Abstract
Serial undiluted passage of a porcine rotavirus in MA104 cells yielded three distinct virus populations, each of which bore different rearranged genes. Sequencing revealed that each of two populations bore a distinct intragenic recombinant NSP3 gene consisting of a partial duplication in a head-to-tail orientation without altering the NSP3 open reading frame and the third population carried both an intragenic recombinant NSP3 gene and an intergenic recombinant gene (1,647 nucleotides in length) which contained a truncated NSP2 gene inserted into the NSP5 gene at residue 332. The former two populations were viable, whereas the latter population was defective and interfering.
Group A rotaviruses, members of the Reoviridae family, are responsible for approximately 600,000 deaths annually among infants and young children, predominantly in developing countries (5, 16). Although deaths due to rotavirus diarrhea occur much less frequently in industrialized countries, the rate of hospitalizations and associated health care costs in such countries is enormous (5). Thus, the development and implementation of a safe and efficacious vaccine have been important global public health goals.
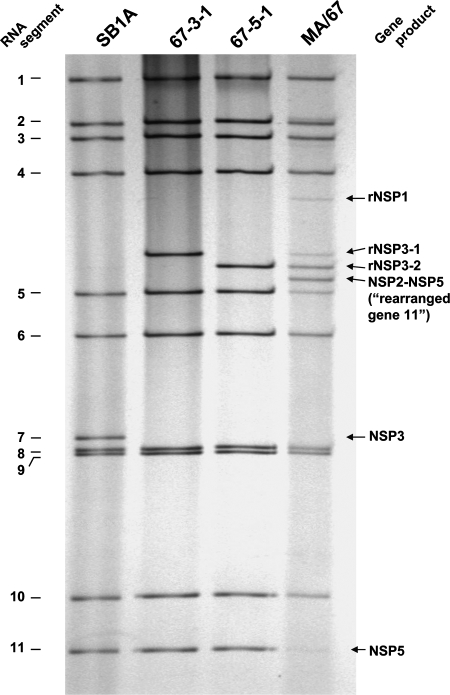
The rotavirus genome consists of 11 segments of double-stranded RNA (dsRNA) and displays a characteristic 4-2-3-2 distribution pattern of the segments in polyacrylamide gel electrophoresis (PAGE) (9), as shown for strain SB1A in Fig. 1. Rotaviruses bearing a rearranged genome segment(s), which can be readily identified by regular PAGE analyses, have been (i) recovered from humans and various animal species (4) and (ii) generated by serial passages of the virus at a high multiplicity of infection (MOI) in cell culture (8). In most cases, the rearrangement results from a partial head-to-tail duplication of the gene (intragenic recombination) which starts at various positions after the stop codon (4). Rotaviruses carrying intragenic recombinant genes are not defective, and the rearranged segments can reassort in vitro and replace their normal counterparts structurally and functionally (1, 3, 6). Genome rearrangements have been proposed to play a role in the evolution of rotaviruses and contribute to their diversity (19, 20). So far, no mosaic structures due to an intergenic recombination have been found in rotavirus rearranged genes.
FIG. 1.
Electrophoretic migration patterns of genomic RNAs of the porcine rotavirus SB1A strain, passage 67 lysate (MA/67), and two plaque-purified viruses (67-3-1 and 67-5-1) derived from the MA/67 lysate in a 10% polyacrylamide gel. Each of the 67-3-1 and 67-5-1 viruses bears a distinct intragenic recombinant NSP3 gene (i.e., rNSP3-1 and rNSP3-2, respectively). In addition to the 67-3-1 and 67-5-1 populations, the MA/67 lysate contains a virus population bearing both an intergenic recombinant NSP2-NSP5 gene and an intragenic recombinant NSP3 gene. The fourth faint extra band located right below gene segment 4 is an intragenic recombinant NSP1 gene (data not shown).
Defective interfering (DI) virus particles, which do not possess the complete viral genome in comparison to that of the standard virus, can infect cells but need the helper function of standard virus particles to complete their replication cycle and interfere with the replication of infectious particles (2). Serial passage of a virus in vitro at a high MOI frequently gives rise to DI particles, a property which has been demonstrated for many virus species, including reoviruses (14). Despite many attempts, however, rotavirus DI particles have not been generated.
To analyze the underlying mechanisms of the formation of viruses bearing an intragenic recombinant gene and of their effects and to pursue a possible use of such viruses as vaccine candidates, we initiated serial undiluted passage of a porcine rotavirus in cell culture.
Detection of rotaviruses bearing rearranged genes.
The rotavirus SB1A strain, which was isolated from a piglet with diarrhea at Ohio State University, Wooster, bears a G4P9[7] specificity (7). The first passage of the virus was initiated in MA104 cells in a culture glass roller tube (approximately 2 × 105 cells/tube) with an MOI of approximately 80. Following adsorption for 1 h, the inoculum was removed, 2 ml of maintenance medium (Eagle's minimal essential medium supplemented with 0.5 μg/ml trypsin and antibiotics) was added to each tube, and the tubes were rolled on a roller drum at 37°C. When the cells exhibited an approximately 50 to 75% cytopathic effect, the cultures were frozen and thawed once and then harvested. An undiluted virus suspension (500 μl/tube) was then used for the next passage on fresh MA104 cells (MOIs used throughout the serial passages ranged from 1 to 9). We found that around passage 40, segment 7 of the virus lost its intensity and two new bands became visible in the silver nitrate-stained gel, and around passage 67, segment 11 became barely visible and a new band became visible. The electropherotype of viruses in the passage-67 lysate (designated MA/67) displayed an unusual pattern of 14 visible dsRNA bands, including 10 normal (segments 1 to 6 and 8 to 11) and 4 aberrant segments located between segments 4 and 5 (Fig. 1). Notably, segment 7 was invisible, and the intensity of segment 11 was low. Such an RNA profile indicated that the MA/67 material contained several subpopulations of viruses, with some bearing rearranged genes. Attempts to separate constituent virus subpopulations comprising the MA/67 group by plaque purification yielded the following: (i) two distinct populations (designated 67-3-1 and 67-5-1), each of which bore 10 normal genes, including gene 11, and one rearranged gene, gene 7 (Fig. 1); and (ii) no virus population bearing the normal gene 7 or “rearranged gene 11.” Figure 1 shows the fourth extra band just below gene segment 4, which was shown later to be an intragenic NSP1 gene (i.e., a rearranged genome segment 5). However, since, we could not plaque isolate the virus bearing the rearranged NSP1 gene (data not shown), we did not include it in the current study. Unexpectedly, although the virus population that lacked the normal gene 11 (and supposedly bore “rearranged gene 11”) was dominant as judged by the intensity of the “rearranged gene 11” band relative to the normal gene 11 band (in Fig. 1, the “rearranged gene 11” band is correctly labeled the “NSP2-NSP5” gene, as demonstrated by sequence analysis shown below), repeated attempts by plaque purification or a limiting dilution to isolate such a population were unsuccessful. We picked more than 300 plaques, amplified each of those plaques in cell culture, and analyzed dsRNAs of amplified viruses by PAGE; each one of them was shown to have the normal gene 11 (data not shown). In addition, we performed plaque purification at 30°C on the assumption that viruses bearing the intergenic recombinant gene might bear a temperature-sensitive phenotype; however, we could not isolate viruses lacking the normal gene 11 (data not shown). These findings suggested that viruses bearing the “rearranged gene 11” may be nonviable. Furthermore, when the MA/67 viruses were passaged at an MOI of approximately 0.01 twice serially in cell culture, a virus population bearing “rearranged gene 11” (i.e., lacking the normal gene 11) became invisible and virus populations bearing the normal gene 11 (i.e., 67-3-1 and 67-5-1) became predominant in PAGE (data not shown). These findings suggested that the virus population lacking the normal gene 11 not only was nonviable (i.e., defective) but also was interfering with the replication of viable viruses bearing the normal gene 11 during serial undiluted passage in cell culture.
Virus population lacking normal gene 11 bore a rearranged gene with dual NSP2 and NSP5 specificities.
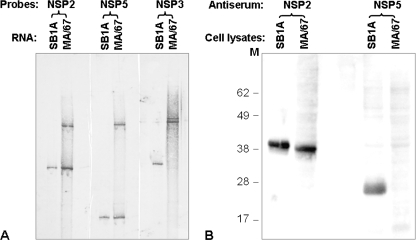
To analyze the structure and gene specificity of the rearranged genes, we performed a Northern blot analysis of the fractionated RNA segments of the passage-67 viruses with NSP2, NSP3, and NSP5 cDNA probes. The analysis revealed (Fig. 2A) that (i) virus populations bearing the normal NSP2 gene and the normal NSP5 gene were present, (ii) populations bearing the normal NSP3 gene were undetectable, (iii) populations comprising MA/67 bore one of two distinct rearranged NSP3 genes, and (iv) populations which were defective did not bear the rearranged gene 11 but carried a rearranged gene bearing both NSP2 and NSP5 specificities.
FIG. 2.
Analysis of rotavirus RNAs and expression of proteins in MA/67-infected MA104 cells. Confluent MA104 cell monolayers in a six-well plate infected with either the undiluted MA/67 (0.5 ml/well) or parental SB1A (10 PFU/cell) strain were harvested at 14 h p.i. (A) Viral genomic dsRNAs extracted from the cell lysates were fractionated on a 10% polyacrylamide gel for 10 h at 12 mA and then blotted onto a BrightStar Plus membrane (Ambion Inc., Austin, Tx). The blots were probed with SB1A NSP2-specific (XbaI/EcoRI fragment, 249 bp), NSP3-specific (SpeI/PstI fragment, 597 bp), or NSP5-specific (full-length, 667 bp) cDNA labeled with digoxigenin-11-UTP. Signals were detected by using the DIG High Prime DNA labeling and detection starter kit I (Roche Diagnostics) according to the manufacturer's instructions. (B) Proteins from the harvested cell lysates described for panel A were electrophoresed in NuPAGE gel (Invitrogen) at 200 V for 50 min and then blotted onto nitrocellulose. The blot was probed with guinea pig anti-NSP5 polyclonal antiserum (1:2,000 dilution) or guinea pig anti-NSP2 polyclonal antiserum (1:1,500), followed by treatment with goat anti-guinea pig horseradish peroxidase-conjugated antibody (1:10,000). Molecular mass markers (in kilodaltons) are shown on the left of the gel.
Rearranged segment 7 bore a head-to-tail concatemerization sequence structure.
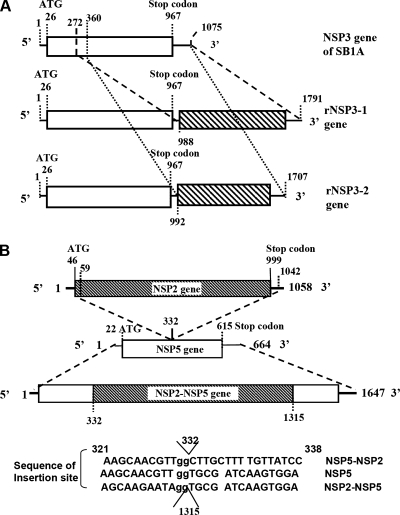
The full-length gene 7, encoding NSP3 of the parental SB1A virus or the MA/67 virus, was amplified using a modified sequence-independent dsRNA amplification method (10, 18). Sequencing revealed that each rearranged gene 7 had a partial head-tail duplication of the nucleotide sequence (Fig. 3A); one (rNSP3-2; 1,707 nucleotides [nt] in length), from residues 360 to 1075 after the stop codon at residue 992, and another (rNSP3-1; 1,791 nt in length) from residues 272 to 1075 at residue 988. Both rearranged NSP3 genes retained a normal open reading frame (ORF) with a long 3′ noncoding region.
FIG. 3.
(A) Schematic diagram of the sequence organization of the normal NSP3 gene and two distinct intragenic recombinant NSP3 genes (i.e., rNSP3-1 and rNSP3-2) of 67-3-1 and 67-5-1 viruses. The region of duplicated sequence of the gene is hatched. Numbers denote nucleotide positions. (B) Schematic diagram of the sequence organization of an intergenic recombinant NSP2-NSP5 gene. Numbers indicate nucleotide positions.
Sequence organization of a rearranged gene with dual NSP2 and NSP5 specificities.
Sequencing of the third rearranged segment revealed that this segment was 1,647 nt in length, contained residues 59 to 1042 of the NSP2 gene inserted into the NSP5 gene at residue 332 (Fig. 3B); and had two potential ORFs. The first ORF could encode a partial NSP6 protein of 58 amino acids (residues 25 to 83), as well as the NSP2 protein, which was 4 amino acids (i.e., MAEL) shorter than the full-length protein. The second ORF could encode the N-terminal half of the NSP5 protein and an 11-amino-acid peptide encoded by the NSP2 gene. An intersegmental recombination, demonstrated for the first time for rotavirus in this study, has been detected in other virus species, including influenza viruses (12) and bacteriophage phi 6 (15). This study demonstrated that an intergenic recombination, albeit rare, could play a role in rotavirus evolution.
Intergenic recombinant NSP2-NSP5 gene exists in virus particles.
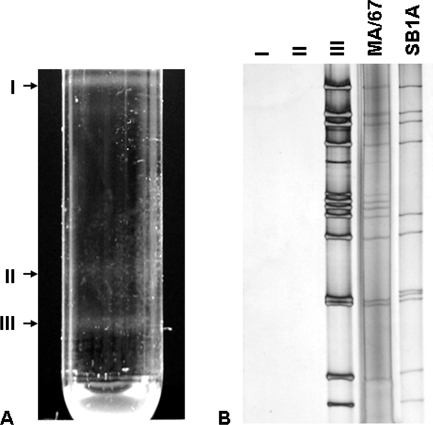
Following extraction of the MA/67-infected MA104 cell lysates with trichlorotrifluoroethane, the virus particles were pelleted by centrifugation at 90,000 × g in a Beckman SW28 rotor for 2 h at 4°C. Resuspended pellets in Tris-buffered saline were mixed with CsCl to achieve a density of 1.370 g/cm3 and centrifuged at 110,000 × g in a Beckman SW55 Ti rotor for 20 h at 4°C. The resulting three bands (Fig. 4A) were separated and analyzed by PAGE. As shown in Fig. 4B, we could not separate a noninfectious virus population bearing the intergenic recombinant NSP2-NSP5 gene and an infectious virus population bearing normal gene 11; fraction III contained both infectious and noninfectious virus particles. Fraction III was also examined by electron microscopy and was shown to consist of virus particles (data not shown). Thus, these findings have shown that the intergenic recombinant NSP2-NSP5 gene is indeed incorporated into virus particles.
FIG. 4.
Fractionation by cesium chloride isopycnic centrifugation of MA/67 viruses (A) and PAGE analysis of fractionated virus populations I, II, and III (B). The dsRNAs of the MA/67 viruses and SB1A were also included in PAGE analysis. Noninfectious DI particles and infectious virus particles were inseparable by this method.
Rotavirus bearing recombinant NSP2-NSP5 gene displayed an interfering ability against parental SB1A virus.
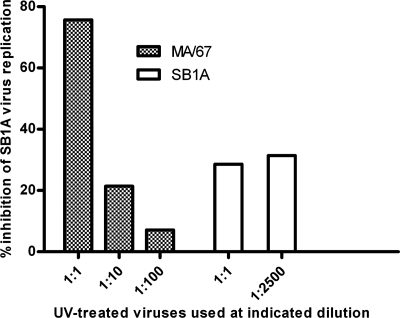
The interfering ability of the inactivated MA/67 virus against infectious parental SB1A virus was measured according to a method reported previously (13). Briefly, the undiluted MA/67 virus (4 × 103 PFU/ml before UV irradiation; the majority of viruses bear the NSP2-NSP5 gene, as judged by PAGE in Fig. 1) was exposed to an 8-W germicidal lamp at a distance of 20 cm for 5 min to inactivate its infectivity. Monolayers of MA104 cells (in a six-well plate) were infected with SB1A (1 PFU/cell) or coinfected with SB1A (1 PFU/cell) and undiluted UV-inactivated MA/67. UV-inactivated SB1A virus (5 × 107 PFU/ml before UV irradiation) was used similarly as a control. After incubation for 30 min at 37°C, maintenance medium was added without removing the inoculum, and the cell cultures were incubated at 37°C. At 2.5 h postinfection (p.i.), the cultures were treated with a 1:1,000 dilution of antiserum to SB1A virus for 20 min at 37°C, washed three times with L15 medium, and incubated in fresh maintenance medium at 37°C. Progeny viruses were harvested at 14 h p.i., and virus yield was determined by plaque assay. As shown in Fig. 5, the undiluted UV-inactivated MA/67 virus inhibited the replication of its parental SB1A virus up to 75.7%, and this interfering ability of the MA/67 virus was dose dependent, whereas the UV-inactivated SB1A virus inhibited homologous SB1A virus replication by 28.6%, and this interfering ability did not appear to be dose dependent.
FIG. 5.
UV-treated MA/67 viruses (the majority of which were DI particles) displayed a strong dose-dependent interfering effect against replication of parental SB1A virus.
Expression of NSP5 protein in MA/67 rotavirus-infected cells appeared to be subdued.
Although the normal NSP5 gene was detectable by Northern blotting in the MA/67 lysates (Fig. 2A), the expression of the NSP5 protein was too low to be detected by Western blotting with an NSP5-specific polyclonal antiserum (Fig. 2B). In addition, fusion proteins were not detectable in the MA/67 lysates. Since the presence of the NSP5 protein is essential for the formation of a viroplasm (the site of viral RNA replication and packaging of genome segments into progeny virions) (17), our findings suggested that virus bearing the recombinant NSP2-NSP5 gene was defective in cell culture in the absence of viable viruses and interfered with the replication of viable viruses. It is noteworthy that the virus population bearing normal gene 11 (which could supply the NSP5 protein, essential for the replication of viruses bearing the recombinant NSP2-NSP5 gene), albeit very minor in population, could never be eliminated during an additional 10 serial undiluted passages of the MA/67 material in cell culture (data not shown), indicating that the presence of infectious viruses is essential for the survival of DI viruses.
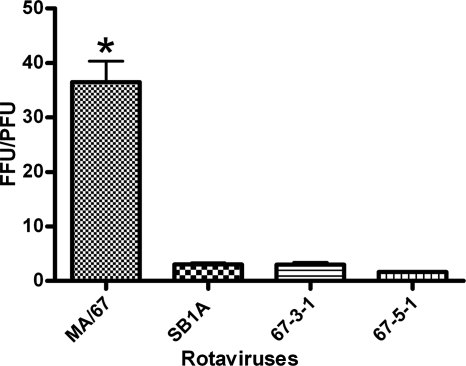
We compared the titers of four viruses (i.e., MA/67, SB1A, 67-3-1, and 67-5-1) determined by two assay systems: numbers of PFU, determined by plaque assay, and focus-forming units (FFU), determined by fluorescent focus-forming assay. As shown in Fig. 6, an FFU/PFU ratio of virus SB1A, 67-3-1, or 67-5-1 ranged from 2 to 3 whereas that of MA/67 virus was approximately 37, and this difference in FFU/PFU ratios between the two groups was statistically significant. This observation suggests that DI particles, after infection, undergo certain steps in the replication cycle and express some viral proteins that can be detected by fluorescent focus-forming assay. More-precise analyses of the replication of DI particles need to be performed.
FIG. 6.
FFU/PFU ratio of virus MA/67, SB1A, 67-3-1, or 67-5-1. Statistical significance was analyzed by one-way analysis of variance (GraphPad Prism, version 5.00 for Windows; GraphPad Software, San Diego, CA), followed by Tukey's posttest analysis. The asterisk indicates a significant difference between two groups (P < 0.05).
Recently intranasally administered DI influenza A virus vaccine was shown to be safe and protective against virulent virus challenge with a ferret model (the best model for human influenza) (11). In an attempt to rescue the porcine SB1A DI population in cell culture, further characterize viruses bearing the NSP2-NSP5 gene, and pursue a possible use of such viruses as a vaccine, efforts are being made to establish a cell line that expresses the NSP2 protein or the NSP5 protein.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study are available from GenBank under accession numbers EU169869 to EU169874.
Acknowledgments
We thank Ronald W. Jones for expert technical assistance, Albert Z. Kapikian for the continuing support of our work, and Edward H. Bohl for providing us with rotavirus strain SB1A.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
There is no conflict of interest to declare.
Footnotes
Published ahead of print on 16 April 2008.
REFERENCES
- 1.Allen, A. M., and U. Desselberger. 1985. Reassortment of human rotaviruses carrying rearranged genomes with bovine rotavirus. J. Gen. Virol. 662703-2714. [DOI] [PubMed] [Google Scholar]
- 2.Bangham, C. R., and T. B. Kirkwood. 1993. Defective interfering particles and virus evolution. Trends Microbiol. 1260-264. [DOI] [PubMed] [Google Scholar]
- 3.Biryahwaho, B., F. Hundley, and U. Desselberger. 1987. Bovine rotavirus with rearranged genome reassorts with human rotavirus. Brief report. Arch. Virol. 96257-264. [DOI] [PubMed] [Google Scholar]
- 4.Desselberger, U. 1996. Genome rearrangements of rotaviruses. Arch. Virol. Suppl. 1237-51. [DOI] [PubMed] [Google Scholar]
- 5.Glass, R. I., U. D. Parashar, J. S. Bresee, R. Turcios, T. K. Fischer, M. A. Widdowson, B. Jiang, and J. R. Gentsch. 2006. Rotavirus vaccines: current prospects and future challenges. Lancet 368323-332. [DOI] [PubMed] [Google Scholar]
- 6.Graham, A., G. Kudesia, A. M. Allen, and U. Desselberger. 1987. Reassortment of human rotavirus possessing genome rearrangements with bovine rotavirus: evidence for host cell selection. J. Gen. Virol. 68115-122. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino, Y., R. G. Wyatt, H. B. Greenberg, J. Flores, and A. Z. Kapikian. 1984. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J. Infect. Dis. 149694-702. [DOI] [PubMed] [Google Scholar]
- 8.Hundley, F., B. Biryahwaho, M. Gow, and U. Desselberger. 1985. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology 14388-103. [DOI] [PubMed] [Google Scholar]
- 9.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 10.Lambden, P. R., S. J. Cooke, E. O. Caul, and I. N. Clarke. 1992. Cloning of noncultivatable human rotavirus by single primer amplification. J. Virol. 661817-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mann, A., A. C. Marriott, S. Balasingam, R. Lambkin, J. S. Oxford, and N. J. Dimmock. 2006. Interfering vaccine (defective interfering influenza A virus) protects ferrets from influenza, and allows them to develop solid immunity to reinfection. Vaccine 244290-4296. [DOI] [PubMed] [Google Scholar]
- 12.Moss, B. A., and G. G. Brownlee. 1981. Sequence of DNA complementary to a small RNA segment of influenza virus A/NT/60/68. Nucleic Acids Res. 91941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayak, D. P., K. Tobita, J. M. Janda, A. R. Davis, and B. K. De. 1978. Homologous interference mediated by defective interfering influenza virus derived from a temperature-sensitive mutant of influenza virus. J. Virol. 28375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nonoyama, M., Y. Watanabe, and A. F. Graham. 1970. Defective virions of reovirus. J. Virol. 6226-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onodera, S., X. Qiao, P. Gottlieb, J. Strassman, M. Frilander, and L. Mindich. 1993. RNA structure and heterologous recombination in the double-stranded RNA bacteriophage φ6. J. Virol. 674914-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parashar, U. D., C. J. Gibson, J. S. Bresse, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patton, J. T., L. S. Silvestri, M. A. Tortorici, R. Vasquez-Del Carpio, and Z. F. Taraporewala. 2006. Rotavirus genome replication and morphogenesis: role of the viroplasm. Curr. Top. Microbiol. Immunol. 309169-187. [DOI] [PubMed] [Google Scholar]
- 18.Potgieter, A. C., A. D. Steele, and A. A. van Dijk. 2002. Cloning of complete genome sets of six dsRNA viruses using an improved cloning method for large dsRNA genes. J. Gen. Virol. 832215-2223. [DOI] [PubMed] [Google Scholar]
- 19.Ramig, R. F. 1997. Genetics of the rotaviruses. Annu. Rev. Microbiol. 51225-255. [DOI] [PubMed] [Google Scholar]
- 20.Tian, Y., O. Tarlow, A. Ballard, U. Desselberger, and M. A. McCrae. 1993. Genomic concatemerization/deletion in rotaviruses: a new mechanism for generating rapid genetic change of potential epidemiological importance. J. Virol. 676625-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]