Abstract
Development of a successful hepatitis C virus (HCV) vaccine requires the definition of neutralization epitopes that are conserved among different HCV genotypes. Five human monoclonal antibodies (HMAbs) are described that cross-compete with other antibodies to a cluster of overlapping epitopes, previously designated domain B. Each HMAb broadly neutralizes retroviral pseudotype particles expressing HCV E1 and E2 glycoproteins, as well as the infectious chimeric genotype 1a and genotype 2a viruses. Alanine substitutions of residues within a region of E2 involved in binding to CD81 showed that critical E2 contact residues involved in the binding of representative antibodies are identical to those involved in the binding of E2 to CD81.
The role of neutralizing antibodies in the resolution of hepatitis C virus (HCV) infection is increasingly recognized. Early animal studies showed that gamma globulin therapy delayed the onset of acute infection (13) and that this is correlated with antibody capable of inhibiting E2 binding to CD81 (20) and blocking the infection of cells with retroviral pseudotype particles expressing the HCV E1 and E2 glycoproteins (HCVpp) (1). The protective effect of neutralizing antibodies is further supported by observations that patients with a strong and progressive neutralizing HCVpp antibody response demonstrate decreased viremia and better control of viral replication (14, 19). Thus, it is likely that any successful HCV vaccine will need to be capable of inducing a protective antibody response. However, a significant challenge for vaccine development is defining conserved epitopes that are recognized by protective antibodies.
We previously described a panel of neutralizing and nonneutralizing human monoclonal antibodies (HMAbs) to conformational epitopes on HCV E2 that were derived from the peripheral B cells of an individual infected with genotype 1b HCV. Cross-competition analyses delineated three immunogenic clusters of overlapping epitopes with distinct functions and properties (11, 12). All nonneutralizing antibodies fell within one cluster designated antigenic domain A (11). Neutralizing HMAbs segregated into two clusters designated domains B and C; domain B HMAbs have greater potency than domain C HMAbs in blocking infection with genotype 2a cell culture-infectious virus (HCVcc) (10). All domain B and domain C HMAbs inhibit E2 binding to CD81, a receptor for HCV that is essential for HCVpp and HCVcc entry into host cells (7, 8, 21). Although four different HMAbs directed to overlapping epitopes within domain B were isolated from one HCV-infected individual, it remains unclear whether the domain B epitopes on E2 are dominant targets of the immune response. This report describes the isolation of five new HMAbs from a genotype 1a HCV-infected individual that cross-compete with domain B antibodies in the earlier panel (6) that were isolated from a genotype 1b patient. Analysis of these new antibodies has expanded the number of overlapping epitopes within this domain and, moreover, has shown that antibody recognition of this domain is a conserved feature of these two prevalent HCV subgenotypes.
Peripheral blood B cells were isolated from an individual with chronic HCV genotype 1a infection who had a high serum antibody binding titer to E2 and high neutralizing activity (>1:10,000 titer) against genotype 1a HCVpp. The B cells were activated by Epstein-Barr virus and used to produce human hybridomas, as described previously (6). Both HCV 1a and 1b recombinant E2 proteins expressed in HEK293 cells were used as the target antigens. Five hybridomas, designated HC-1, HC-2, HC-11, HC-12, and HC-13, were identified that secreted antibodies that bound to the E2 proteins, using an immunofluorescence assay (11). Monoclonality was confirmed by sequencing of the immunoglobulin G (IgG) genes isolated from 10 individual cell clones derived from each hybridoma. The cell lines HC-1 and HC-2 produced IgG2 antibodies, and cell lines HC-11, HC-12, and HC-13 produced IgG1 antibodies. All of the secreted IgG possessed κ light chains, and all of the cell lines secreted approximately 20 to 60 μg of human IgG per ml in spent cultured supernatant. Each of the five antibodies immunoprecipitated genotype 1b E2 (Fig. 1) but did not bind E2 under reducing conditions, as found by either enzyme-linked immunosorbent assay (ELISA) or Western blotting analyses (data not shown), indicating that the HC HMAbs recognize conformational epitopes on the HCV E2 glycoprotein.
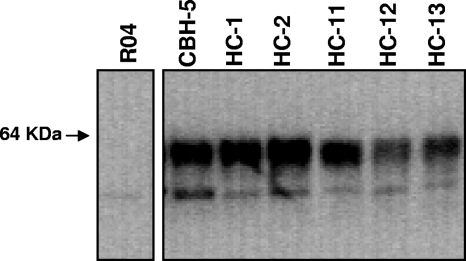
FIG. 1.
HC HMAbs immunoprecipitate genotype 1b HCV E2. Antibodies used for the immunoprecipitation of E2 expressed in 293T cells are indicated at the top of the panel. CBH-5 was used as a positive control, and RO4, an isotype-matched CMV HMAb, was used as a negative control. The apparent molecular mass (in kDa) is shown at the left. The immunoprecipitation pellet was separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis under reducing conditions, and immunoblots were analyzed with an anti-E2 murine MAb, AP33 (3).
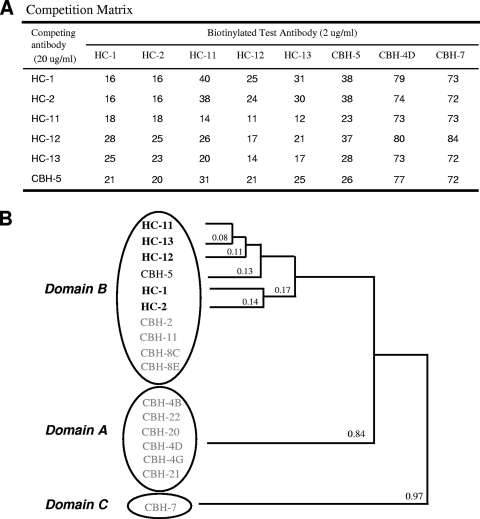
To define the relationship of these new antibodies to those in the earlier antibody panel (6), we carried out a competition analysis using representative biotin-labeled domain A (CBH-4D)-, B (CBH-5)-, and C (CBH-7)-specific HMAbs (12) (Fig. 2). Each of the five HC HMAbs showed minimum or no competition with domain A- and C-specific antibodies but did show 62 to 89% competition with CBH-5 (Fig. 2A), indicating that the epitopes recognized by these new antibodies are located within domain B. Note that Fig. 2A lists the percentages of residual binding of biotinylated test HMAbs in the presence of the competing HMAb to HCV 1b E2. The same competition analysis using HCV 1a E2 suggested a domain allocation for these antibodies similar to that in 1b E2, indicating that domain A, B, and C segregation is not a subtype-specific phenomenon (data not shown). The new group of antibodies was tested further by cross-competition, and their degree of relatedness was plotted in a hierarchical fashion using a dendrogram (Fig. 2B). In this analysis, the most closely related antibodies among the domain B HMAbs are placed next to one another as determined by an algorithm described previously (12). HC-1 is most closely related to HC-2, and HC-11, HC-12, and HC-13 are more closely related to each other as a subgroup. CBH-5 is closer to HC-11, HC-12, and HC-13 than to HC-1 and HC-2. The small distance apparent between the antibodies points to a series of overlapping epitopes.
FIG. 2.
Analysis of competition between the five HC HMAbs (bold type) and other HCV HMAbs to domain B. (A) The cross-competition matrix is based on the binding of a biotinylated test antibody to genotype 1b HCV E2 (expressed in 293T cells), captured onto Galanthus nivalis lectin-coated microtiter plates in the presence of various concentrations of competing antibodies (12). The bound biotinylated test antibody was detected by using alkaline phosphatase-conjugated streptavidin and p-nitrophenyl phosphate as the substrate. The numbers shown represent the percentages of residual binding. (B) Hierarchical clustering of HCV HMAbs based on relatedness in a competitive binding assay. Solid lines with numbers indicate the relatedness of the two adjoining antibodies. In this analysis, the cross-competition percentages of any two antibodies were averaged and expressed as a fraction; the smaller the fraction, the greater the cross-competition (12). Circles are clusters of antibodies in a specific domain. Three domains are indicated at the left. The relatedness of domain A and C HMAbs has been described previously (12).
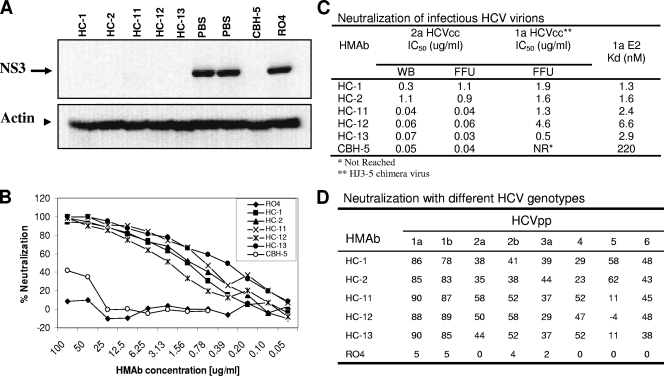
Since antibodies recognizing overlapping epitopes within a domain might be expected to possess similar functional activities (12), the five HC HMAbs were evaluated for their neutralization activities via blocking E2 binding to CD81. Neutralization was tested with infectious genotypes 1a and 2a HCVcc. For 2a HCVcc, neutralizing activity was measured by two assays, the inhibition of the NS3 protein expression in infected Huh7.5 cells, using Western blotting analysis (Fig. 3A), and a focus-forming unit (FFU) reduction assay (Fig. 3C). The effect of each HMAb on 1a HCVcc infectivity was determined by the FFU reduction assay (Fig. 3B and C). As shown in Fig. 3A, neutralization of 2a HCVcc was complete with each HC HMAb at 10 μg/ml, as assessed by immunoblotting for HCV NS3 expression. In contrast, the infectivity of 2a HCVcc was unaffected by the negative control antibody, RO4, in comparison to that of the no antibody control (phosphate-buffered saline [PBS]). The levels of β-actin protein used as an internal loading control for the immunoblots were comparable between different samples. More quantitative measures of neutralization potency were determined by calculating the 50% inhibitory concentration (IC50). HCVcc 2a virions (HCV titer at 105 FFU/ml) were incubated for 1 h with a range of concentrations for each HMAb (0.001 to 100 μg/ml) before Huh7.5 cells were infected. Neutralizing activity was again determined by NS3 protein expression in the infected cells, using β-actin protein as an internal control. As shown in Fig. 3C, HC HMAbs neutralized 2a HCVcc with IC50 values ranging from 0.04 to 1.1 μg/ml. Results obtained with the FFU reduction assay for 2a HCVcc (where much smaller quantities of virus were incubated with antibody) were comparable, with IC50 values in this assay ranging from 0.03 to 1.1 μg/ml. This is remarkable, since these two assays measure the neutralization of amounts of the 2a HCVcc genotype that differ by many orders of magnitude and, moreover, use very different methodologies. An issue in comparing neutralization potency from laboratory to laboratory is the relative equivalency using different assays. Neutralization of the 1a HCVcc genotype (Fig. 3B and C) was measured only in the FFU reduction assay, using concentrations of each HC HMAb ranging from 0.05 to 100 μg/ml. The IC50 values ranged from 0.5 to 4.6 μg/ml.
FIG. 3.
Neutralization with genotypes 1a and 2a infectious HCV virions and different HCVpp genotypes. (A) Virus neutralization of genotype 2a HCVcc infection as determined by NS3 expression. Genotype 2a HCVcc was incubated with each HMAb as indicated at 10 μg/ml prior to the infection of Huh7.5 cells. At 3 h postinfection, the HCV/antibody-containing medium was removed, and the cells were washed with PBS. Samples were harvested at 72 h postinfection for Western blotting analysis. The HCV NS3 protein expression was determined by using a murine MAb to NS3 (2). The β-actin protein used as an internal control was detected by using an anti-β-actin monoclonal antibody. R04 was used as a negative control. PBS was used as a reagent control. (B) Virus neutralization of genotype 1a HJ3-5 HCVcc infection as determined by FFU reduction (22, 23). Infectious genotype 1a HJ3-5 chimeric virus inoculum was incubated with each HMAb at different concentrations, as indicated, for 1 h at 37°C prior to inoculation of Huh7.5 cells preseeded onto eight-well tissue culture chamber slides. Cells were fixed and stained with a MAb to core antigen at day 4 postinfection and enumerated by FFU assay. (C) The inhibitory antibody concentrations reducing HCV infection by 50%, IC50, of HC HMAbs with genotypes 2a and Ia infectious virions. Neutralization was assessed by two methods: the reduction of NS3 expression as monitored by Western blotting (WB) analysis (10) and reduction in focus-forming units (FFU) as described previously (22, 23). Antibody affinity measurements were performed by ELISA with Galanthus nivalis-captured 1a E2 as described previously (11). Data were analyzed by nonlinear regression to ascertain the antibody disassociation constant, Kd, using Prism software (GraphPad). (D) Neutralization with HCVpp pseudotyped with the E1 and E2 sequences of genotypes 1 to 6. Each antibody was tested at 20 μg/ml. The numbers are percentages of neutralization compared to that of the no-antibody control. RO4 is an isotype-matched HMAb to HCMV. The E1 and E2 sequences used were those described previously (16).
Importantly, each of the antibodies showed significantly greater potency than CBH-5 for neutralizing 1a HCVcc, as CBH-5 was not capable of causing a 50% reduction in FFU, even at 100 μg/ml (Fig. 3B). This may reflect subgenotype-specific differences in their respective epitopes, since CBH-5 was isolated from a 1b-infected patient and the new HC antibodies were isolated from a 1a-infected patient. Thus, although domain B is conserved as an antigenic site recognized by neutralizing antibodies in both of these subgenotypes, there appear to be significant differences in some of the epitopes that comprise this cluster. Among the expanded panel of domain B HMAbs, neutralization potency was correlated with antibody binding affinity. CBH-5 has a lower antibody binding affinity for the 1a E2 genotype, with a Kd (dissociation constant of 2.2 × 10−7 M) (12), compared to the affinities of the HC HMAbs, with Kds 1.3 × 10−9 and 6.6 × 10−9 M (Fig. 3C).
The breadth of cross-neutralization was investigated by measuring the neutralization activities of each antibody at 20 μg/ml against the genotypes 1a, 1b, 2a, 2b, 3a, 4, 5, and 6 (16) HCVpp (Fig. 3D). The production and use of HCVpp neutralization assays have been described previously (10). As shown in Fig. 3D, each of the HC HMAbs reduced 1a and 1b HCVpp infection by over 80%. Reduction of genotype 2a, 2b, 3a, 4, 5, and 6 HCVpp infection ranged from 30 to 60% compared to that of R04, an isotype-matched control antibody. However, HC-11, HC-12, and HC-13 failed to neutralize genotype 5 HCVpp infection. These results suggest that the HC HMAbs targeting domain B are, in general, capable of mediating broad virus neutralization across multiple genotypes, although there are differences in both the specificity and the efficiency among the individual HMAbs. One possibility is that antibody affinities to these genotypes are less than those to genotype 1a and 1b, since all the HC HMAbs bound to E2 from each of these genotypes, as detected by immunofluorescence assay, which suggests that the cognate epitopes are conserved among different HCV genotypes and subtypes (data not shown).
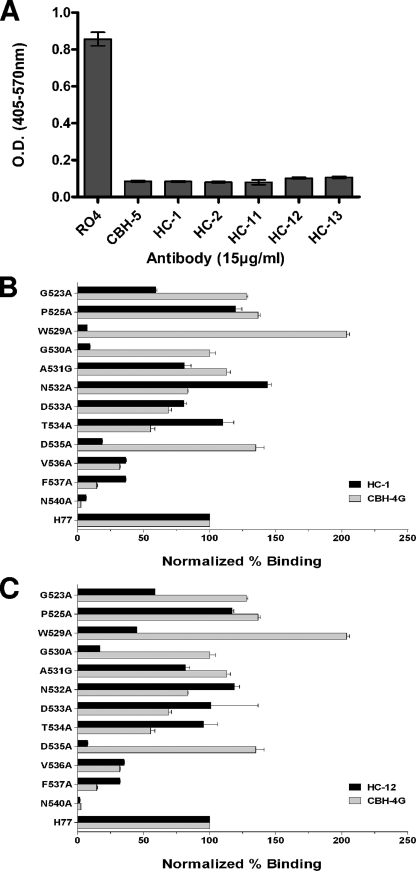
Previous work suggests that the domain B HMAbs described previously neutralize virus by inhibiting the binding of E2 to CD81. We used a CD81 capture assay to determine if the HC HMAbs shared this potential neutralization mechanism. As shown in Fig. 4A, preincubation of E2 with 15 μg/ml of each HC HMAb or CBH-5 reduced E2 binding to CD81 by over 90%, compared to that of the RO4 negative control. Thus, similar to other domain B HMAbs, the HC HMAbs appear to mediate neutralization by inhibiting an intermediate step in virus entry that involves the binding of the E2 glycoprotein to the cell surface protein CD81. Taken together, these results are consistent with a model in which antibodies that recognize overlapping conformational epitopes within a distinct immunogenic domain on the E2 glycoprotein have similar functions. The isolation of these antibodies to domain B from patients infected with different virus subgenotypes confirms that the domain B epitopes are highly immunogenic and elicit potent neutralizing antibody responses in HCV-infected persons.
FIG. 4.
Inhibition of E2 binding to CD81 by HC HMAbs and location of critical residues involved in their epitopes. (A) Genotype 1a H77c E1E2 expressed in 293T cells containing 1 μg/ml E2 was incubated with each test HMAb at 15 μg/ml, and the antibody-antigen complex was then added onto CD81-precoated wells. Detection of E2 bound to CD81 was measured with biotinylated CBH-4D (12). CBH-5 was used as a positive control and RO4 as a negative control. The experiments were performed twice in triplicate. Error bars indicate one standard deviation from the mean. (B and C) Epitope mapping of the two representative HC HMAbs, HC-1 and HC-12, by alanine replacement. Mutated E2 proteins were expressed in 293T cells and analyzed by ELISA as described previously (11). The mutated amino acids are shown on the y axis. The numbers at the beginning of the peptides correspond to the position in the polyprotein of reference strain H (GenBank accession no. AF009606). HC HMAb binding to each mutant is expressed as the percentage of the binding value normalized by the binding of CBH-17 (6) and divided by HC HMAb binding to the wild type on the x axis.
Next, we carried out alanine scanning mutagenesis to define the residues within the HC HMAb epitopes that are engaged in E2-CD81 interactions. Since HC-1 is more cross-competitive with HC-2 and HC-11, HC-12, and HC-13 are more competitive with each other than with HC-1 or HC-2 (Fig. 2), HC-1 and HC-12 were selected as representative antibodies for epitope mapping studies. A series of E2 proteins mutated as point mutations between residue 523 and 540 were generated by site-directed mutagenesis (Stratagene). This region contains Trp529, Gly530, and Asp535, which are known to be essential contact point residues for E2 binding to CD81 (17). In addition, the residues Gly523, Pro525, Gly530, and Asp535 are known to be involved in the CBH-5 epitope (16). Mutated E2 proteins were expressed in HEK 293 cells, and the binding of antibodies to cell lysates analyzed by ELISA (Fig. 4B and C). To account for potential differences in E2 abundance in cell lysates, results were normalized according to the detection of E2 by an antibody to a linear epitope outside of domain B, CBH-17 (6). CBH-4G, a nonneutralizing HMAb (12), was included as a control to identify residues that are specific to HC-1 or HC-12. A lack of binding by both CBH-4G and the HC HMAb would suggest that an alanine substitution had caused either an overall change in E2 conformation or that there is some degree of overlap between the neutralizing and the nonneutralizing epitopes (Fig. 4B). Compared to that of the wild type, HC-1 binding was significantly reduced (by 90% or more) by the replacement of Trp529, Gly530, and Asn540 with alanine. However, since Asn540 is modified by N-linked glycosylation, it is not likely to be part of an epitope. In addition, substitution of this residue alters the folding of E2 (5). A mutation at position Asp535 also specifically decreased HC-1 binding by >80%. For HC-12, a significant decrease in binding (>90%) was observed when Asp535 and Asn540 were replaced by alanine, as shown in Fig. 4C. A mutation at Gly530 also decreased HC-1 binding by >80%. In contrast to HC-1, a mutation at Trp529 reduced HC-12 binding by only 44% compared to that of the wild type.
Collectively, these results show that Trp529, Gly530, and Asp535 are specific contact residues for HC-1, while Gly530 and Asp535 are contact residues for HC-12. These two contact patterns suggest that the HC-12 epitope is closer to the CBH-5 epitope, which also involves Gly530 and Asp535 but with at least one different contact point at Gly523 (16). Indeed, competition studies also suggest that the HC-12 epitope is spatially closer to CBH-5 than to either HC-1 or HC-2 (Fig. 2). Both HC-1 and HC-12 bind to E2 with alanine substitutions at Gly523, in contrast to CBH-5, which showed no binding. Thus, the ability of HMAbs HC-1 and HC-12 to neutralize HCVpp and HCVcc infections is associated with interactions with residues that are also contact points for E2 binding to CD81. The differences in cross-reactivity and neutralizing potency for different isolates suggest that HMAbs HC-1 and HC-12 each bind to discontinuous epitopes involving other contact points that have yet to be defined. These putative residues may or may not be involved directly in E2 binding to CD81. Recent studies show that other broadly neutralizing human antibodies recovered from combinatorial libraries isolated from three HCV-infected individuals, two with genotype 1a or 2b virus, also recognize epitopes containing Gly523, Trp529, Gly530, and Asp535 (9, 15, 18).
In aggregate, these results suggest that domain B is an immunodominant region on HCV E2, containing multiple overlapping neutralization epitopes, and induces potent neutralizing antibodies in patients infected with different genotype 1 subtypes. Previous studies show that HCVpp with an alanine substitution mutation at Asn532 demonstrates enhanced sensitivity for neutralization by sera obtained from individuals infected with genotype 1a, 1b, 2b, 3, 4, or 5 virus (4). This suggests that these sera contain antibodies directed at residues near Asn532, in close proximity to residues contributing to domain B. These results thus confirm that the epitopes comprising domain B are frequent targets of the humoral immune response in patients with chronic hepatitis C infection. However, from a vaccine perspective, additional studies are required to determine which of the domain B epitopes may be prone to accumulate mutations under immune pressure that lead to virus escape from neutralization, as observed with the antibody response to HVR1, and which if any domain B epitopes must remain relatively invariant to accommodate the interaction of E2 with CD81 required for viability of the virus.
Acknowledgments
We thank Guangxiang Luo for the anti-NS3 antibody and Shoshana Levy for the CD81 construct.
This work was supported in part by National Institutes of Health grants HL079381 to S.K.H.F. and U19-AI40035 to S.M.L. M.G.-T. was supported by a McLaughlin postdoctoral research fellowship.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 10014199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 7913963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 767672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkowska, E., F. Kajumo, E. Garcia, J. Reinus, and T. Dragic. 2007. Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J. Virol. 818072-8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goffard, A., N. Callens, B. Bartosch, C. Wychowski, F. L. Cosset, C. Montpellier, and J. Dubuisson. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 798400-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 7410407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helle, F., and J. Dubuisson. 2007. Hepatitis C virus entry into host cells. Cell. Mol. Life Sci.: CMLS 653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 1007271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson, D. X., C. Voisset, A. W. Tarr, M. Aung, J. K. Ball, J. Dubuisson, and M. A. Persson. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. USA 10416269-16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keck, Z., J. Xia, Z. Cai, T. Li, A. M. Owsianka, A. H. Patel, G. Luo, and S. K. H. Foung. 2007. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J. Virol. 811043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keck, Z. Y., T. K. Li, J. Xia, B. Bartosch, F. L. Cosset, J. Dubuisson, and S. K. Foung. 2005. Analysis of a highly flexible conformational immunogenic domain a in hepatitis C virus E2. J. Virol. 7913199-13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keck, Z. Y., A. Op De Beeck, K. G. Hadlock, J. Xia, T. K. Li, J. Dubuisson, and S. K. Foung. 2004. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J. Virol. 789224-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krawczynski, K., M. J. Alter, D. L. Tankersley, M. Beach, B. H. Robertson, S. Lambert, G. Kuo, J. E. Spelbring, E. Meeks, S. Sinha, and D. A. Carson. 1996. Effect of immune globulin on the prevention of experimental hepatitis C virus infection. J. Infect. Dis. 173822-828. [DOI] [PubMed] [Google Scholar]
- 14.Lavillette, D., Y. Morice, G. Germanidis, P. Donot, A. Soulier, E. Pagkalos, G. Sakellariou, L. Intrator, B. Bartosch, J. M. Pawlotsky, and F. L. Cosset. 2005. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J. Virol. 796023-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law, M., T. Maruyama, J. Lewis, E. Giang, A. W. Tarr, Z. Stamataki, P. Gastaminza, F. V. Chisari, J. M. Jones, R. Fox, J. K. Ball, J. McKeating, N. Kneteman, and D. R. Burton. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 1425-27. [DOI] [PubMed] [Google Scholar]
- 16.Owsianka, A. M., A. W. Tarr, Z. Keck, T. Li, J. Witteveldt, R. Adair, S. Foung, J. K. Ball, and A. H. Patel. 2008. Broadly neutralizing human monoclonal antibodies to Hepatitis C Virus E2 glycoprotein. J. Gen. Virol. 89653-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owsianka, A. M., J. M. Timms, A. W. Tarr, R. J. Brown, T. P. Hickling, A. Szwejk, K. Bienkowska-Szewczyk, B. J. Thomson, A. H. Patel, and J. K. Ball. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 808695-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perotti, M., N. Mancini, R. A. Diotti, A. W. Tarr, J. K. Ball, A. Owsianka, A. H. Patel, M. Clementi, and R. Burioni. 2007. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the HCV E2 Protein. J. Virol. 821047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pestka, J. M., M. B. Zeisel, E. Blaser, P. Schurmann, B. Bartosch, F. L. Cosset, A. H. Patel, H. Meisel, J. Baumert, S. Viazov, K. Rispeter, H. E. Blum, M. Roggendorf, and T. F. Baumert. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. USA 1046025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 931759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time-and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 801734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 1032310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]