Abstract
The yellow fever virus (YFV) 17D strain is one of the most effective live vaccines for human use, but the in vivo mechanisms for virulence attenuation of the vaccine and the corresponding molecular determinants remain elusive. The vaccine differs phenotypically from wild-type YFV by the loss of viscerotropism, despite replicative fitness in cell culture, and genetically by 20 amino acid changes predominantly located in the envelope (E) protein. We show that three residues in E protein domain III inhibit spread of 17D in extraneural tissues and attenuate virulence in type I/II interferon-deficient mice. One of these residues (Arg380) is a dominant glycosaminoglycan-binding determinant, which mainly accounts for more rapid in vivo clearance of 17D from the bloodstream in comparison to 17D-derived variants with wild-type-like E protein. While other mutations will account for loss of neurotropism and phenotypic stability, the described impact of E protein domain III changes on virus dissemination and virulence is the first rational explanation for the safety of the 17D vaccine in humans.
The live, attenuated yellow fever virus (YFV) strain 17D vaccine has been used safely and effectively in over 500 million individuals over the past 70 years. This excellent safety record also underpins its potential application as a platform for chimeric vaccines against other medically important flaviviruses (dengue, Japanese encephalitis, and West Nile viruses), which are currently in clinical trials (reviewed in reference 13). Despite knowledge of the complete genome sequences of 17D (33) and its virulent parent (10), the Asibi strain isolated in Africa in 1927, the in vivo mechanism(s) and molecular determinants for virulence attenuation of the YFV vaccine are still not completely resolved. YFV strain 17D was the first human vaccine derived from repeated passage in tissue culture (reviewed in reference 39). More than 230 passages in embryonic mouse tissue (18 passages), chicken embryo tissue (50 passages), and chicken embryo tissue from which the head and spinal chord had been removed (152 passages) separate the 17D from the Asibi strain. The adaptation of 17D to growth in mouse and chicken embryonal tissues culture has resulted in loss of viscerotropism, a property which accounts for the major disease manifestations of yellow fever in primates: high viremia, hepatitis, renal dysfunction, hemorrhagic fever, and circulatory shock (reviewed in reference 28); it also resulted in reduced neurotropism and loss of tropism for mosquitoes, the obligatory vector in the natural transmission cycle of YFV. The complex passage history of YFV strain 17D has given rise to as many as 68 nucleotide mutations and 32 amino acid changes in the viral RNA genome (10). However, when the Asibi genome is compared to the consensus sequence of different 17D vaccine strains derived from it, the number of differences potentially contributing to the attenuated virulence phenotype can be reduced to 20 amino acids and 4 nucleotides in the 3′ untranslated region (8).
The majority of genetic differences separating the 10,862-nucleotide-long genome of Asibi from that of the 17D vaccine strains are found in the envelope (E) protein gene (10), consistent with the function of this protein in receptor binding and entry and the likelihood that changes in receptor usage were, at least in part, the driving force during host cell adaptation of the vaccine. The E protein is the major flavivirus transmembrane protein and covers most of the surface of the virion in the form of 90 E protein dimers. X-ray crystallography structures suggest that the E protein is organized into three domains: a central domain (dI), which is flanked on one side by an elongated dimerization domain (dII) and on the other by an immunoglobulin-like domain (dIII) which makes the highest protrusion from the otherwise smooth particle surface (26, 31). The receptor-binding site is thought to be located on dIII. Analyses of neurovirulent variants of YFV-17D, which have acquired E protein reversions to the Asibi sequence, provide support for a role of dIII residues 305, 325, and 380 as well as dI/II hinge region residues 52 and 173 in neurovirulence (6, 29, 34, 35, 37). While the location of the dIII residues is consistent with their involvement in receptor binding, the dI/II hinge region changes are speculated to affect fusion based on studies of other flaviviruses (reviewed in reference 27). However, experimental proof for these putative mechanisms of neurovirulence attenuation has not yet been provided. It is currently also not possible to pinpoint which mutations in the E protein, or elsewhere in the genome, account for the attenuation of viscerotropism of YFV-17D, owing to the absence of an appropriate animal model for the viscerotropic disease. Accordingly, the contribution of E protein changes, relative to those in the nonstructural proteins, to virulence attenuation of 17D vaccines in the vertebrate host remain elusive, despite the availability of the full-length cDNA clones of Asibi (25) and 17D (32) that allows this question to be addressed. This has practical implications for the suitability of the YFV 17D genome as a backbone for chimeric flavivirus vaccine, which is based on the premise that (so-far-unidentified) attenuation markers in the nonstructural polyprotein region confer the well-established safety profile of 17D to the chimeric vaccine candidates (reviewed in references 13 and 27).
The acquisition of high binding affinity for glycosaminoglycans (GAG) following serial propagation in tissue culture appears to be a common mechanism for virulence attenuation of flaviviruses (14-16, 18, 21, 23) and members of other virus families (3, 5, 11, 36). GAG are sulfated polysaccharides ubiquitously expressed on cellular surfaces and extracellular matrices of multicellular organisms (reviewed in reference 30). Virus binding to GAG involves primarily the electrostatic interaction of clusters of basic amino acids on the virus with negatively charged sulfate groups on the polysaccharide. Accordingly, GAG-binding variants typically display amino acid substitutions that result in an increase in net positive charge on the virion surface. The selective advantage associated with a high GAG-binding affinity for virus growth in cell culture is frequently accompanied by a reduced efficiency of growth in the vertebrate host because it prevents efficient virus spread through the circulation (3, 5, 14, 15, 18). Given that the attenuation of YFV-17D is associated with basic amino acid substitutions on the E protein, one at a known GAG-binding determinant of the encephalitic flaviviruses corresponding to YFV residue 380 (14, 16, 21), and in view of the efficient heparan sulfate binding property of YFV-17D (9), we speculated that at least one of the in vivo mechanisms for virulence attenuation of the 17D vaccine involves the poor ability to spread in the animal. Here we identify E protein determinants of 17D, absent in the Asibi strain, which enhance GAG binding and reduce virus dissemination.
MATERIALS AND METHODS
Viruses and cells.
African green monkey kidney (Vero), baby hamster kidney (BHK-21), and Aedes albopictus mosquito (C6/36) cells were grown as described elsewhere (14, 18). The 17D-204 substrain of YFV was from the American Type Culture Collection and was plaque purified twice; working stocks for YFV-17D and 17D variants were BHK-21 cell supernatants.
Plasmid constructs.
The plasmid pACNR/FLYF, which contains the full-length genome of YFV-17D (204 substrain) downstream of an SP6 RNA polymerase promoter (4), was used for generating mutant viruses. For ease of manipulation, pACNR/FLYF was digested with KpnI and XbaI to remove a 7-kb fragment from nucleotides 3262 to 10708, flush ended with T4 DNA polymerase (New England Biolabs), and religated with T4 DNA ligase (Roche) to generate the shuttle plasmid pYFdel. For introduction of mutations at E protein residues 52, 173, and 200 in domains I/II and residues 305, 326, and 380 in domain III, fusion-PCR was performed using upstream primers p347 (5′-GGC TTG GCT GTT CTA AGG-3′) or p1562 (P5′-GAG ATG GAA ACA GAG AGC TG-3′) and downstream primers p1989 (5′-TAT TGA TTG CCG CTG TAA GAT-3′) or p2567 (5′-CAC TAT TGA TGC AAG CTT CAC AGG-3′), respectively. First-round PCR was performed using Pfu Ultra DNA polymerase (Stratagene), pACNR/FLYF plasmid, and the mutagenesis primers (sequences provided by upon request) along with the appropriate domain I/II or domain III upstream and downstream primers in a volume of 25 μl in accordance with the manufacturer's recommendations. PCR mixtures were incubated for 1 cycle of 5 min at 94°C, 5 cycles of 30 s at 94°C, 30 s at 48°C, and 90 s at 70°C, 25 cycles of 30 s at 94°C, 30 s at 54°C, and 90 s at 70°C, and held at 70°C for 5 min before electrophoresis on a 1% agarose gel to isolate fragments of 0.8 kb and 0.9 kb for R52G, 1.2 kb and 0.5 kb for I173T and T200K, 0.3 kb and 0.7 kb for F305S, 0.4 kb and 0.6 kb for S325P, and 0.5 kb and 0.5 kb for R380T changes using the Wizard SV gel and PCR cleanup system (Promega). Second-round fusion-PCR was performed using gel-isolated pairs of fragments for each mutation, either domain I/II or domain III upstream and downstream primer pairs, the Pfu Ultra enzyme, and the conditions for thermal cycling described above. The domain I/II mutagenized fragments of 1.7 kb were digested with NdeI and ApaI, while the domain III mutagenized fragments of 1.0 kb were digested with ApaI and SacI, gel isolated as above, and inserted into appropriately restriction enzyme-digested pYFdel DNA. Domain I/II and domain III mutations were combined by ligations of three fragments: the NdeI-ApaI and ApaI-SacI mutagenized fragments with the NdeI-SacI fragment from pYFdel. Ligation mixtures were transformed into Escherichia coli MC1061.1 cells, and the prM and E gene sequences in the mutagenized pYFdel plasmids were verified by sequence analysis using BigDye v. 3 (Applied Biosystems) at the ACRF Biomolecular Resources Facility in the John Curtin School of Medical Research, Canberra, Australia, in accordance with the manufacturer's protocol. Plasmids with the expected mutations and no additional changes in prM and E were digested with NotI and MluI, and fragments of 3.2 kb were ligated into appropriately digested pACNR/FLYF to generate full-length mutant plasmids for SP6 RNA transcription. Incorporation of multiple mutations in domain I/II or domain III was achieved by use of mutated versions of pYFdel plasmids with confirmed single or double changes as templates in the first round of fusion-PCR.
RNA transcription and electroporation of BHK-21 cells.
Plasmids containing the full-length YFV cDNAs were digested with XhoI and transcribed using SP6 RNA polymerase, and full-length RNA transcripts were electroporated into BHK-21 cells as described elsewhere (18), but with the following modifications. For determination of electroporation efficiency, 200 μl, 20 μl, or 2 μl of electroporated cells (25 ml at 4 × 105 cells/ml) was plated onto BHK-21 and Vero cell monolayers in six-well trays (3.5 × 105 cells/well; Nunc) and incubated for 1 h, and agar overlay medium (M199 medium plus 1% agar and 2% fetal calf serum) was added. Vero cells were stained by addition of neutral red (0.02%) at 4 days postinfection (p.i.), and plaques were counted after overnight incubation. BHK-21 cells were stained at 5 days p.i. by addition of crystal violet (0.2% [wt/vol] in 20% ethanol) after removal of agar. Plaque-purified stocks were prepared from plaques derived from electroporated BHK-21 cells and amplified once in BHK-21 cells.
The prM and E protein genes of plaque-purified, recombinant virus stocks were completely sequenced as follows: total cellular RNA from infected BHK-21 cells was extracted and reverse transcription-PCR (RT-PCR) was performed as described elsewhere (18) using random hexamers in the RT step, the YFV-specific primer pair p347 and p2567 for PCR, and additional primers P992 (5′-ACT GAC AGG GAT TTC ATT GAG′), P1562, and P1989 for sequencing of YFV prM and E genes.
Heparin inhibition and heparin-Sepharose binding assays.
Virus binding to heparin-Sepharose was determined as described previously (14). Inhibition of infectivity of YFV-17D and variant stocks by heparin was examined by incubating virus (1 × 106 to 5 × 106 PFU) with heparin (0.2, 2, 20, or 200 μg/ml) for 15 min prior to its addition to BHK-21 cell monolayers (4 × 105 cells/well in six-well plates) treated with similar concentrations of heparin (Sigma) as described elsewhere (17). Mock-treated controls were performed by preincubation of the same amount of virus in Hanks' balanced salt solution-bovine serum albumin (HBSS-BSA; 0.25% [wt/vol]) in the absence of heparin. Cells were harvested at 20 h p.i. using trypsin, and the percentage of infected cells was determined by flow cytometry following staining for nonstructural protein NS1 expression with the monoclonal antibody 4G4 (20).
Analysis of fusion-related properties.
Fusion-from-within (FFWI) assays were performed in C6/36 cells infected in replicates of 10 in 48-well plates at a multiplicity of infection (MOI) of 0.5 and incubated in growth medium with the pH held at ≥7.6. At 4 days p.i., cells were washed once in phosphate-buffered saline; a series of 0.3-ml aliquots of fusion medium comprising minimal essential medium lacking serum, buffered at pH 6.0, 6.2, 6.4, 6.6, 6.8, 7.0, 7.2, 7.4, 7.6, and 7.8 using 20 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH < 6.8) or HEPES (pH ≥ 6.8) was then added to each replicate series and incubated at 40°C for 1 h. After removal of fusion medium, cells were fixed and stained using Quick Dip solutions (Fronine) and examined by light microscopy. The fusion index was determined using the following formula: 1− (number of cytoplasts/number of nuclei), based on counting five fields of 200 cells. Acid inactivation assays were performed by addition of 90 μl of HBSS-BSA, adjusted to pH 5.5, 5.75, 6.0, 6.25, 6.5, 6.75, 7.0, and 8.0 with 20 mM MES (pH < 6.8) or HEPES (pH ≥ 6.8), to a replicate series of 10-μl virus samples, incubation at 37°C for 15 min, followed by the addition of 900 μl of HBSS-BSA adjusted to pH 8.0. Titration of treated virus samples was performed by plaque formation on Vero cell monolayers. Assays for sensitivity to ammonium chloride, NH4Cl, were performed by pretreating BHK-21 cell monolayers in six-well plates (4 × 105 cells per well) for 30 min with growth medium containing 0 or 10 mM NH4Cl and addition of virus samples in duplicates at an MOI of ∼0.5 for 1 h. After removal of virus inocula, cells were washed once with PBS and then incubated under 0.5 ml HBSS-BSA adjusted to pH 5.5 for 3 min to remove all extracellular residual infectivity. Growth medium was added and cells incubated for 21 h before harvesting using trypsin and detection of infected cells by fluorescence-activated cell sorter (FACS) analysis.
Mouse virulence and pathogenesis assays.
All mice were obtained from the Animal Breeding Facility at the John Curtin School of Medical Research, Canberra, Australia. For virulence determinations, 6-week-old IFN-α/γ-R−/− mice (40) were used. Virus inocula were diluted in HBSS-BSA and injected intravenously (100 μl) or by the footpad route (20 μl) into mice anesthetized with ketamine-xylazol. Mortality and morbidity were monitored over a period of 28 days. Clearance of virus from the circulation was assayed as described elsewhere (14). Titration of virus content in blood, spleen, liver, popliteal lymph nodes, and brain was performed by plaque assay on Vero cells as described previously (20) and by quantitative RT-PCR (qRT-PCR).
Virion RNA quantitation by RT-PCR.
For determination of viral genome content of mouse tissue samples, total RNA in clarified tissue homogenates (50 μl; 10% [wt/vol] for spleen, liver, and brain; 0.1% [wt/vol] for lymph node suspension) was extracted using Trizol (18), and virion RNA content was determined by qRT-PCR. Standard curves were generated from RNA transcripts derived from XhoI-linearized pACNR/FLYF DNA, using SP6 RNA polymerase followed by digestion with DNase I (1 unit; Roche) for 30 min at 37°C, extraction with phenol, phenol-chloroform, and chloroform, precipitation with sodium acetate and ethanol, one wash with 70% ethanol, and resuspension in nuclease-free water at 109 copies/μl (6 ng/μl). Serial dilutions of the in vitro-transcribed YFV RNA were performed to yield standards of 106, 105, 104, and 103 RNA copies/μl. Reverse transcription was performed at 43°C for 90 min in a 10-μl mixture containing 1 μl sample RNA, Expand reverse transcriptase (Roche), 10 mM deoxynucleoside triphosphates, 10 pmol primer (YFVqPCRP2; 5′-CTC CTC CTT TCC TTC TTC CTT CAC-3′), 10 mM dithiothreitol, and the manufacturer's recommended buffer conditions. Real-time PCR was performed using TaqMan qPCR mixtures (Bio-Rad), 0.2 nM concentrations of YFVqPCRP2 and YFVqPCRP1 (5′-GTG GCA CTT CAG GAT CTC CTA TTG-3′), and 0.1 nM TaqMan probe primer (5′-ATG GCA TCC TTG TCG GTG ACA ACT CCT TCG TGT-3′), under cycling conditions of 95°C for 3 min for 1 cycle and 40 cycles of 95°C for 30 s, 63°C for 30 s, and 72°C for 60 s. Each RNA sample was tested in duplicate, and virion RNA content was determined by extrapolation from the standard curves generated within each experiment.
RESULTS
GAG-binding property of YFV-17D.
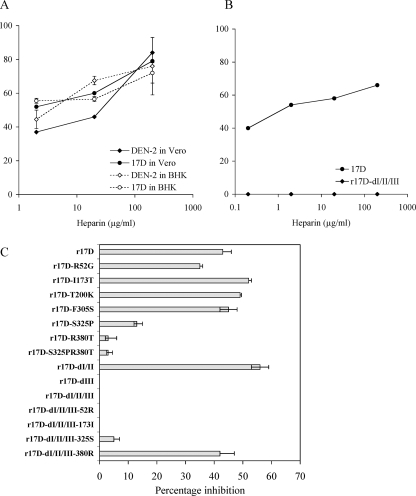
Acquisition of GAG-binding determinants often accompanies virulence attenuation of laboratory-adapted virus strains generated by serial passage in cultured cells or tissues from nonnatural hosts (3, 14, 18, 23, 36). The YFV-17D vaccine was generated through extensive adaptation to mouse and chick embryonal tissues and has been shown to exhibit sensitivity toward inhibition by heparin, a representative GAG (9). To explore a putative role of GAG-binding adaptations in virulence attenuation of the YFV vaccine, we first confirmed its high GAG-binding affinity. Using the YFV-17D 204 strain, heparin-mediated inhibition of virus infectivity in BHK-21 and Vero cells and of virus binding to heparin-Sepharose beads was tested in comparison to the mouse brain-adapted dengue 2 virus (DEN-2; NGC strain), which exhibits high-affinity GAG interactions (7, 18). The infectivity of YFV-17D in Vero and BHK-21 cells was significantly inhibited by heparin, with 50% effective concentration values of ≤2 μg/ml (Fig. 1A). This susceptibility to inhibition was comparable to that of DEN-2 in Vero cells at high doses of heparin and greater in the low-dose range, while in BHK cells the DEN-2 strain was more susceptible to the antiviral effect of heparin at high doses (also see reference 17) than YFV-17D. In addition, both YFV-17D and DEN-2 NGC strains displayed strong binding to heparin-Sepharose (99.8% and 99.7%, respectively) in an in vitro assay which distinguishes high-affinity GAG-binding viruses from those that bind poorly (14, 18).
FIG. 1.
Inhibition by heparin of infectivity of YFV-17D and r17D variants with wt E protein changes. A. The percent inhibition of infectivity of DEN-2 NGC and YFV 17D strains in BHK-21 and Vero cells at heparin concentrations of 2 to 200 μg/ml was determined by FACS analysis at 21 h p.i. Error bars show the standard errors determined from two independent experiments. B. Percent inhibition of infectivity of r17D and r17D-dI/II/III in BHK-21 cells using 0.2 to 200 μg/ml heparin. C. Percent inhibition of infectivity of r17D variants in BHK-21 cells using 20 μg/ml heparin. Error bars show the standard errors determined from two independent experiments.
Molecular determinants for GAG interaction in the YFV E protein.
To assess whether 17D differs from wild-type (wt) YFV in GAG-binding affinity and to identify the molecular determinants responsible, nonconservative amino acid differences distinguishing the E proteins of the 17D and Asibi strains of YFV were reverted to the wt sequence in the 17D background. Given that the E protein constitutes most of the surface of the flavivirus particle and that flavivirus GAG-binding determinants have so far only been mapped to the E protein (14, 16, 18, 23), we speculated that putative differences in GAG-binding between wt and 17D YFV would result from one or more of these nonconservative amino acid differences. Of the 12 amino acid differences in the E protein between 17D (204 substrain) and Asibi (10), 7 are nonconservative changes (Table 1). Among these, residues 52, 173, and 200 in domains I/II and 305, 325, and 380 in domain III of the YFV E protein are predicted to be localized to the top layer of the virion envelope by analogy with the crystal structures of tick-borne encephalitis and dengue viruses (12, 26, 31) and may therefore influence GAG interaction individually or in combination. We introduced reversal changes by site-directed mutagenesis using the YFV-17D full-length infectious cDNA clone at the six sites individually as well as in combinations of domain I/II clusters, domain III cluster, or domain I/II/III clusters combined. The nine mutant rYFV-17D viruses (r17D-R52G, r17D-I173T, r17D-T200K, r17D-F305S, r17D-S325P, r17D-R380T, r17D-dI/II, r17D-dIII, and r17D-dI/II/III) showed similar electroporation efficiencies in BHK cells (1 × 104 to 3 × 104 PFU/μg RNA) and formed plaques of comparable sizes on Vero (1 to 2 mm), BHK (2 to 3 mm), and SW13 (1 to 2 mm) cell monolayers in comparison to each other and infectious clone-derived 17D.
TABLE 1.
Amino acid differences between YFV 17D-204 and Asibi strains
| E residuea | Amino acid at indicated E residue in strain
|
Localization | Presence in all 17D substrainsd | |
|---|---|---|---|---|
| Asibi | 17D-204 | |||
| 52b | Gly | Argc | dII | + |
| 56 | Ala | Val | dII | − |
| 170 | Ala | Val | dI | + |
| 173b | Thr | Ilec | dI | + |
| 200b | Lys | Thrc | dII | + |
| 299 | Met | Ile | dIII | + |
| 305b | Ser | Phec | dIII | + |
| 325b | Pro | Serc | dIII | − |
| 331 | Lys | Arg | dIII | + |
| 380b | Thr | Argc | dIII | + |
| 407 | Ala | Val | Stem-anchor | + |
| 416 | Ala | Thrc | Stem-anchor | − |
Residue numbering is from the amino terminus of E.
Changes analyzed in this study.
Nonconservative change.
Changes common to 17D-204, 17D-213, and 17DD substrains are indicated by a +, while those absent from one or more or the three are indicated by a −.
Comparison of the sensitivity to heparin inhibition of recombinant 17D (r17D) and r17D-dI/II/III at concentrations of 0.2, 2, 20, and 200 μg/ml showed >50% inhibition of r17D at ∼2 μg/ml heparin and above and no inhibition of r17D-dI/II/III at any of the concentrations tested (Fig. 1B). This result clearly shows that 17D with an Asibi-like E protein displays markedly poorer interaction with GAG than the 17D vaccine. The remaining r17D variants were tested in parallel with r17D using 20 μg/ml heparin (Fig. 1C): the S325P and R380T substitutions gave rise to significantly reduced sensitivity to heparin inhibition (13% and 3% inhibition, respectively), while changes at residues 52, 173, 200, and 305 did not show a marked impact relative to r17D. Consistent with this result, r17D-dI/II also did not differ from r17D in heparin sensitivity, while r17D-dIII, which is altered at amino acids 305, 325, and 380, showed no inhibition by heparin. These results suggest that the domain III differences at residues 325 and 380 contribute predominantly to the heparin-sensitive phenotype of 17D.
The sole contribution of E protein residues S325 and R380 to the high GAG-binding affinity of the 17D vaccine was confirmed by using the converse approach of reverting the two residues singly in the context of an Asibi-like E protein (r17D-dI/II/III). An Arg at residue 380 almost completely restored heparin sensitivity to that of the 17D phenotype (42% inhibition), while a Ser at residue 325 had only a minor influence (5% inhibition). Residues 52 (r17D-dI/II/III-52R) and 173 (r17D-dI/II/III-173I) were tested in a similar fashion in parallel but did not impact on heparin sensitivity (Fig. 1C).
Growth of 17D E protein variants in BHK and Vero cells.
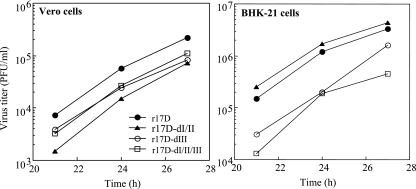
A comparison of the growth properties of r17D-dI/II, -dIII, and -dI/II/III with those of r17D was performed in BHK and Vero cells to determine if the altered heparin sensitivity impacted viral replication. In previous studies on Murray Valley encephalitis, Japanese encephalitis, West Nile, and DEN-2 viruses, heparin sensitivity correlated with increased specific infectivity for BHK and a second cell line (SW13), but not for Vero cells (14, 15, 18). Single-step growth curves in Vero cells showed that r17D and r17D-dI/II gave the best and poorest replication kinetics, respectively, with a ∼5-fold difference throughout, and that the growth of r17D-dIII and r17D-dI/II/III was intermediate (Fig. 2A). Single-step growth curves in BHK cells showed that r17D and r17D-dI/II grew to similarly high titers, while r17D-dIII and r17D-dI/II/III variants gave ∼5- to 10-fold-lower titers (Fig. 2B), consistent with the beneficial effect of high GAG-binding affinity on growth in that cell line noted for other viruses.
FIG. 2.
Growth assays of r17D, r17D-dI/II, r17D-dIII, and r17D-dI/II/III in Vero and BHK-21 cells. Virus titers from supernatant harvests over 3-h intervals at 21, 24, and 27 h p.i. (Vero) or 18, 21, and 24 h p.i. (BHK-21) were determined by plaque assays on Vero or BHK-21cells, respectively.
Analysis of fusion-related properties.
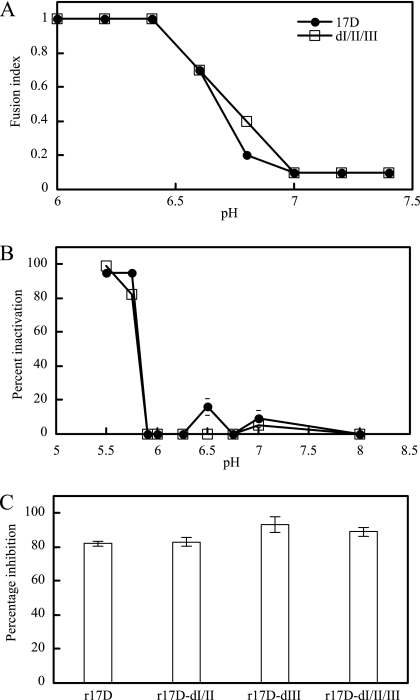
Domain I and II E protein differences between Asibi and 17D strains of YFV have been proposed to alter acid-induced fusion properties required for virus uptake by receptor-mediated endocytosis based on the location of residues 52, 173, and 200 in a hinge region undergoing conformational change during fusion, but there is an absence of supporting experimental data (reviewed in reference 27). Here we have tested for the first time fusion-related properties of 17D and wt-like E proteins of YFV in assays examining low-pH-induced syncytium formation in infected mosquito cells (C6/36) (FFWI), acid-induced inactivation of virus infectivity, and sensitivity of virus infectivity to the acidotropic agent NH4Cl. Comparison between r17D and r17D-dI/II/III in FFWI assays showed that the threshold for induction of syncytium formation in C6/36 cells occurred between pH 6.6 and 6.8 for both viruses (Fig. 3A). Both viruses showed no inactivation of infectivity at pH 5.9 and 97 to 99% inactivation at pH 5.5; at pH 5.75, r17D and r17D-dI/II/III showed 82% and 97% inactivation, respectively (Fig. 3B). Treatment of BHK cells with 10 mM NH4Cl, which neutralizes the endosomal pH, caused a similarly dramatic inhibition of infectivity of r17D and the three variants tested (r17D-dI/II, r17D-dIII, and r17D-dI/II/III), ranging from 82 to 93% (Fig. 3C). Given the lack of marked difference between r17D and r17D-dI/II/III in the three assays, it is unlikely that the E protein changes at residues 52, 173, 200, 305, 325, and 380 exerted an impact on virus fusion with cellular membranes of sufficient magnitude to influence in vivo virulence.
FIG. 3.
Fusion-related properties of r17D and r17D-dI/II/III. A. Fusion-from-within assays were performed in C6/36 cells, infected at an MOI of ∼0.1 for 4 days, in a pH range of 6.0 to 7.4. The fusion index was determined using the following formula: [1− (number of cytoplasts)/(number of nuclei)]. B. Sensitivity of virus to acid-mediated inactivation in a pH range of pH 5.5 to 7.0 was determined; percent inactivation was calculated by the formula 100 − [(virus titers of treated samples)/(virus titers of samples held at pH 8.0) × 100]. C. Sensitivity to NH4Cl was determined in BHK-21 cells, pretreated for 30 min with 0 or 10 mM NH4Cl and infected with r17D viruses at an MOI of ∼0.5 in the presence of NH4Cl. Nonadsorbed virus was inactivated after 1 h by addition of medium adjusted to pH 5.0. The assay for infectivity was performed by FACS, and percent inactivation was calculated by the formula 100 − [(percent infected cells treated with NH4Cl)/(percent infected cells without NH4Cl treatment) × 100].
Contribution of E protein changes to attenuation of virulence of YFV-17D vaccine.
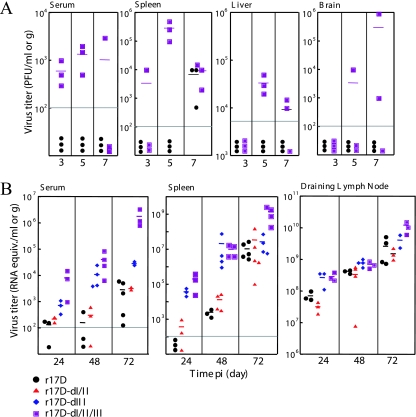
The pathogenesis of YFV in humans and other primates is predominantly a consequence of efficient virus replication in visceral organs. Neurological manifestations are rare in human infections with YFV (28). In contrast, both wt and 17D strains of YFV are neurotropic in laboratory mice but lack viscerotropism and are hence lethal for mice only by direct CNS infection (2). We show here that mice defective in type I and type II interferon (IFN) responses (IFN-α/γ-R−/−) are uniformly susceptible to YFV inoculation into the footpad at a low dose (103 Vero PFU), and we make use of this animal model to examine the impact of E protein changes on growth properties of 17D. YFV-17D and 17D variants with E protein changes present in the Asibi strain mostly gave rise to morbidity and mortality in IFN-α/γ-R−/− mice, albeit with distinct kinetics, and shared signs of illness suggesting that neuroinvasion and subsequent mortality due to encephalitis had occurred. The r17D-dI/II/III variant showed virus entry into the brain in 50% of infected mice by day 7 p.i. (Fig. 4A) and 100% mortality, with an average survival time (AST) of 12 days (Table 2). In contrast, r17D was not yet detectable in the brain at day 7 p.i. (Fig. 4A) and produced 88% mortality with a significantly prolonged AST (18.1 days) (Table 2). The 17D variants with single amino acid changes in E at residue 52, 200, 305, 325, or 380 showed little difference in AST (16.1 to 18.1 days) from r17D, and only r17D-I173T showed a slight but significant reduction in AST to 15.1 days (Table 2). However, infection with the r17D-dIII, which displays reduced GAG binding relative to 17D, resulted in significantly earlier signs of disease and mortality (AST, 12.9 days). Increased virulence comparable to that associated with domain III changes was not found for r17D-dI/II (AST, 17.0 days).
FIG. 4.
Dissemination of YFV-17D and r17D variants with wt E protein changes in IFN-α/γ-R−/− mice. A. Distribution of r17D and r17D-dI/II/III between 3 and 7 days p.i. in serum, spleen, liver, and brain of 6-week-old IFN-α/γ-R−/− mice inoculated with 1,000 PFU via the hind footpad. B. Distribution of r17D, r17D-dI/II, r17D-dIII, and r17D-dI/II/III at 24, 48, and 72 h p.i. in serum, spleen, and draining popliteal lymph nodes of 6-week-old IFN-α/γ-R−/− mice inoculated with 105 PFU via the hind footpad. Virus content was measured by plaque assay in Vero cells (A) or qRT-PCR (B). Each symbol corresponds to an individual mouse, and horizontal bars indicate mean titers.
TABLE 2.
Virulence properties of YFV-17D variants in IFN-α/γ-R−/− mice
| Virus | E residue(s) different from 17D | Mortalitya | AST (days) (SD) | P valueb |
|---|---|---|---|---|
| r17D | − | 14/16d | 18.1 (2.4) | − |
| r17D-R52G | 52 | 10/10 | 17.7 (1.6) | 0.9 |
| r17D-I173T | 173 | 10/10 | 15.1 (2.3) | 0.01 |
| r17D-T200K | 200 | 9/10 | 16.7 (1.2) | 0.2 |
| r17D-F305S | 305 | 8/8 | 16.3 (2.8) | 0.1 |
| r17D-S325P | 325 | 9/10 | 16.1 (1.5) | 0.07 |
| r17D-R380T | 380 | 9/9 | 18.0 (2.5) | 1 |
| r17D-dI/II/III | 52, 173, 200, 305, 325, 380 | 9/9 | 11.6 (1.3) | <0.0001 |
| r17D-dIII | 305, 325, 380 | 9/9 | 12.9 (2.6) | 0.0006 |
| r17D-dI/II | 52, 173, 200 | 8/8 | 17.0 | 0.2c |
| r17D-S325P/R380T | 325, 380 | 0/8 | NAe | |
| r17D-dI/II/III-305F | 52, 173, 200, 325, 380 | 10/10 | 11.5 (2.1) | <0.0001 |
Mortality over 4 weeks is shown as the fraction of the total number of mice inoculated with 103 Vero PFU via the footpad; the specific infectivity of YFV-17D and variants corresponds to 2 × 103 to 5 × 103 genome equivalents/PFU.
The P value was calculated using the Mann-Whitney ranking U test on survival times versus r17D; values under 0.05 are shown in bold.
A P value of 0.001 was calculated when the survival times were tested against those of r17D-dI/II/III using the Mann-Whitney ranking U test.
Combined data from three virulence determinations are shown.
NA, not applicable.
Interestingly, we found that r17D-S325P/R380T, in which both GAG-binding determinants identified for the 17D vaccine strain were reverted to the low-GAG-binding phenotype of the Asibi E protein, was not lethal following footpad inoculation into the IFN-deficient mice. The reason for this unexpected finding was that the virus had lost neurovirulence, which became evident in view of the absence of mortality following intracerebral inoculation (104 PFU) into 4-week-old BALB/c mice (data not shown). YFV-17D, tested in parallel, is uniformly lethal in this challenge model (see also reference 2). However, when the two GAG reversal changes were combined with the domain I/II changes, as seen in the variant rYFVdI/II/III-F305, virulence was restored and footpad inoculation of IFN-α/γ-R−/− mice with this recombinant virus gave rise to a reduced AST (11.5 days) similar to that of the virulent r17D-dIII and r17D-dI/II/III variants. Therefore, it appears that the loss of neurovirulence accompanying the GAG reversal changes could be compensated by either the F305S change or the domain I/II changes.
Contribution of E protein changes to attenuation of viscerotropism of YFV-17D vaccine.
To define the events accompanying the earlier mortality of mice infected with r17D-dI/II/II relative to r17D, virus loads in serum, spleen, and liver of IFN-α/γ-R−/− mice were examined at 3, 5, and 7 days after infection via the footpad (103 PFU). Mice infected with r17D-dI/II/III showed viremia and virus titers in the spleen and liver at 3, 5, and 7 days p.i., with peak levels occurring on day 5 p.i. (Fig. 4A). In contrast, mice infected with r17D showed no detectable virus in serum and liver, but low virus titers were found in the spleen on day 7 p.i., suggesting delayed virus spread (Fig. 4A).
We also compared the spread of YFV-17D, and variants with wt E protein reversions derived from it, in IFN-α/γ-R−/− mice by qRT-PCR following inoculation of a high virus dose (105 PFU) into the hind footpad. A comparable and increasing presence of all virus strains tested (r17D, r17D-dI/II, r17D-dIII, and r17D-dI/II/III) was found in the draining lymph nodes at 24, 48, and 72 h p.i. This indicates that transfer of virus from the inoculation site into the draining lymph node, mediated most likely by Langerhans and/or dendritic cells, and virus growth in the lymphatic tissue were not differentially influenced by the E protein changes. However, the r17D-dIII and r17D-dI/II/III strains displayed markedly better viscerotropic spread, exemplified by viremia titers and presence of virus in the spleen, than the r17D and r17D-dI/II strains. Mean viremia titers of r17D-dI/II/III were 50- to 1,000-fold higher than those of r17D and r17D-dI/II strains during the first 3 days of infection; viremia induced by the r17D-dIII virus was in the intermediate range. Virus load in the spleen for r17D-dIII and r17D-dI/II/III was 100-fold higher at 1 and 2 days p.i. than that of r17D and r17D-dI/II. It is notable that following delayed spread of the latter two strains to the spleen (and probably other visceral organs), virus replication, like in the draining lymph node, was efficient, given that the day 3 virus titers in the spleen reached that of the 17D-dIII virus; however, this relatively efficient growth contrasted with a persistently low viremia of r17D and r17D-dI/II strains.
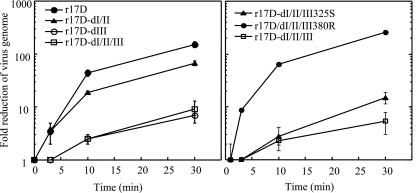
In vivo mechanism for loss of viscerotropism of YFV-17D. One of the key in vivo effects of high GAG-binding affinity acquired during laboratory adaptation of animal viruses, including flaviviruses, is the detrimental impact on virus spread in the animal host (3, 5, 14, 15, 18). This is thought to be due to the rapid removal of virus from the bloodstream, most likely as the result of nonproductive binding of virus to extracellular matrix components, which are rich in GAG, or readsorption of progeny virus to infected cells. The kinetics of in vivo virus clearance from the blood was determined for r17D and variants with the Asibi E protein changes to assess the effect of the differential GAG-binding phenotypes (Fig. 5). Mice were injected intravenously with 107 virus particles, and virus genome content in the serum over 30 min was determined by qRT-PCR. The YFV-17D vaccine showed significantly more rapid loss of virus from the bloodstream (150-fold reduction at 30 min) than r17D-dI/II/III (9-fold reduction at 30 min) or r17D-dIII (7-fold reduction at 30 min). The variant r17D-dI/II was also cleared rapidly (67-fold reduction at 30 min), consistent with a GAG-binding phenotype similar to that of r17D. To assess the impact of single GAG-binding determinants on in vivo clearance, the variants r17D-dI/II/III-325S and r17D-dI/II/III-380R were compared with r17D-dI/II/III. The GAG-binding motif Arg380 was shown to exert a dramatic impact on virus clearance, with a >200-fold reduction in virus genome content from circulation after 30 min, whereas E residue 325S only slightly increased clearance (15-fold versus 5-fold reduction of r17D-dI/II/III).
FIG. 5.
Virus clearance in vivo. Virus genome content in the serum was determined over 30 min after inoculation of a dose of r17D, r17D-dI/II, r17D-dIII, r17D-dI/II/III, r17D-dI/II/III-325S, or r17D-dI/II/III-380R equivalent to 107 genome copies. The reduction at each time point was calculated as (input virus)/(virus content in serum sample), assuming a blood volume of 7% of body weight of each mouse. Standard errors shown were calculated using results from two mice for each virus sample.
DISCUSSION
Insight into the in vivo mechanism(s) for virulence attenuation of the widely used and highly successful YFV-17D vaccine has remained elusive since its production by Theiler and Smith 70 years ago. Here we have demonstrated that loss of the ability to efficiently disseminate in the mammalian host accounts, at least in part, for the safety of the live vaccine without markedly compromising its immunogenicity. Furthermore, we have defined key residues in the YFV E protein which determine this attenuation mechanism.
The 17D vaccine diverged from the virulent Asibi strain during 200 passages in mouse and chicken embryo tissue culture; of a total of 32 amino acid differences, 6 nonconservative changes are located in the E protein ectodomain. These changes were engineered into the 17D 204 vaccine substrain to generate a 17D variant with a wt-like E protein. A key discovery was the substantially reduced sensitivity of this variant (r17D-dI/II/III) to inhibition of infectivity by heparin treatment, in comparison to the vaccine. We have shown in previous studies that variants of DEN and encephalitic flaviviruses that display high GAG-binding affinities are rapidly cleared from the bloodstream, preventing effective dissemination from the infection site to target organs and the manifestation of disease. Supporting the generality of this mechanism of virulence attenuation among flaviviruses, the 17D vaccine was removed from the circulation almost 20 times faster than r17D-dI/II/III and displayed a markedly poorer ability to spread to visceral organs following footpad infection; it also showed delayed entry into the CNS of infected mice to produce mortality significantly later than the 17D variants with Asibi E protein changes. It is notable that three live flavivirus vaccines derived from repeated passaging in nonnatural host tissues and proven to be immunogenic and safe in humans, namely, the Sabin DEN-2 NGC strain (its wider use was prevented by the acquisition of neurotropism), the Japanese encephalitis virus 14-14-2 vaccine, and the YFV-17D vaccine, are GAG-binding viruses deficient in dissemination (15, 18). While it is not entirely clear why these live vaccines fail to produce viremia of an adequate magnitude and duration to infect the target tissues and produce disease, we speculate that attachment of virus with high GAG-binding affinity to the extracellular matrix, a structural entity that is enriched in GAG (30), prevents seeding of virus from infected cells into the bloodstream as well as facilitating its clearance.
This study defines key determinants on the surface of YFV-17D involved in GAG binding. Of the six nonconservative amino acid differences in the E proteins of 17D and Asibi, only two residues, Ser325 and Arg380, both located in E protein domain III, accounted for the GAG-binding phenotype of 17D. Residue Arg380 clearly exerted the dominant effect, which is consistent with the previous identification of the corresponding region on the lateral edge of domain III of flaviviruses belonging to the Japanese encephalitis virus serocomplex as a GAG-binding domain (14). Given that the Ser325 acquired by 17D did not alter the E protein net positive charge thought to enhance GAG-binding affinity, we suggest that Ser325 exerts its minor effect by orienting the neighboring basic residue, Arg326, to interact optimally with GAG. Notably, a 17D variant with an R326E mutation did not show binding to heparin-Sepharose but was also defective in replication in Vero cells (data not shown). Given that the P325S mutation is not present in the 17DD vaccine substrain (8), it is presumably not essential to the attenuation process.
When Asibi E protein residues distinguishing it from 17D were substituted into the 17D E protein according to their location in one of the three E protein domains, we found that the combination of three domain III changes produced a blood clearance, viscerotropic spread, and virulence phenotype resembling that of r17D-dI/II/III. The domain III cluster of changes in r17D-dIII included the two GAG-binding determinants, in addition to a third change (F305S) that contributed to neurovirulence (see below). Importantly, the combination of Asibi domain I and II residues substituted into the 17D E protein (r17D-dI/II) did not alter blood clearance kinetics, in vivo spread, and virulence markedly from that of 17D. On this basis we conclude that, at least in the mouse model available for this study, the mutations in E protein domain III acquired by the vaccine are vital for reducing viscerotropism and virulence of the vaccine. It should be emphasized that despite the striking impact of domain III changes on viscerotropism and virulence found in this study, additional contributions to the safety of the 17D vaccine may be provided by other mutations impacting on viral properties such as cell tropism, growth efficiency, and susceptibility to antiviral defense.
Assessment of virulence of YFV is achieved most reliably using monkeys, given their susceptibility to viscerotropic disease (reviewed in reference 28). Viremia is an excellent correlate of viscerotropism of YFV in monkeys (and humans), since 17D produces very low viremia, in contrast to the high viremia associated with wt YFV infection of primates. Choice of a small animal model suitable for analysis of YFV virulence is hampered by the inefficient extraneural replication of nonadapted strains (reviewed in reference 28). In contrast, direct CNS infection of most YFV strains in mice is lethal (2). This obstacle can be overcome with the use of viscerotropic strains of YFV in hamsters (24, 38) or of immunodeficient (e.g., scid) mice with increased sensitivity to YFV by peripheral routes of infection (6). We show in this study that type I and II interferons are crucial in restricting viral replication in the mouse visceral organs, hence adult IFN-α/γ-R−/− mice are uniformly susceptible to low-dose inoculation of 17D variants into the footpad and succumb following virus spread into the CNS. Thus, mortality of IFN-α/γ-R−/− mice is influenced by viscerotropic properties as well as neurovirulence of a given YFV strain. Importantly, viremia and extraneural virus growth can be measured in this mouse strain as correlates of viscerotropism. This has allowed us to identify three residues in E protein domain III of the 17D vaccine, two of which account for enhanced GAG binding, as important determinants of viscerotropic attenuation. The impact of E protein changes on mouse neurovirulence, in particular residues 52, 173, and 305, is also evident from this and other studies (29, 34, 35). Notably, the Ser305Phe change may exert a dominant effect in attenuating neurovirulence and thus influence virulence in IFN-α/γ-R−/− mice observed for r17D variants with single or double amino acid substitutions in domain III. Contrary to the suggestion by others (reviewed in reference 27), we found no effect of these neurovirulence determinants on fusion; thus, the mechanism by which neurovirulence of YFV is modulated by these E protein residues remains elusive.
The success of the YFV-17D vaccine is due to a combination of excellent immunogenicity and safety. Accordingly, loss of virulence should not substantially impact on the ability of the vaccine to establish a primary infection following inoculation and to elicit the broad cellular and humoral immune responses characteristic of live viral infections. Our results show comparable growth of 17D and 17D variants with Asibi E protein changes in the lymph node draining the footpad infection site. Transport of intradermally inoculated virus to the draining lymph node is thought to be mediated by Langerhans or dendritic cells (22) and would therefore not be subject to GAG-dependent inhibition of spread. We also found better replication of 17D in Vero and BHK cells as well as in ex vivo mouse macrophage cells (data not shown), compared with 17D variants with Asibi E protein changes, while others have reported more efficient growth of 17D in human liver HepG2 cells than the Asibi parent, as well as efficient replication of 17D in immature and mature human dendritic cells (1, 19). Collectively, this demonstrates that the overall replication efficiency of the vaccine strain in a diverse range of cell types is not lowered. Importantly, the efficient growth of 17D in lymphoid tissue, which is specialized in antigen presentation for the establishment of adaptive immune responses, may explain the excellent immunogenicity of the vaccine.
Acknowledgments
We thank Roy Hall for provision of the monoclonal antibody 4G4. We are grateful to Charles Rice for provision of the infectious cDNA clone of YFV.
This work was supported by a project grant (NHMRC366736) from the National Health and Medical Research Council of Australia.
Footnotes
Published ahead of print on 9 April 2008.
REFERENCES
- 1.Barba-Spaeth, G., R. S. Longman, M. L. Albert, and C. M. Rice. 2005. Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J. Exp. Med. 2021179-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, A. D., and E. A. Gould. 1986. Comparison of neurovirulence of different strains of yellow fever virus in mice. J. Gen. Virol. 67631-637. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, K. A., W. B. Klimstra, and R. E. Johnston. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 27693-103. [DOI] [PubMed] [Google Scholar]
- 4.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 841261-1268. [DOI] [PubMed] [Google Scholar]
- 5.Byrnes, A. P., and D. E. Griffin. 2000. Large-plaque mutants of Sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J. Virol. 74644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, T. J., and M. Nickells. 2001. Neuroadapted yellow fever virus 17D: genetic and biological characterization of a highly mouse-neurovirulent virus and its infectious molecular clone. J. Virol. 7510912-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3866-871. [DOI] [PubMed] [Google Scholar]
- 8.dos Santos, C. N., P. R. Post, R. Carvalho, I. I. Ferreira, C. M. Rice, and R. Galler. 1995. Complete nucleotide sequence of yellow fever virus vaccine strains 17DD and 17D-213. Virus Res. 3535-41. [DOI] [PubMed] [Google Scholar]
- 9.Germi, R., J. M. Crance, D. Garin, J. Guimet, H. Lortat-Jacob, R. W. Ruigrok, J. P. Zarski, and E. Drouet. 2002. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 292162-168. [DOI] [PubMed] [Google Scholar]
- 10.Hahn, C. S., J. M. Dalrymple, J. H. Strauss, and C. M. Rice. 1987. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proc. Natl. Acad. Sci. USA 842019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 727357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, C. J., and T. P. Monath. 2003. Chimeric flaviviruses: novel vaccines against dengue fever, tick-borne encephalitis, and Japanese encephalitis. Adv. Virus Res. 61469-509. [DOI] [PubMed] [Google Scholar]
- 14.Lee, E., R. A. Hall, and M. Lobigs. 2004. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J. Virol. 788271-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, E., and M. Lobigs. 2002. Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. J. Virol. 764901-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, E., and M. Lobigs. 2000. Substitutions at the putative receptor-binding site of an encephalitic flavivirus alter virulence and host cell tropism and reveal a role for glycosaminoglycans in entry. J. Virol. 748867-8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, E., M. Pavy, N. Young, C. Freeman, and M. Lobigs. 2006. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antivir. Res. 6931-38. [DOI] [PubMed] [Google Scholar]
- 18.Lee, E., P. J. Wright, A. Davidson, and M. Lobigs. 2006. Virulence attenuation of Dengue virus due to augmented glycosaminoglycan-binding affinity and restriction in extraneural dissemination. J. Gen. Virol. 872791-2801. [DOI] [PubMed] [Google Scholar]
- 19.Lefeuvre, A., H. Contamin, T. Decelle, C. Fournier, J. Lang, V. Deubel, and P. Marianneau. 2006. Host-cell interaction of attenuated and wild-type strains of yellow fever virus can be differentiated at early stages of hepatocyte infection. Microbes Infect. 81530-1538. [DOI] [PubMed] [Google Scholar]
- 20.Licon Luna, R. M., E. Lee, A. Mullbacher, R. V. Blanden, R. Langman, and M. Lobigs. 2002. Lack of both Fas ligand and perforin protects from flavivirus-mediated encephalitis in mice. J. Virol. 763202-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobigs, M., R. Usha, A. Nestorowicz, I. D. Marshall, R. C. Weir, and L. Dalgarno. 1990. Host cell selection of Murray Valley encephalitis virus variants altered at an RDG sequence in the envelope protein and in mouse virulence. Virology 176587-595. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandl, C. W., H. Kroschewski, S. L. Allison, R. Kofler, H. Holzmann, T. Meixner, and F. X. Heinz. 2001. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J. Virol. 755627-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McArthur, M. A., M. T. Suderman, J. P. Mutebi, S. Y. Xiao, and A. D. Barrett. 2003. Molecular characterization of a hamster viscerotropic strain of yellow fever virus. J. Virol. 771462-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McElroy, K. L., K. A. Tsetsarkin, D. L. Vanlandingham, and S. Higgs. 2005. Characterization of an infectious clone of the wild-type yellow fever virus Asibi strain that is able to infect and disseminate in mosquitoes. J. Gen. Virol. 861747-1751. [DOI] [PubMed] [Google Scholar]
- 26.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 1006986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monath, T. P. 2005. Yellow fever vaccine. Expert Rev. Vaccines 4553-574. [DOI] [PubMed] [Google Scholar]
- 28.Monath, T. P., and A. D. Barrett. 2003. Pathogenesis and pathophysiology of yellow fever. Adv. Virus Res. 60343-395. [DOI] [PubMed] [Google Scholar]
- 29.Nickells, M., and T. J. Chambers. 2003. Neuroadapted yellow fever virus 17D: determinants in the envelope protein govern neuroinvasiveness for SCID mice. J. Virol. 7712232-12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman, R., V. Sasisekharan, and R. Sasisekharan. 2005. Structural insights into biological roles of protein-glycosaminoglycan interactions. Chem. Biol. 12267-277. [DOI] [PubMed] [Google Scholar]
- 31.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375291-298. [DOI] [PubMed] [Google Scholar]
- 32.Rice, C. M., A. Grakoui, R. Galler, and T. J. Chambers. 1989. Transcription of infectious yellow fever virus RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1285-296. [PubMed] [Google Scholar]
- 33.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: Implications for flavivirus gene expression and evolution. Science 229726-733. [DOI] [PubMed] [Google Scholar]
- 34.Ryman, K. D., T. N. Ledger, G. A. Campbell, S. J. Watowich, and A. D. Barrett. 1998. Mutation in a 17D-204 vaccine substrain-specific envelope protein epitope alters the pathogenesis of yellow fever virus in mice. Virology 24459-65. [DOI] [PubMed] [Google Scholar]
- 35.Ryman, K. D., H. Xie, T. N. Ledger, G. A. Campbell, and A. D. Barrett. 1997. Antigenic variants of yellow fever virus with an altered neurovirulence phenotype in mice. Virology 230376-380. [DOI] [PubMed] [Google Scholar]
- 36.Sa-Carvalho, D., E. Rieder, B. Baxt, R. Rodarte, A. Tanuri, and P. W. Mason. 1997. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J. Virol. 715115-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlesinger, J. J., S. Chapman, A. Nestorowicz, C. M. Rice, T. E. Ginocchio, and T. J. Chambers. 1996. Replication of yellow fever virus in the mouse central nervous system: comparison of neuroadapted and non-neuroadapted virus and partial sequence analysis of the neuroadapted strain. J. Gen. Virol. 771277-1285. [DOI] [PubMed] [Google Scholar]
- 38.Tesh, R. B., H. Guzman, A. P. da Rosa, P. F. Vasconcelos, L. B. Dias, J. E. Bunnell, H. Zhang, and S. Y. Xiao. 2001. Experimental yellow fever virus infection in the Golden hamster (Mesocricetus auratus). I. Virologic, biochemical, and immunologic studies. J. Infect. Dis. 1831431-1436. [DOI] [PubMed] [Google Scholar]
- 39.Theiler, M., and H. H. Smith. 2000. The use of yellow fever virus modified by in vitro cultivation for human immunization (J. Exp. Med. 65787-800, 1937.) Rev. Med. Virol. 10 6-16. [PubMed] [Google Scholar]
- 40.van den Broek, M. F., U. Muller, S. Huang, M. Aguet, and R. M. Zinkernagel. 1995. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J. Virol. 694792-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]