Abstract
The cellular proprotein convertase site 1 protease (S1P) has been implicated in the proteolytic processing of the glycoproteins (GPs) of Old World arenaviruses. Here we report that S1P is also involved in the processing of the GPs of the genetically more-distant South American hemorrhagic fever viruses Guanarito, Machupo, and Junin. Efficient cleavage of Guanarito virus GP, whose protease recognition sites deviate from the reported S1P consensus sequence, indicates a broader specificity of S1P than anticipated. Lack of GP processing of Junin virus dramatically reduced production of infectious virus and prevented cell-to-cell propagation. Infection of S1P-deficient cells resulted in viral persistence over several weeks without the emergence of escape variants able to use other cellular proteases for GP processing.
Several arenaviruses cause severe hemorrhagic fevers (HFs) in humans, posing a serious public health problem (5). The Old World arenavirus Lassa virus (LASV) is estimated to infect several hundred thousand individuals yearly in its regions of endemicity in West Africa, resulting in significant mortality and high morbidity (9). The New World arenavirus Junin virus (JUNV) causes Argentine HF, a severe illness with hemorrhagic and neurological manifestations and a case fatality of 15 to 30%, whereas the arenaviruses Machupo virus (MACV) and Guanarito virus (GTOV) emerged as causative agents of HF in Bolivia and Venezuela, respectively (10). High mortality caused by human-pathogenic arenaviruses is associated with the failure of the host's immune system to control viral replication, leading to an unchecked viremia associated with hemorrhagic disease.
The genome of arenaviruses consists of two single-stranded RNA species, a large segment encoding the virus polymerase (L) and a small zinc finger motif protein (Z) and a small segment encoding the virus nucleoprotein (NP) and glycoprotein precursor (GPC) (3). Proteolytic processing of GPC yields the N-terminal GP1 that is implicated in receptor binding and the C-terminal transmembrane GP2, which is structurally similar to the fusion-active portions of other viral GPs. Previous studies demonstrated that the GPs of the Old World arenaviruses lymphocytic choriomeningitis virus (LCMV) and LASV are proteolytically cleaved by the cellular proprotein convertase site 1 protease (S1P), also known as subtilisin-kexin-isozyme 1 (SKI-1), to yield the two mature virion glycoproteins GP1 and GP2 (1, 6, 7, 8). S1P also mediates processing of the GP precursor of Crimean-Congo HF virus (20). In the host cell, S1P is involved in the proteolytic processing of a defined set of cellular proteins, including the brain-derived neurotrophic factor precursor protein (18), the sterol regulatory element-binding proteins (SREBP-1 and SREBP-2), membrane-associated transcription factors that regulate genes involved in lipid metabolism (2, 15), and the activating transcription factor 6, which is a key player in the cellular response to endoplasmic reticulum stress (17, 23).
S1P is required for the proteolytic processing of the GPs of human-pathogenic New World arenaviruses.
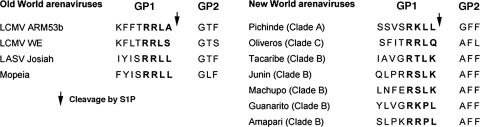
Mutation function studies identified the consensus sequence R-(R/K/H)-L-(A/L/S/T/F) as the S1P recognition site of LCMV GP (1). This consensus motif is also found in the GP sequences of LASV, the Old World viruses Mopeia and Mobala viruses and the New World virus Pichinde virus, but it is absent in the GPs of JUNV, GTOV, MACV, Amapari virus (AMPV), and Tacaribe virus (Fig. 1). In the predicted cleavage sites of GTOV (pathogenic) and the closely related AMPV (nonpathogenic), the highly conserved L residue in position −2 undergoes the nonconservative change to P. This observation led us to investigate whether S1P mediated the GP processing of these other arenaviruses.
FIG. 1.
Amino acid sequences at the sites of proteolytic processing of the GP precursor (GPC) into GP1 and GP2 for selected Old World and New World arenaviruses. The putative recognition sequences of S1P protease are indicated in bold, and the site of S1P cleavage is marked with an arrow.
Due to the lack of specific antibodies for the detection of some of the viral GPs of interest, we used C-terminally flag-tagged versions of LASV, GTOV, JUNV, MACV, and AMPV GPs to facilitate their detection (Fig. 2A). C-terminally flag-tagged versions of JUNV and LASV have been described elsewhere (12). The flag-tagged GTOV, MACV, and AMPV GP variants consist of full-length GPs in which the normal stop codon has been replaced by a spacer sequence (GGGS) followed by the flag tag (DYKDDDDK). The C-terminal GGGS sequence followed by the flag epitope and a new stop codon were introduced into the GPs of GTOV, MACV, and AMPV at the transcription level by PCR amplification of 3′-end cDNA fragments that were then inserted into expression constructs for full-length GTOV, MACV, and AMPV GPs (14).
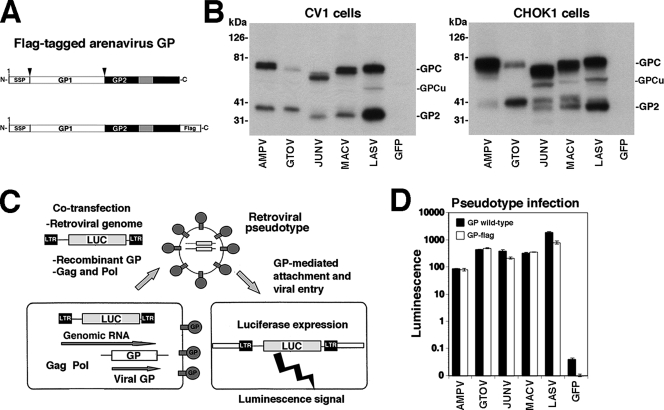
FIG. 2.
Expression and function of flag-tagged arenavirus GPs. (A) Schematic representation of flag-tagged arenavirus GPs: gray boxes and boxes labeled SSP correspond, respectively, to the transmembrane domain and stable signal peptide. Arrows indicate sites of proteolytic processing. (B) The expression of C-terminally flag-tagged GPs of AMPV, GTOV, JUNV, MACV, and LASV as well as a green fluorescent protein (GFP) control in lysates of transfected HEK293T cells was examined by Western blotting using an antiflag antibody. The positions of the fully glycosylated (GPC) and underglycosylated (GPCu) GPC species and mature GP2 are indicated. (C) Generation of recombinant retroviral vectors pseudotyped with arenavirus GPs. The packaging cell line GP2293 stably transfected with murine leukemia virus (MLV) gag and pol was cotransfected with a plasmid containing the packable MLV genomic plasmid pLZRS-Luc-gfp, carrying a luciferase and a GFP reporter (22), and an expression plasmid for the heterologous recombinant GP. Retroviral pseudotypes are released into the cell supernatant. LTR, long terminal repeat. (D) Infection of cells with retroviral pseudotypes. Retroviral pseudotypes were generated with wild-type and flag-tagged GPs of AMPV, GTOV, JUNV, MACV, and LASV and, as a control, GFP. Retroviral pseudotype-containing supernatants were added to Vero E6 cells, and 48 h later, infection was assessed by a Steady Glo luciferase assay (n = 3; error bars indicate standard deviations).
All flag-tagged GPs were expressed to similar levels in transfected primate (CV1) and hamster (CHOK1) cell lines (Fig. 2B), but we noticed differences in extent of GPC processing (e.g., compare the results for pathogenic GTOV and the closely related nonpathogenic AMPV). In addition to the bands corresponding to the unprocessed glycosylated GP precursor (GPC) and mature GP2, a specific relatively sharp band with an apparent molecular mass of 50 to 60 kDa, which may correspond to the underglycosylated precursor GPC, was detected in some samples (Fig. 2B). While mature GP2 of JUNV and MACV expressed in CV1 cells appeared as one distinct band, GP2 of JUNV and MACV expressed in CHOK1 cells separated into two distinct bands, likely due to differential glycosylation or proteolytic degradation.
To verify the biological function of the GP-flag versions in viral entry, flag-tagged GPs and the corresponding wild-type forms were incorporated into recombinant retroviruses containing a luciferase reporter by use of the strategy shown in Fig. 2C (14). As shown in Fig. 2D, we obtained similar infectious retroviral pseudotype titers with wild-type and flag-tagged GPs. These results ruled out the possibility that the flag tag had a negative impact on GP function in the process of cell entry. Consistent with our previous findings (14), infectious titers of LASV pseudotypes were routinely higher than those obtained with New World arenavirus pseudotypes.
To address the role of S1P in the processing of the GPs of New World arenaviruses, we took advantage of the mutant CHO cell line SRD12B, which is deficient in S1P (11). All GPs expressed in CHOK1 cells showed the expected processing, whereas no detectable cleavage of any of the GPs was observed in the S1P-deficient SRD12B cells (Fig. 3A). Complementation of SRD12B with S1P, via transfection of an expression construct for recombinant S1P, confirmed the critical role of S1P for GP processing (Fig. 3B).
FIG. 3.
S1P is required for processing and function, but not trafficking, of the GPs of human-pathogenic arenaviruses. (A) Expression of arenavirus GPs in S1P-deficient cells. S1P-deficient SRD12B cells and wild-type CHOK1 cells were transfected with flag-tagged GPs of AMPV, GTOV, JUNV, MACV, and LASV or with GFP as a control, and 48 h later, cell lysates were analyzed by Western blotting with an antiflag antibody. (B) Complementation of SRD12B cells with recombinant S1P. SRD12B cells were cotransfected with the indicated flag-tagged GPs and either empty control vector (SRD12B cells) or an expression plasmid for S1P (SRD12B cells + S1P). Viral GPs were detected by Western blotting as described for panel A. (C) S1P processing is not required for cell surface expression of arenaviral GPs. SRD12B cells and CHOK1 cells were transfected with the indicated GPs or, as a control, an empty plasmid (mock). After 48 h, cells were detached by nonenzymatic treatment and live, nonpermeabilized cells were stained with MAb 83.6 to AMPV GP2 and LASV GP2 (21) and MAb BE08 to JUNV GP1 (16). Primary antibody was detected with a phycoerythrin (PE)-labeled secondary antibody and analyzed by flow cytometry using a FACSCalibur flow cytometer (13). Data were acquired and analyzed using Cell Quest and FloJo software packages. In dot plots, the y axis represents forward scatter and the x axis represents PE fluorescence intensity. (D) S1P-mediated processing is required for the function of arenavirus GPs. SRD12B cells and CHOK1 cells were cotransfected with a plasmid expressing MLV gag and pol, the MLV genomic plasmid pLZRS-Luc-gfp, and expression plasmids for the indicated GPs, and 48 h later, conditioned supernatants were harvested, cleared, and added to Vero E6 monolayers. Infection was detected by a luciferase assay as described in the legend for Fig. 2D. Note that the y axis has a log scale.
Next, we assessed the role of processing by S1P in GP trafficking. For this we transfected SRD12B and CHOK1 cells with flag-tagged GPs of AMPV, JUNV, and LASV; 48 h later, cell surface expression levels of the viral GPs were determined by immunostaining with anti-GP antibodies by using flow cytometry. Despite the lack of proteolytic processing, similar cell surface expression levels of the GPs of AMPV, JUNV, and LASV were detected in SRD12B and CHOK1 cells (Fig. 3C), indicating that S1P processing is dispensable for trafficking of the viral GPs to the cell surface.
To test the biological activity of the GPs of AMPV, GTOV, JUNV, and MACV produced in S1P-deficent cells, we generated the corresponding retroviral pseudotypes in SRD12B cells and wild-type CHOK1 cells. For a control, we included the GP of vesicular stomatitis virus, which does not require S1P-mediated processing. After 48 h, retroviral pseudotypes were harvested and used to infect Vero E6 cells. Infection was determined by a luciferase assay. While high levels of infectious pseudotypes decorated with arenavirus GPs were produced in CHOK1 cells, no infectivity was detected when these pseudotypes were produced in SRD12B cells (Fig. 3D). High levels of infectious vesicular stomatitis virus pseudotypes were produced in both cell lines, indicating that the lack of infectivity was not due to a problem in the assembly or release of the retroviral particles in SRD12B cells but was related to a functional defect in the arenavirus GP. Together, our data indicate that proteolytic processing by S1P is crucial for the biosynthesis of functional GPs of the New World arenaviruses AMPV, GTOV, JUNV, and MACV.
To confirm the putative S1P cleavage sites of the GPs of JUNV and AMPV, which each represent a subgroup within the clade B New World arenaviruses, we determined the N-terminal amino acid sequence of their GP2 parts. We included the GP of LASV as a representative Old World arenavirus. Flag-tagged versions of JUNV GP, AMPV GP, and LASV GP were transiently expressed in HEK293T cells (two T175 flasks each) by using Superfect. After 48 h, the cells were lysed and flag-tagged recombinant GPs were isolated by immunoprecipitation as described previously (12) with 0.1% (wt/vol) sodium dodecyl sulfate (SDS) included in the wash buffer to increase the stringency. Immunocomplexes were separated by reducing SDS-polyacrylamide gel electrophoresis (PAGE) and blotted onto polyvinylidene difluoride membranes. Proteins were stained with Coomassie brilliant blue, membranes were destained extensively, and the bands corresponding to the GP2 parts of the recombinant GPs were excised. Protein sequencing was performed by Edman degradation using an Applied Biosystems Procise sequencer model 494 at the Center of Protein Sciences at the Scripps Research Institute. The sequences obtained confirmed the predicted N termini of the GP2 parts of AMPV (AFF), JUNV (AFF), and LASV (GTF), as shown in Table 1.
TABLE 1.
Determination of the N-terminal sequences of the GP2 parts of AMPV, JUNV, and LASVa
| Arenavirus | Amino acid at indicated position
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Predicted N terminus of GP2
|
Determined N-terminal sequence
|
||||||||
| Major sequence
|
Minor signals
|
||||||||
| +1 | +2 | +3 | +1 | +2 | +3 | +1 | +2 | +3 | |
| LASV | G | T | F | G | T | F | |||
| JUNV | A | F | F | A | F | F | G | L | |
| AMPV | A | F | F | A | F | F | G | ||
The predicted N-terminal sequences of AMPV, JUNV, and LASV are as reported previously (1). The residue +1 indicates the N terminus of GP2 after proteolytic processing. The N-terminal sequences of the GP2 parts of recombinant, flag-tagged GPs of AMPV, JUNV, and LASV were determined by Edman degradation (for details, see the text). The amino acids of the major sequences correspond to the major peaks obtained in the sequencing cycle at the given position. Minor signals observed in some cases are also indicated.
Our sequencing data revealed the N-terminal amino acid sequence AFF for JUNV GP2, indicating cleavage between residues K251 and A252, suggesting the sequence RSLK to be the primary site of cleavage at least in HEK293T cells. However, in contrast to the other arenavirus GPs, the GP of JUNV contains another R residue one position upstream of the sequence RSLK, opening the possibility of cleavage at the related site RRSL, yielding a GP2 form with the N-terminal sequence KAFF as a minor product. To address this possibility, we performed site-directed mutagenesis of JUNV GP. In the JUNV GP R247E mutant, the residue R247 was replaced by E, resulting in the sequence ERSLK at the putative cleavage site of the closely related MACV (Fig. 4A). In the JUNV GP R248A mutant, the critical R248 residue of the apparent primary cleavage site RSLK was mutated. Alanine replacements of both residues R247 and R248 in the R247A/R248A mutant resulted in a noncleavable form of JUNV GP, as previously described (24). Amino acid replacements were introduced into JUNV GP by PCR amplification of cDNA fragments using synthetic primers spanning the putative S1P recognition site. The cDNA fragments were then introduced into flag-tagged full-length JUNV GP (12), and sequences were verified by DNA sequencing.
FIG. 4.
Mutagenesis of the putative S1P cleavage site of JUNV GP. (A) Sequence of wild-type and mutant JUNV GPs at the putative S1P cleavage site (arrow) with mutated amino acids show in bold. The sequence of MACV is shown for comparison. (B) Wild-type and mutant JUNV flag-tagged GPs were expressed in HEK293T cells, and total protein lysates were analyzed by Western blotting as described for Fig. 2B. The positions of GPC, the putative underglycosylated GPC form GPCu, and mature GP2 are indicated. (C) Densitometric analysis of the blot in panel B. Densitometry was performed as described previously (6), and the ratios of the signal intensities for GP2 (IGP2) to GPC (IGPC) were calculated (GP2/GPC) for wild-type and mutant JUNV GP-flag.
When expressed in HEK293T cells and analyzed by Western blotting using an antibody to the flag epitope, all mutants showed high levels of expression with significant differences in proteolytic processing, as assessed by determining the ratio of mature GP2 to GPC (Fig. 4B). The mutation R247E resulted in a circa 50% reduction in cleavage, while >90% reduction was observed with R248A. Consistent with previous studies (24), no detectable cleavage was observed with the R247A/R248A mutant. The marked reduction in processing of the R248A mutant is consistent with RSLK being the primary site of processing, as indicated by our protein sequencing data. However, the low residual cleavage observed with the R248A mutant suggests that the sequence RRSL may represent an additional cleavage site used by S1P to process JUNV GP, albeit much less efficiently. The reduction of JUNV GP cleavage caused by the mutation R247E is a little surprising, given the fact that the MACV GP contains an E at the corresponding position (Fig. 4A) and is cleaved with an efficiency comparable to JUNV GP in the cells tested so far (Fig. 2B). A similar negative effect of the adjacent E residue on the processing of MACV GP may have been compensated for by another mutation.
S1P-mediated processing of JUNV GP is critical for production and cell-to-cell propagation of infectious virus.
To study the role of S1P processing in infection with a live pathogenic New World arenavirus, we used the safe JUNV vaccine strain Candid 1 that can be used at biosafety level 2 (4). The GPC sequences of JUNV XJ (GenBank accession no. U70799) and JUNV Candid 1 (GenBank accession no. U70801) differ only in two amino acids close to the GP2 transmembrane domain, F427I and T446S, without affecting GPC processing.
To compare the susceptibilities of SRD12B and CHOK1 cells to JUNV Candid 1 infection, we used a low multiplicity of infection (MOI) for infection and examined the expression of virus antigen by immunofluorescence using a monoclonal antibody (MAb) to JUNV NP (16). We observed comparable numbers of infectious foci per well in SRD12B and CHOK1 cells at 24 h postinfection, indicating similar efficiencies of virus-to-cell infection in the two cell lines (Fig. 5A) However, while infectious foci in SRD12B cells were small and involved only cells immediately adjacent to each other, infectious foci in CHOK1 cells tended to be significantly larger, likely due to efficient cell-to-cell propagation of virus (Fig. 5B). This markedly reduced cell-to-cell propagation of JUNV Candid 1 in SRD12B cells correlated with a >3-log reduction of infectious virus titers in the supernatant as assessed by plaque assay on Vero cells (Fig. 5C). Therefore, as with LASV (8) and LCMV (6), the processing of GP by S1P is also critical for infectious virus production and cell-to-cell propagation of the pathogenic New World arenavirus JUNV.
FIG. 5.
S1P processing is required for the production of infectious JUNV and cell-to-cell propagation. (A) Infection of SRD12B and CHOK1 cells with JUNV Candid 1. Cells (96-well plates) were infected with 100 PFU of JUNV Candid 1. At the indicated times, cells were fixed and stained with MAb BG12 for JUNV NP. The number of infectious foci was determined in each well (n = 3; error bars indicate standard deviations [SD]). (B) Representative infectious foci in SDR12B and CHOK1 cells at 48 h postinfection. Bar = 20 μm. (C) Production of infectious JUNV in SRD12B and CHOK1 cells. Cells were infected with JUNV Candid 1 at an MOI of 1. Cell supernatants were harvested at the indicated times, and infectious virus titers were determined by a plaque assay on Vero E6 cells. A compilation of three independent experiments is shown (means ± SD). (D to F) Analysis of the composition of JUNV particles produced in SRD12B and CHOK1 cells. Triplicate samples of 5 × 106 SRD12B (SRD) and CHOK1 (CHO) cells grown in T75 flasks were infected with JUNV Candid 1 at an MOI of 1. (D) Cell culture supernatants were harvested after 48 h, and infectious virus titers were determined by a plaque assay on Vero E6 cells. (E) Detection of viral proteins by Western blotting. Supernatants were subjected to ultracentrifugation through a 20% sucrose cushion (6). Pellets were solubilized in hot SDS-PAGE loading buffer, and viral proteins were separated by SDS-PAGE under nonreducing conditions. Blots were probed with MAbs GB03-BE08 (anti-JUNV GP1) and SA02-BG12 (anti-JUNV NP) (16) combined with a biotinylated secondary antibody and horseradish peroxidase-conjugated streptavidin. Signals were detected using the Super Signal West Femto chemiluminescence detection kit from Pierce. (F) Densitometric analysis of the blots in panel E. Densitometry was performed as described in the legend to Fig. 4C, and the ratios of the signal intensities for GP1 (IGP1) to NP (INP) were calculated (GP1/NP1) for virion particles from SRD12B cells (SRD) and CHOK1 cells (CHO). (G) Persistent infection of SRD12B cells with JUNV Candid 1 does not result in escape variants. SRD12B (three independent cell cultures) and CHOK1 cells were infected with JUNV Candid 1 at an MOI of 1 and passaged every 3 days. At the indicated times, virus titers were determined by a plaque assay. The results of one representative example out of three independent experiments is shown.
While our data indicate a crucial role for S1P processing of JUNV GP in the production of infectious progeny virus from infected cells, we observed low but measurable virus titers in the supernatants of S1P-deficient SRD12B cells (Fig. 5C). Similarly low levels of LASV and LCMV production have been reported in SRD12B cells (6, 8). The reason for this is currently unclear and may be due to either the activation of arenavirus GPS by another protease in the absence of S1P or, less likely, the ability of uncleaved (i.e., fusion-inactive) GP to mediate viral entry.
Since the JUNV titers obtained from infected SRD12B cells were consistently more than 3 orders of magnitude lower than those obtained from CHOK1 cells (Fig. 5C), we addressed the question of whether noninfectious virus particles were released from these S1P-deficent cells. SRD12B and CHOK1 cells were infected with JUNV at an MOI of 1, culture supernatant was harvested after 48 h, and infectious virus titers were determined (Fig. 5D). Supernatants were subjected to ultracentrifugation, and isolated particles were analyzed by Western blotting for the presence of JUNV NP and JUNV GP1 using MAbs GB03-BE08 (anti-JUNV GP1) and SA02-BG12 (anti-JUNV NP) as described previously (16). While JUNV NP was detected in pellets from supernatants of SRD12B and CHOK1 cells (Fig. 5E), JUNV GP1 was detected only in particles from supernatants of CHOK1 cells, not in those isolated from supernatants of SRD12B cells. Consistent with earlier reports (16), only cleaved GP1 was detected in virions released from infected CHOK1 cells (Fig. 5E). The amount of viral NP detected in supernatants from SRD12B cells was circa fivefold lower than the amount detected in supernatants from CHOK1 cells. This may be due to less cell-to-cell spread of JUNV in cultures of SRD12B cells and possibly lower levels of viral replication and/or to NP gene expression over time in S1P-deficent cells. However, as our detection for GP was more sensitive than that for NP (compare signal intensities for GP and NP in the blots), the absence of detectable GP from particles released by SRD12B cells indicated a significant reduction in their GP/NP ratio compared to the particles released by CHOK1 cells, as confirmed by densitometric quantification of the signals (Fig. 5F). The reduced GP levels in JUNV particles released from S1P-deficient cells observed here are in line with similar observations made earlier with LASV (8) and LCMV (6). Since cell surface expression levels of arenavirus GPs are not reduced by lack of cleavage (1, 6; this study [Fig. 3C]), the reduced GP/NP ratio in virions released from S1P-deficient cells suggest that cleavage of GP is required for its efficient incorporation into virus particles at some point during the virus assembly process.
Emergence of arenavirus variants capable of growing independently of S1P-mediated processing.
As with most, if not all, RNA viruses, arenaviruses have an error-prone replication machinery that confers on them high mutation rates and the potential for rapid evolution. This, in turn, provides these viruses with a remarkable plasticity to adapt to different types of environmental selection pressures (19). We therefore expected that the absence of S1P activity in SRD12B cells would facilitate the emergence of viral escape variants capable of using other cellular proteases for the processing of their GPs. A rapid appearance of such arenavirus variants would pose difficulties for the implementation of an antiviral strategy aimed at targeting S1P-mediated processing of the arenavirus GP. To address this issue, we exploited the fact that arenaviruses can readily establish long-term persistent infections in mammalian cells in culture, allowing selection of viral variants over time. Cultures of S1P-deficient SRD12B and control CHOK1 cells were infected with JUNV Candid 1 at a high MOI, resulting in >90% infected cells after 48 h. Infected cells were then passaged every 72 h for 24 days, and virus titers in supernatants were determined by a plaque assay on Vero E6 cells. Infection of CHOK1 cells resulted in a persistently infected cell line that produced high levels of infectious virus over time (Fig. 5G). In contrast, despite the persistence of JUNV in SRD12B cells, revealed by the continued presence of intracellular viral antigen (NP) as determined by immunofluorescence detection (data not shown), only negligible viral infectivity was detected in the supernatants of several independent cell lineages for the duration of the experiment (Fig. 5G). The inability to detect significant viral infectivity in persistently infected SRD12B cultures suggests that the emergence of viral escape variants that have acquired mutations rendering them S1P independent has not occurred under our experimental conditions.
Our present study implicates the cellular proprotein convertase S1P in the proteolytic processing of the GPs of the South American HF viruses JUNV, MACV, GTOV and the closely related nonpathogenic AMPV. Efficient cleavage of GTOV and AMPV GP, whose protease recognition sites deviate from the reported S1P consensus sequence, indicates a broader specificity of S1P than anticipated. Lack of GP processing dramatically reduced production of infectious JUNV and prevented cell-to-cell propagation. Infection of S1P-deficient cells resulted in viral persistence without the emergence of escape variants able to use other cellular proteases for GP processing. Together with previous reports (1, 6, 8), our data show that S1P is crucial for GP processing for all major human-pathogenic arenaviruses. The key role of S1P in productive infection and cell-to-cell spread of human-pathogenic arenaviruses make S1P a promising cellular target for the development of novel and potent antiarenaviral drugs.
Acknowledgments
This is publication number 19219 from the Molecular and Integrative Neurosciences Department (MIND) of the Scripps Research Institute (TSRI).
We thank Michael B. A. Oldstone (TSRI), Juan-Carlos de la Torre (TSRI), and Amalio Telenti (University Hospital Center and University of Lausanne) for their generous support and insightful discussions. We further acknowledge Michael Buchmeier (TSRI) for the Junin Candid 1 virus, Jack Nunberg (University of Montana, Missoula) for anti-Junin virus antibodies, and Bruce Beutler (TSRI) for the recombinant S1P cDNA. We are grateful to J. L. Goldstein for kindly providing the S1P-deficient cell line SRD12B. The retroviral construct pLZRs-Luc-gfp was kindly provided by Gary Nabel.
This research was supported by U.S. Public Health Service grants AI065560 and 1U54 AI065359. Stefan Kunz was supported by a Medical Research Position Award of the Foundation Max Cloetta (Switzerland).
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Beyer, W. R., D. Popplau, W. Garten, D. von Laer, and O. Lenz. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 772866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89331-340. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier, M. J., J. C. de la Torre, and C. J. Peters. 2007. Arenaviridae: the viruses and their replication, p. 1791-1828. In D. L. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, PA.
- 4.Enria, D. A., and J. G. Barrera Oro. 2002. Junin virus vaccines. Curr. Top. Microbiol. Immunol. 263239-261. [DOI] [PubMed] [Google Scholar]
- 5.Geisbert, T. W., and P. B. Jahrling. 2004. Exotic emerging viral diseases: progress and challenges. Nat. Med. 10S110-S1121. [DOI] [PubMed] [Google Scholar]
- 6.Kunz, S., K. H. Edelmann, J.-C. de la Torre, R. Gorney, and M. B. A. Oldstone. 2003. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314168-178. [DOI] [PubMed] [Google Scholar]
- 7.Lenz, O., J. ter Meulen, H. Feldmann, H. D. Klenk, and W. Garten. 2000. Identification of a novel consensus sequence at the cleavage site of the Lassa virus glycoprotein. J. Virol. 7411418-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenz, O., J. ter Meulen, H. D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 9812701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormick, J. B., and S. P. Fisher-Hoch. 2002. Lassa fever. Curr. Top. Microbiol. Immunol. 26275-109. [DOI] [PubMed] [Google Scholar]
- 10.Peters, C. J. 2002. Human infection with arenaviruses in the Americas. Curr. Top. Microbiol. Immunol. 26265-74. [DOI] [PubMed] [Google Scholar]
- 11.Rawson, R. B., D. Cheng, M. S. Brown, and J. L. Goldstein. 1998. Isolation of cholesterol-requiring mutant Chinese hamster ovary cells with defects in cleavage of sterol regulatory element-binding proteins at site 1. J. Biol. Chem. 27328261-28269. [DOI] [PubMed] [Google Scholar]
- 12.Rojek, J. M., K. P. Campbell, M. B. Oldstone, and S. Kunz. 2007. Old World arenavirus infection interferes with the expression of functional α-dystroglycan in the host cell. Mol. Biol. Cell 184493-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojek, J. M., C. F. Spiropoulou, K. P. Campbell, and S. Kunz. 2007. Old World and clade C New World arenaviruses mimic the molecular mechanism of receptor recognition used by α-dystroglycan's host-derived ligands. J. Virol. 815685-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojek, J. M., C. F. Spiropoulou, and S. Kunz. 2006. Characterization of the cellular receptors for the South American hemorrhagic fever viruses Junin, Guanarito, and Machupo. Virology 349476-491. [DOI] [PubMed] [Google Scholar]
- 15.Sakai, J., A. Nohturfft, J. L. Goldstein, and M. S. Brown. 1998. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J. Biol. Chem. 2735785-5793. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez, A., D. Y. Pifat, R. H. Kenyon, C. J. Peters, J. B. McCormick, and M. P. Killet. 1989. Junin virus monoclonal antibodies: characterization and cross-reactivity with other arenaviruses. J. Gen. Virol. 701125-1132. [DOI] [PubMed] [Google Scholar]
- 17.Schroder, M., and R. J. Kaufman. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74739-789. [DOI] [PubMed] [Google Scholar]
- 18.Seidah, N. G., S. J. Mowla, J. Hamelin, A. M. Mamarbachi, S. Benjannet, B. B. Toure, A. Basak, J. S. Munzer, J. Marcinkiewicz, M. Zhong, J. C. Barale, C. Lazure, R. A. Murphy, M. Chretien, and M. Marcinkiewicz. 1999. Mammalian subtilisin/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. USA 961321-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sevilla, N., E. Domingo, and J. C. de la Torre. 2002. Contribution of LCMV towards deciphering biology of quasispecies in vivo. Curr. Top. Microbiol. Immunol. 263197-220. [DOI] [PubMed] [Google Scholar]
- 20.Vincent, M. J., A. J. Sanchez, B. R. Erickson, A. Basak, M. Chretien, N. G. Seidah, and S. T. Nichol. 2003. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J. Virol. 778640-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber, E. L., and M. J. Buchmeier. 1988. Fine mapping of a peptide sequence containing an antigenic site conserved among arenaviruses. Virology 16430-38. [DOI] [PubMed] [Google Scholar]
- 22.Yang, Z., R. Delgado, L. Xu, R. F. Todd, E. G. Nabel, A. Sanchez, and G. J. Nabel. 1998. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 2791034-1037. [DOI] [PubMed] [Google Scholar]
- 23.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 61355-1364. [DOI] [PubMed] [Google Scholar]
- 24.York, J., V. Romanowski, M. Lu, and J. H. Nunberg. 2004. The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 7810783-10792. [DOI] [PMC free article] [PubMed] [Google Scholar]