is cleaved by 2APro in PV-infected cells more slowly than eIF4GI (15), was also cleaved more slowly than eIF4GI in cell-free reaction mixtures (Fig. 4B). eIF4GII was cleaved between 60 and 120 min of incubation in reaction mixtures containing wild-type PV mRNA (Fig. 4B), whereas eIF4GI was cleaved between 20 and 60 min of incubation (Fig. 4A). As with eIF4GI (Fig. 4A), eIF4GII was not cleaved during the translation of PV mRNA encoding a 2ACys109Ser mutation (Fig. 4B). Likewise, eIF4GII cleavage was slightly delayed in reaction mixtures containing PV mRNA encoding a 3CHis39Glu mutation (Fig. 4B). eIF4GII remained intact during the incubation of mock reactions (Fig. 4B). Therefore, the results in Fig. 4 demonstrate that eIF4GI and eIF4GII were cleaved coincident in time with the decreased PV mRNA translation associated with the 2ACys109Ser mutation (Fig. 1) and with the decreased PV mRNA stability associated with the 2ACys109Ser mutation (Fig. 2).
Polysome formation by PV mRNAs with mutations in the viral proteases.
PV polysomes form coordinate in time with the elongation of ribosomes across the viral ORF. Under the conditions used for these experiments, ribosomes require approximately 20 min to traverse the ORF of PV RNA2 (as demonstrated in Fig. 1C and in reference 23). Ribosome density on PV mRNAs then increases modestly between 20 and 60 min of incubation (23).
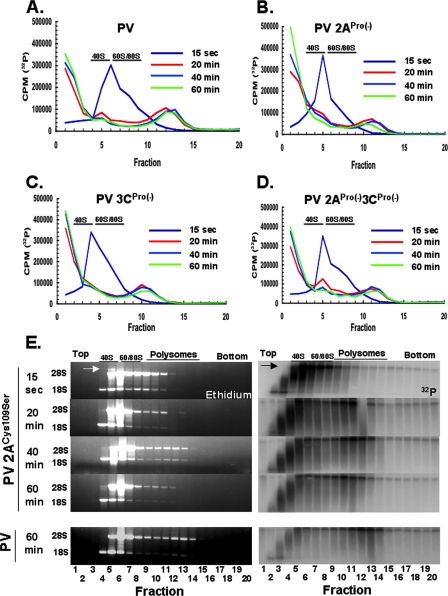
PV mRNAs were programmed into cell-free translation reactions as in Fig. 1 to 4, and polysomes within the reaction mixtures were measured at the indicated time points (Fig. 5). Reaction mixtures without PV mRNA had no detectable polysomes (Fig. 5A to D, Mock). In contrast, reaction mixtures containing PV RNA2 formed polysomes with 5 to 15 ribosomes per ORF after 60 min of incubation (Fig. 5A to D, PV RNA2). De novo polysome formation and maturation by PV mRNAs with mutations in the viral proteases were examined by observing polysomes after 15 seconds and 20 min, 40 min, and 60 min of incubation (Fig. 5). Both wild-type and protease mutant PV mRNAs formed polysomes with 1 to 5 ribosomes per ORF in the first 15 seconds of incubation (Fig. 5, 15 sec) and ribosome density per ORF increased by 20 min of incubation (Fig. 4A to D, 20 min). Polysomes containing wild-type PV mRNA typically increased in size between 20 and 60 min of incubation (Fig. 5A). In contrast, polysomes containing PV mRNAs expressing mutant 2APro failed to mature after 20 min of incubation, while polysomes with PV mRNA expressing the mutant 3CPro alone matured in a manner resembling polysomes with wild-type PV (Fig. 5B and C, 40- and 60-min samples). Rather than increasing ribosome density between 20 and 60 min of incubation, PV 2APro(−) and PV 2APro(−)3CPro(−) polysomes decreased in size and magnitude (Fig. 5B and D). Thus, PV polysomes formed coordinate with the elongation of ribosomes across the ORF; however, PV polysomes without 2APro activity were unstable and did not mature or persist.
PV mRNA stability within polysomes.
We used 32P-labeled PV mRNAs and sucrose gradient fractionation to coordinately examine polysome formation and viral mRNA stability (Fig. 6). PV mRNAs were programmed into cell-free translation reactions as in Fig. 1 to 4, and the distribution and integrity of mRNAs were determined at the indicated time points (Fig. 6). PV mRNAs programmed into the reactions comigrated with ribosomal subunits and small nascent polysomes after 15 seconds of incubation (Fig. 6A to E, 15 sec, fractions 4 to 10). Agarose gel electrophoresis indicated that the PV 2APro(−) mRNA was largely intact at this initial time point (Fig. 6E, top right gel detecting 32P-labeled PV 2APro(−) mRNA) and very little radiolabel was detected at the top of the gradients initially (Fig. 6A to D, 15 sec, fractions 1 to 3). As polysomes formed and matured, a portion of radiolabel cofractionated with polysomes (Fig. 6A to D, fractions 8 to 15), while another portion of radiolabel corresponding to degraded PV mRNA fractionated at the top of the gradient (Fig. 6A to D, fractions 1 to 3). Agarose gel electrophoresis revealed that the PV 2APro(−) mRNA within polysomes was intact and that the radiolabel at the top of the gradient was degraded PV mRNA (Fig. 6E). Analogous data were obtained for wild-type PV mRNA (Fig. 6E, PV mRNA at 60 min) (23). The amounts of intact PV 2APro(−) mRNA associated with polysomes decreased over time as the amounts of degraded PV 2APro(−) mRNA at the top of the gradients increased (Fig. 6B). A large portion of the radiolabel in fractions 1 to 3 was not retained in the agarose gel, especially the radiolabel in fraction 1, indicating that many mRNAs were degraded into mononucleotides or very small oligonucleotides. The size of partially degraded PV mRNA increased from fractions 1 to 4 as would be expected by centrifugation in sucrose (Fig. 6E, lanes 1 to 4).
FIG. 6.
PV 2ACys109Ser mRNA was degraded as unstable polysomes disassembled. (A to D) Cell-free translation reaction mixtures containing 50 μg/ml of the indicated PV mRNAs were incubated for 15 seconds (0.25 min) or 20, 40, and 60 min: PV RNA2 mRNA (A), PV 2APro(−) mRNA (B), PV 3CPro(−) mRNA (C), and PV 2APro(−)3CPro(−) mRNA (D). Products from the reactions were fractionated by sucrose gradient centrifugation. Twenty fractions were collected as the sucrose gradients were fractionated. The amount (cpm) of 32P in each fraction was determined by Cerenkov counting and plotted versus fraction number in panels A to D. (E) RNAs from each fraction of the PV 2APro(−) mRNA gradients were fractionated by electrophoresis in 1% agarose. PV RNA2 mRNA incubated for 60 min is included for comparison (bottom gels). RNAs were detected by ethidium bromide and UV light (left gels) and by phosphorimaging (right gels). The migration positions of rRNAs and intact 32P-labeled PV mRNAs (arrows) are indicated.
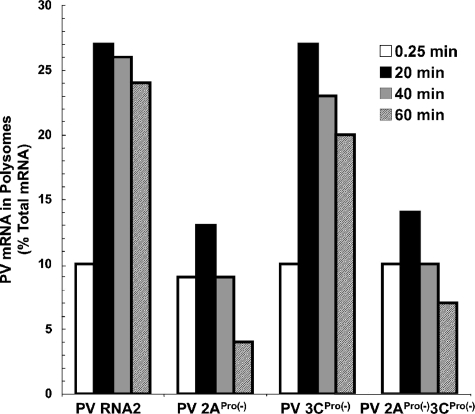
The amounts of PV mRNAs associated with polysomes (Fig. 6A to D, sucrose fractions 8 to 15) relative to the total amount of PV mRNA programmed into the reactions was plotted versus time (Fig. 7). Twenty-five percent of the wild-type PV mRNA programmed into the reactions was associated with polysomes after 20 min of incubation, and this amount of wild-type PV mRNA remained associated with the polysomes for 60 min (Fig. 7, PV RNA2). Comparable amounts of PV 3CPro(−) mRNA associated with polysomes for 20 to 60 min (Fig. 7, PV 3CPro(−)). In contrast, only 13 to 14% of PV mRNAs expressing mutant 2APro cofractionated with polysomes after 20 min of incubation, and the amounts of these mRNAs associated with polysomes decreased to 4 to 7% after 60 min of incubation (Fig. 7, PV 2APro(−) RNA and PV 2APro(−)3CPro(−) RNA). Thus, 2APro activity was required as PV polysomes formed.
FIG. 7.
Amounts of PV mRNA within polysomes. The amounts of 32P-labeled PV mRNAs present within polysomes (fractions 8 to 15 of gradients in Fig. 6A to D) after 0.25, 20, 40, and 60 min of incubation relative to the initial (total) amounts of PV mRNA programmed into the reaction mixtures.
5′ caps restored stability to C24A and 2APro(−) PV mRNAs.
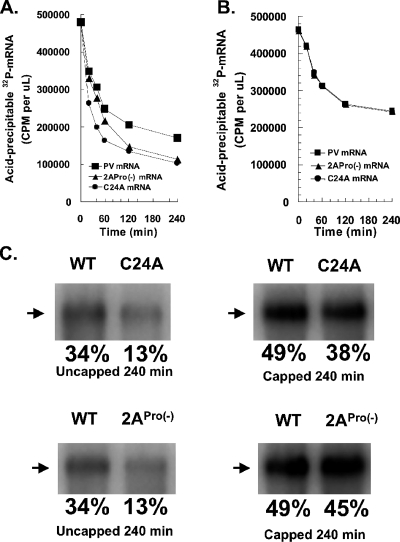
A C24A mutation that blocks the binding of PCBP to the cloverleaf RNA structure at the 5′ end of uncapped PV mRNA rendered PV mRNA unstable (Fig. 8A, note kinetics of C24A mRNA stability relative to wild-type PV mRNA). C24A mRNA was immediately less stable than wild-type PV mRNA, being degraded as polysomes formed (Fig. 8A) (23). A 2ACys109Ser mutation rendered PV mRNA unstable coordinate in time with the cleavage of eIF4G, between 20 and 120 min of incubation as polysomes matured. A 5′ cap on both C24A and 2ACys109Ser mRNAs restored stability to wild-type mRNA levels (Fig. 8A and B). A 5′ cap also modestly increased the stability of wild-type PV mRNA (Fig. 8). Because 5′ caps block degradation of mRNA by 5′ exonuclease, these data implicate a 5′ exonuclease in the degradation of uncapped PV mRNA and further indicate that the C24A and 2ACys109Ser mutations increased the susceptibility of uncapped PV mRNA to degradation by 5′ exonuclease.
FIG. 8.
5′ caps restored stability to C24A and 2APro(−) PV mRNAs. (A and B) Cell-free translations were programmed with 50 μg/ml of uncapped (A) or 5′-capped (B) 32P-labeled PV RNA2, C24A, or 2APro(−) mRNA. Acid-precipitable 32P-labeled RNA (32P-RNA) (in cpm per μl) was measured after 0, 20, 40, 60, 120, and 240 min of incubation. (C) The amount of intact PV mRNA after 240 min of incubation was also determined by fractionation of RNA by electrophoresis in 1% agarose and quantification by phosphorimaging. The migration positions of intact PV mRNAs are indicated by arrows to the left of the gels. WT, wild type.
eIF4GCOOH cofractionated with PV polysomes.
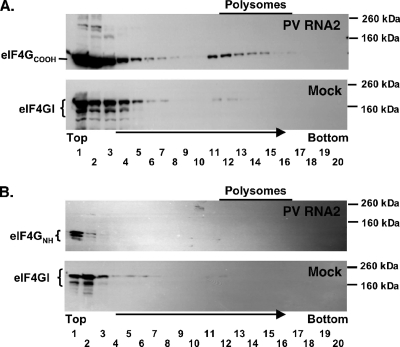
PV mRNA translation requires either intact eIF4G or eIF4GCOOH following 2APro-mediated cleavage of eIF4G, whereas eIF4GNH is not required for PV mRNA translation (35). Therefore, we predicted that eIF4GCOOH would cofractionate with PV polysomes and that eIF4GNH would not cofractionate with PV polysomes. To test this prediction, we fractionated PV polysomes using sucrose gradients and examined the distribution of eIF4GCOOH and eIF4GNH in the gradients by Western blotting (Fig. 9). Consistent with our predictions, eIF4GCOOH was found to cofractionate with PV polysomes (Fig. 9A, sucrose fractions 11 to 17). A large portion of eIF4GCOOH was also detected at the top of the sucrose gradients (Fig. 9, sucrose fractions 1 to 7). In contrast, eIF4GNH was not detected within sucrose fractions corresponding to those containing PV polysomes (Fig. 9B, fractions 11 to 17), whereas eIF4GNH was detected at the top of the sucrose gradients (Fig. 9, sucrose fractions 1 to 7). It is important to note that eIF4GNH Western blotting may not have sufficient sensitivity to establish whether eIF4GNH is absolutely excluded from PV polysomes. Nonetheless, these results are consistent with the possibility that eIF4GCOOH is present within PV polysomes and that eIF4GNH is released from PV polysomes following cleavage of eIF4G by 2APro.
FIG. 9.
eIF4GCOOH cofractionated with PV polysomes. Cell-free translation reaction mixtures containing 50 μg/ml PV RNA2 mRNA or no mRNA (Mock) were incubated for 60 min. Reaction products were then separated by centrifugation in sucrose gradients, and the gradients were fractionated (20 0.5-ml fractions [fractions 1 to 20] per gradient from top to bottom). Polysomes were detected by UV absorbance during fractionation. Proteins from the sucrose gradient fractions were separated by SDS-PAGE and probed by Western blotting with antibodies as described in Materials and Methods. Proteins from sucrose gradient fractions 1 to 10 were diluted 1:10 relative to proteins in sucrose gradient fractions 11 to 20 due to the bulk of material near the top of the gradients. Polysomes were present in fractions 12 to 16. Proteins were probed by Western blotting with eIF4GCOOH-specific antibody (A) and eIF4GNH-specific antibody (B). In panels A and B, PV RNA2 reaction products are shown in the top gel, and mock reaction products are shown in the bottom gel. Note that eIF4GNH in the PV RNA2 reaction mixture and intact eIF4GI in the mock reaction mixture were detected at the top of the gradient.
PV mRNA translation complemented defective PV 2APro(−) mRNA stability but did not complement defective PV C24A mRNA stability.
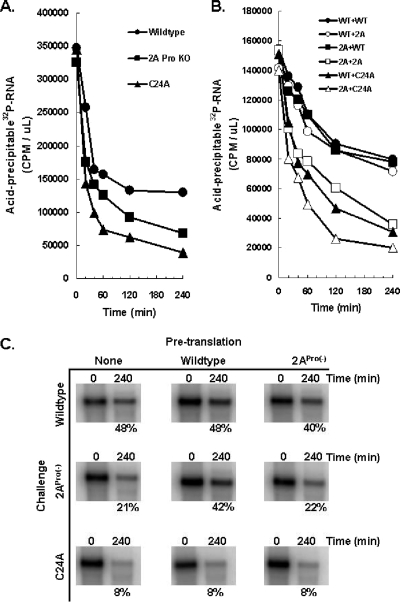
In order to further explore the correlation between 2APro expression and PV mRNA stability, we determined whether expression of wild-type 2APro complemented the defective stability of 2APro(−) and C24A mutant PV mRNAs (Fig. 10). As previously established, wild-type PV mRNA was more stable than either PV C24A or 2APro(−) mRNA incubated in HeLa S10 translation reaction mixtures (Fig. 10A and C, first column of samples labeled “None” for no pretranslation). PV C24A mRNA was most unstable, exhibiting nearly complete instability by 60 min of incubation (Fig. 10A and C with 8% of PV C24A mRNA present after 240 min of incubation). PV 2APro(−) mRNA was progressively less stable than wild-type PV mRNA between 20 min and 240 min of incubation (Fig. 10A and C with 21% of PV C24A mRNA present after 240 min of incubation in comparison to 48% of wild-type PV mRNA).
FIG. 10.
Expression of 2APro from wild-type PV mRNA restored stability to PV 2APro(−) mRNA but not PV C24A mRNA. (A) Cell-free translation reactions were programmed with 50 μg/ml of 32P-labeled PV RNA2, C24A, or 2APro(−) mRNA. Acid-precipitable 32P-labeled RNA (32P-RNA) (in cpm per μl) was measured after 0, 20, 40, 60, 120, and 240 min of incubation. 2A Pro KO, 2APro knockout. (B) Cell-free translation reactions were programmed with 50 μg/ml of PV RNA2 (wild type [WT) or 2APro(−) (2A) mRNA. After 1 h of incubation at 34°C, 32P-labeled PV RNA2, C24A, or 2APro(−) mRNA was added to a final concentration of 25 μg/ml. Acid-precipitable 32P-labeled RNA (32P-RNA) was measured after 0, 20, 40, 60, 120, and 240 min of incubation. (C) The amounts of input PV mRNAs present after 240 min of incubation from the reaction mixtures in panels A and B were determined by electrophoresis in 1% agarose and phosphorimaging. The percentage of radiolabeled input PV mRNA after 240 min of incubation is indicated below the gels.
Complementation of PV mRNA stability was tested by preincubating reaction mixtures for 1 h with unlabeled wild-type or 2APro(−) PV mRNAs followed by the incubation of radiolabeled “challenge” PV mRNAs for an additional 240 min (Fig. 10B and C). The percentage of intact challenge PV mRNAs under each condition were determined by acid-precipitable radioactivity (Fig. 10B) and by 1% agarose gel electrophoresis comparing input mRNA with mRNA present after 240 min of incubation (Fig. 10C). One hour of preincubation with wild-type or 2APro(−) PV mRNAs was chosen because this time of preincubation is sufficient to cleave eIF4G to near completion, whereas preincubation of reaction mixtures with PV 2APro(−) mRNA does not result in cleavage of eIF4G (as established in Fig. 4). As revealed in Fig. 10B, expression of 2APro from wild-type PV mRNA for 1 h restored stability to PV 2APro(−) mRNA compared to control reactions (Fig. 10B and C). In contrast, expression of 2APro from wild-type PV mRNA for 1 h did not restore stability to PV C24A mRNA compared to control reactions (Fig. 10B and C, note that PV C24A mRNA was equally unstable under all conditions). These data indicate that wild-type PV mRNA expressing wild-type 2APro can complement in trans-diminished PV 2APro(−) mRNA stability but cannot complement in trans-diminished PV C24A mRNA stability.
DISCUSSION
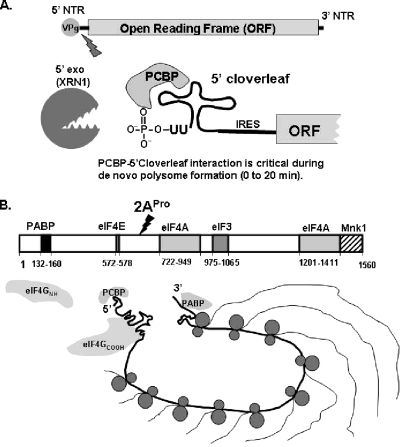
We and others (11) postulate that positive-strand RNA viruses likely evolved mechanisms to uncouple the translation of their viral mRNA from host mRNA turnover pathways because the viral mRNA goes on to become the template for RNA replication (34). Degradation of viral mRNA by host mRNA turnover machinery would preclude subsequent viral RNA replication. Our data demonstrate that 2APro activity was required for the stability of naturally uncapped PV mRNA. 2APro cleaved eIF4GI and eIF4GII as PV polysomes assembled and matured (Fig. 4). A 2ACys109Ser protease active site mutation that prevented cleavage of eIF4GI and eIF4GII coordinately inhibited the stability of viral polysomes (Fig. 5) and the stability of PV mRNA within polysomes (Fig. 6 and 7). Importantly, mRNAs may be translationally repressed before their degradation (reviewed in reference 9), and the data in Fig. 5, 6, and 7 do not address whether the PV 2APro(−) mRNAs were degraded within polysomes or whether they were translationally repressed and then degraded. As shown in Fig. 4, inactivation of 3CPro delayed the cleavage of eIF4GI and II by 2APro. The sequestration of 2APro within PV precursor proteins (due to the inactivation of 3CPro, which is required for liberating 2APro from the P2 precursor polyprotein) appears to impair the speed by which 2APro can cleave eIF4G (Fig. 4). This delayed cleavage of eIF4GI and eIF4GII correlated with small but detectable decreases in PV mRNA translation (Fig. 1) and PV mRNA stability (Fig. 2) associated with the 3CHis39Glu mutation. 2APro expressed from wild-type PV mRNA complemented in trans the stability of PV 2APro(−) mRNA but did not complement in trans defective stability of PV C24A mRNA (Fig. 10). Our findings (23, 31), in conjunction with those from the Flanegan laboratory (22), indicate that uncapped PV mRNA uses two concerted strategies to evade degradation by 5′ exonuclease (Fig. 11). PCBP binding to the 5′ cloverleaf appears to block 5′ exonuclease as ribosomes initially load onto PV mRNA (Fig. 11A, 0 to 20 min of incubation in cell-free reactions), while 2APro activity appears to fortify resistance to 5′ exonuclease soon after its expression (Fig. 11B, after 20 min of incubation and as discussed below).
FIG. 11.
Uncapped PV mRNA uses PCBP and 2APro to mediate PV mRNA and polysome stability. (A) PCBP-5′ cloverleaf interactions block 5′ exonuclease (5′ exo) as de novo polysomes form (as reported by references 23 and 31). (B) 2APro increased PV mRNA stability coincident in time with cleavage of eIF4G (after 20 min of translation as reported herein). The diagram of eIF4G was adapted from reference 40.
PCBP-5′ cloverleaf RNA interactions are critical immediately when uncapped PV mRNA is introduced into HeLa S10 translation reactions, during the first 20 min of incubation as ribosomes initially traverse the viral ORF (as summarized in Fig. 11A and in references 23 and 31). In contrast, 2APro activity fortified uncapped PV mRNA stability after ribosomes traversed the ORF (as reported herein and summarized in Fig. 11B). Because eIF4GI and eIF4GII were cleaved by 2APro coincident in time with the effect of 2APro on increased PV mRNA stability, we speculate that eIF4G cleavage may be important in the polysome and mRNA stability phenotype. The amino terminus of eIF4G (eIF4GNH), either directly or indirectly via interacting proteins, may be involved in the recognition of mRNAs by host mRNA turnover machinery. Domains within the eIF4GNH anchor RNA-protein-protein-RNA interactions between the 5′ and 3′ termini of mRNAs. Interactions between 5′- and 3′-terminal mRNPs may be particularly important for the deadenylation, decapping, and 5′ exonuclease-mediated mRNA degradation pathway (reviewed in references 9, 12, 32, and 37). Importantly, the 5′ exonuclease XRN1 was not degraded by 2APro in our experiments (data not shown), indicating that 2APro does not directly inactivate XRN1 via proteolytic cleavage. Furthermore, PABP associated with PV polysomes was not degraded by 2APro in our experiments (23). Notably, PABP and the COOH-terminal portion of eIF4G cofractionated with PV polysomes (Fig. 9) (23). We speculate that 2APro-mediated cleavage of eIF4G and release of the eIF4GNH may remodel 5′-3′ RNP interactions in a manner that uncouples PV polysomes from cellular mRNA turnover pathways.
Two genes express functionally redundant forms of eIF4G, eIF4GI and eIF4GII (14). eIF4GI and eIF4GII have several domains that interact with specific translation factors (reviewed in reference 40) (Fig. 11B). Near the NH terminus of eIF4G is a domain that binds PABP (20). Within the central portion of eIF4G are separate domains for binding eIF4E, eIF4A, and eIF3. The COOH-terminal portion of eIF4G has domains that bind eIF4A and Mnk1. By simultaneously interacting with eIF4E, the 5′ cap binding protein, and PABP on the 3′ poly(A) tail, eIF4G serves as a protein bridge coordinating interactions between the 5′ and 3′ termini of capped and polyadenylated cellular mRNAs (46). A major mRNA degradation pathway involves 3′ deadenylation coupled to 5′ decapping followed by XRN1-mediated 5′→3′ exonucleolytic degradation of the uncapped mRNA (reviewed in references 9, 12, 32, and 37). The manner in which this mRNA degradation pathway is normally coupled to mRNA translation and 5′-3′ protein bridges is unclear; however, enzymes associated with regulated mRNA turnover appear to be recruited via 5′ cap structures and eIF4E (4, 42, 45). Although eIF4GNH has a domain for interacting with both eIF4E and PABP, the amino terminus of eIF4G has not been demonstrated to participate in the recruitment of mRNA degradation machinery. Nonetheless, because eIF4GI and eIF4GII were cleaved by 2APro coincident in time with the effect of 2APro on increased PV mRNA stability and eIF4GNH was not associated with PV polysomes (Fig. 9), we think it is prudent to consider the potential contribution of eIF4GNH in the recruitment of mRNA turnover machinery to mRNAs. This important possibility requires additional experimental evidence and validation. Furthermore, because the temporal association of eIF4G cleavage with increased PV mRNA stability is only circumstantial evidence of a role in the stability phenotype, it is important to consider other possible mechanistic explanations for 2APro-mediated increases in PV mRNA stability that do not involve cleavage of eIF4G.
Enterovirus and rhinovirus 2A proteases as well as aphthovirus L proteases cleave eIF4Gs into two fragments: an NH-terminal fragment containing the PABP and eIF4E binding domains and a COOH-terminal fragment containing the eIF4A, eIF3, and Mnk1 binding domains (13) (Fig. 11B). The COOH-terminal portion of eIF4G interacts with IRESs to recruit 43S ribosomal subunit complexes for the initiation of translation (35), perhaps via an amino acid motif at the NH terminus of the COOH-terminal fragment that facilitates interactions with viral RNA (41). In contrast, the NH-terminal fragment of eIF4G that mediates bridging between the 5′ and 3′ termini of mRNAs and serves as a potential scaffold for the recruitment of proteins associated with regulated mRNA turnover is not required for translation of uncapped picornavirus mRNAs (35). We found that the COOH-terminal portion of eIF4GI cofractionated with PV polysomes, while the NH-terminal portion did not (Fig. 9) (23). In concert with PCBP-5′ cloverleaf RNA interactions, which protect uncapped PV mRNAs from 5′ exonuclease during de novo polysome formation (23, 31) (Fig. 11A), cleavage of eIF4G may allow PV mRNAs to form mRNP complexes that use the COOH-terminal portion of eIF4G for IRES-mediated translation initiation in the absence of the NH-terminal fragment of eIF4G (Fig. 11B).
2A protease has proven to be a valuable tool to investigate the mechanisms of translation initiation, especially in regard to the role(s) of eIF4G in cap-dependent mRNA translation. Our investigation suggests that 2A protease may be a valuable tool to help reveal how cellular mRNA turnover machinery is coupled to mRNA translation. The absence of a 5′ cap on PV mRNA, the binding of PCBP by the 5′ cloverleaf RNA to block 5′ exonuclease (31), the cleavage of eIF4G by 2APro, and the utilization of an IRES appear to be concerted strategies that allow PV mRNA to form mRNP complexes that evade the cellular machinery associated with regulated turnover of capped and polyadenylated mRNAs (Fig. 11). This process ensures the integrity of PV mRNA, which goes on to become the template for viral RNA replication (34). Perhaps other RNA viruses within and beyond the Picornaviridae family use alternate mechanisms to uncouple viral mRNA translation from cellular mRNA turnover machinery, thereby preserving the integrity of viral mRNAs that become templates in RNA replication. For example, many positive-strand RNA viruses use mRNAs without 5′ 7-methylguanosine caps. Hepatitis C virus mRNA lacks both a 5′ cap and a poly(A) tail and translates in an eIF4E- and eIF4G-independent manner (24). If mRNAs with 5′-3′ interactions mediated by eIF4E, eIF4GNH, and PABP are specifically recognized by components of host mRNA turnover machinery, then perhaps the strategy of HCV to translate in an eIF4E- and eIF4G-independent manner might exist, in part, to evade 5′ exonuclease. Intriguingly, even 5′ capped and polyadenylated positive-strand RNA viruses appear to encode mechanisms to evade host mRNA turnover machinery (11). Elucidating the alternate mechanisms by which positive-strand RNA viruses evade host mRNA turnover may help reveal how normal cellular mRNA turnover is regulated.
Acknowledgments
Kevin L. Durand and Megan E. Filbin provided technical assistance. We thank Jerry Schaack and Richard Davis for critical evaluation of the manuscript.
This work was supported by Public Health Service grants AI42189 (D.J.B.) and T32 AI07537 (B.J.K.) from the National Institutes of Health.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Ambros, V., and D. Baltimore. 1980. Purification and properties of a HeLa cell enzyme able to remove the 5′-terminal protein from poliovirus RNA. J. Biol. Chem. 2556739-6744. [PubMed] [Google Scholar]
- 2.Ambros, V., R. F. Pettersson, and D. Baltimore. 1978. An enzymatic activity in uninfected cells that cleaves the linkage between poliovirion RNA and the 5′ terminal protein. Cell 151439-1446. [DOI] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, D. Trono, and D. Baltimore. 1990. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5′ noncoding region. J. Virol. 64607-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrei, M. A., D. Ingelfinger, R. Heintzmann, T. Achsel, R. Rivera-Pomar, and R. Luhrmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., and J. B. Flanegan. 1993. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 67822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 27535-57. [DOI] [PubMed] [Google Scholar]
- 7.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 224499-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2005. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J. Biol. Chem. 28018610-18622. [DOI] [PubMed] [Google Scholar]
- 9.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73861-890. [DOI] [PubMed] [Google Scholar]
- 10.Collis, P. S., B. J. O'Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 666480-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau, N. L., K. J. Sokoloski, M. Opyrchal, C. P. Neff, C. J. Wilusz, and J. Wilusz. 2008. The 3′ untranslated region of Sindbis virus represses the deadenylation of viral transcripts in mosquito and mammalian cells. J. Virol. 82880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garneau, N. L., J. Wilusz, and C. J. Wilusz. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8113-126. [DOI] [PubMed] [Google Scholar]
- 13.Glaser, W., and T. Skern. 2000. Extremely efficient cleavage of eIF4G by picornaviral proteinases L and 2A in vitro. FEBS Lett. 480151-155. [DOI] [PubMed] [Google Scholar]
- 14.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 9511089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond, M. L., and L. H. Bowman. 1988. Insulin stimulates the translation of ribosomal proteins and the transcription of rDNA in mouse myoblasts. J. Biol. Chem. 26317785-17791. [PubMed] [Google Scholar]
- 17.Harris, K. S., S. R. Reddigari, M. J. Nicklin, T. Hammerle, and E. Wimmer. 1992. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J. Virol. 667481-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellen, C. U., C. K. Lee, and E. Wimmer. 1992. Determinants of substrate recognition by poliovirus 2A proteinase. J. Virol. 663330-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewlett, M. J., J. K. Rose, and D. Baltimore. 1976. 5′-terminal structure of poliovirus polyribosomal RNA is pUp. Proc. Natl. Acad. Sci. USA 73327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 177480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurgens, C. K., D. J. Barton, N. Sharma, B. J. Morasco, S. A. Ogram, and J. B. Flanegan. 2006. 2Apro is a multifunctional protein that regulates the stability, translation and replication of poliovirus RNA. Virology 345346-357. [DOI] [PubMed] [Google Scholar]
- 23.Kempf, B. J., and D. J. Barton. 2008. Poly(rC) binding proteins and the 5′ cloverleaf of uncapped poliovirus mRNA function during de novo assembly of polysomes. J. Virol. 825835-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krausslich, H. G., M. J. Nicklin, C. K. Lee, and E. Wimmer. 1988. Polyprotein processing in picornavirus replication. Biochimie 70119-130. [DOI] [PubMed] [Google Scholar]
- 26.Kundu, P., S. Raychaudhuri, W. Tsai, and A. Dasgupta. 2005. Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: incomplete shutoff of transcription interferes with efficient viral replication. J. Virol. 799702-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 241779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 7510696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marissen, W. E., D. Triyoso, P. Younan, and R. E. Lloyd. 2004. Degradation of poly(A)-binding protein in apoptotic cells and linkage to translation regulation. Apoptosis 967-75. [DOI] [PubMed] [Google Scholar]
- 30.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 774739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray, K. E., A. W. Roberts, and D. J. Barton. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 71126-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newbury, S. F. 2006. Control of mRNA stability in eukaryotes. Biochem. Soc. Trans. 3430-34. [DOI] [PubMed] [Google Scholar]
- 33.Nomoto, A., N. Kitamura, F. Golini, and E. Wimmer. 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. USA 745345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 81726-1737. [DOI] [PubMed] [Google Scholar]
- 35.Ohlmann, T., M. Rau, V. M. Pain, and S. J. Morley. 1996. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 151371-1382. [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios, R., R. D. Palmiter, and R. T. Schimke. 1972. Identification and isolation of ovalbumin-synthesizing polysomes. I. Specific binding of 125 I-anti-ovalbumin to polysomes. J. Biol. Chem. 2472316-2321. [PubMed] [Google Scholar]
- 37.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25635-646. [DOI] [PubMed] [Google Scholar]
- 38.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11121-127. [DOI] [PubMed] [Google Scholar]
- 39.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Translational efficiency of poliovirus mRNA: mapping inhibitory cis-acting elements within the 5′ noncoding region. J. Virol. 622219-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prevot, D., J. L. Darlix, and T. Ohlmann. 2003. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell 95141-156. [DOI] [PubMed] [Google Scholar]
- 41.Prevot, D., D. Decimo, C. H. Herbreteau, F. Roux, J. Garin, J. L. Darlix, and T. Ohlmann. 2003. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 221909-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez, C. V., C. Vilela, K. Berthelot, and J. E. McCarthy. 2002. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J. Mol. Biol. 318951-962. [DOI] [PubMed] [Google Scholar]
- 43.Ruan, H., C. Y. Brown, and D. R. Morris. 1997. Analysis of ribosome loading onto mRNA species: implications for translational control, p. 305-321. In J. D. Richter (ed.), mRNA formation and function. Academic Press, New York, NY.
- 44.Trono, D., R. Andino, and D. Baltimore. 1988. An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol. 622291-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilela, C., C. Velasco, M. Ptushkina, and J. E. McCarthy. 2000. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 194372-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2135-140. [DOI] [PubMed] [Google Scholar]
- 47.Yu, S. F., and R. E. Lloyd. 1992. Characterization of the roles of conserved cysteine and histidine residues in poliovirus 2A protease. Virology 186725-735. [DOI] [PubMed] [Google Scholar]