Abstract
While the selection of amino acid insertions in human immunodeficiency virus (HIV) reverse transcriptase (RT) is a known mechanism of resistance against RT inhibitors, very few reports on the selection of insertions in the protease (PR) coding region have been published. It is still unclear whether these insertions impact protease inhibitor (PI) resistance and/or viral replication capacity. We show that the prevalence of insertions, especially between amino acids 30 to 41 of HIV type 1 (HIV-1) PR, has increased in recent years. We identified amino acid insertions at positions 33 and 35 of the PR of HIV-1-infected patients who had undergone prolonged treatment with PIs, and we characterized the contribution of these insertions to viral resistance. We prepared the corresponding mutated, recombinant PR variants with or without insertions at positions 33 and 35 and characterized them in terms of enzyme kinetics and crystal structures. We also engineered the corresponding recombinant viruses and analyzed the PR susceptibility and replication capacity by recombinant virus assay. Both in vitro methods confirmed that the amino acid insertions at positions 33 and 35 contribute to the viral resistance to most of the tested PIs. The structural analysis revealed local structural rearrangements in the flap region and in the substrate binding pockets. The enlargement of the PR substrate binding site together with impaired flap dynamics could account for the weaker inhibitor binding by the insertion mutants. Amino acid insertions in the vicinity of the binding cleft therefore represent a novel mechanism of HIV resistance development.
Human immunodeficiency virus (HIV) requires a viral protease (PR) for the processing of Gag and Gag-Pol polyprotein precursors into functional enzymes and structural proteins to yield mature virus progeny (19). Therefore, PR has become one of the major targets of anti-HIV treatment (10). Today, nine protease inhibitors (PIs) have been approved by the U.S. Food and Drug Administration and are clinically available. One of the major complications of such treatment is the evolution of drug-resistant PR variants (9). An understanding of the molecular mechanism of resistance is therefore critical for the design of novel, effective PIs that will maintain viral suppression (8, 31, 45).
The development of PI resistance is only partially understood on the molecular level. In most cases it starts with amino acid substitutions in the substrate-binding pocket of PR (primary mutations) that cause decreased inhibitor binding and may also impair the binding of the natural polyprotein substrate, thus affecting virus replication. Consequently, additional (secondary) mutations accumulate outside the substrate binding pockets. These secondary mutations have little effect on inhibitor binding but improve the proteolytic efficiency of the resistant enzyme and increase viral replication (4, 29). In addition to primary and secondary mutations in the PR itself, amino acid changes in the cleavage sites of viral Gag polyprotein are also introduced in order to improve the ability of the mutated enzyme to bind and cleave its substrate (12, 46, 22). Recently, a novel mechanism of PI resistance involving primary mutations in the natural substrate leading to improved polyprotein processing without any prior mutations in the PR coding region was identified (28).
Occasionally, amino acid insertions instead of substitutions are selected during antiretroviral therapy. Insertion mutations are not unusual in HIV reverse transcriptase (RT) isolated from patients treated by RT inhibitors (24, 42). Amino acid insertions in the PR coding region (1 to 6 amino acids) have been recently detected at various sites in the viral PR sequence, e.g., in regions between codons 17 to 18, 22 to 25, 31 to 32, 35 to 38, 70 to 71, and 95 to 96 (43). The prevalence of insertions in the PR coding regions of HIV-positive patients is estimated to be 0.1% (18), and position 35 seems to be most prone to insertions (43). Most of the insertions stem from duplications of neighboring DNA sequences, which could be explained by primer/template slippage during the reverse transcription process.
Since the insertions are usually accompanied by numerous other mutations in the PR coding region, it is difficult to dissect the relative contributions of amino acid insertions in the PR to the overall resistance and/or viral replicative capacity. To our knowledge, neither an enzymological characterization nor a three-dimensional structure of a PR variant bearing amino acid insertions has been provided so far.
In this study, we set out to characterize the role of amino acid insertions in the PR coding region in resistance development. To this end, we used two PR sequences from HIV-positive patients treated with PIs with amino acid insertions at position 33 or 35 of the PR coding region. To analyze the contribution of the insertions in the PR sequence to viral resistance on a molecular level, we characterized their effects on the virus by studying PR resistance and replicative capacity and on the enzyme directly by using enzymological and structural analysis of the recombinant proteins.
MATERIALS AND METHODS
Database mining.
PR amino acid insertions in a U.S. reference laboratory database consisting of over 208,000 HIV type 1 (HIV-1) clinical samples submitted to Quest Diagnostics Nichols Institute (San Juan Capistrano, CA) for genotype analysis between 1999 and May 2007 were tabulated. To account for imprecision in the placement of the inserted residues in the alignments performed in a reference laboratory setting, the insertions were then grouped into three regions, residues 1 to 31, 32 to 41, and 42 to 99. Predicted antiretroviral resistance was assessed from genotypic data according to the Quest Diagnostics resistance algorithm. Amino acid positions with mixed wild-type and mutant amino acids were excluded from the mutational association estimates.
Viral RNA analysis.
Samples from two patients, identified through a screening procedure for a clinical trial, were used for the study. The entry criteria involved repeated failure with a PI-containing regimen. Viral RNA was isolated from 100 μl of plasma according to the method described by Boom et al. (3). Subsequently, the isolated viral RNA was used to reverse transcribe and amplify viral PR and the C terminus of Gag, essentially as previously described (37). Subsequently, the viral PR was sequenced using a BigDye Terminator cycle sequencing kit, versions 1.1 and 3.1 (Applied Biosystems International, Foster City, CA) (37).
Cells.
MT-2 and SupT1 cells were maintained in RPMI 1640 medium with l-glutamine (BioWhittaker, Verviers, Belgium) supplemented with 10% fetal bovine serum (Gibco, Breda, The Netherlands) and 10 μg/ml gentamicin (Gibco). 293T cells were maintained in Dulbecco's modified Eagle's medium (BioWhittaker) supplemented with 10% fetal bovine serum and 10 μg/ml gentamicin. All cells were passaged twice weekly.
Generation of recombinant virus.
The viral DNA fragment including the viral PR and the C-terminal portion of Gag were cloned into an HXB2 reference strain as previously described (36). To do so, the PCR product and the vector (pHXB2Δ CSPR) were both digested with BstEII and MluNI. The PCR product and vector (pHXB2Δ cleavage site protease plasmid) were ligated using a Rapid DNA Ligation Kit (Roche Diagnostics GmbH, Mannheim, Germany). Several clones were sequenced, and a representative clone was used for transfection. A total of 6 × 106 293T cells were seeded the day prior to transfection to achieve 90 to 95% confluence on the day of transfection. For transfection of 10 μg of plasmid DNA, Lipofectamine 2000 (Invitrogen, Breda, The Netherlands) was used according to the manufacturer's protocol. The recombinant virus was harvested 2 days after transfection.
Generation of site-directed mutants in the recombinant virus.
To investigate the impact of the insert in the viral PR on phenotypic PR susceptibility and replication capacity, we generated site-directed mutants without the inserts (recombinant viruses VPR2 and VPR4). Therefore, PCR was performed on the recombinant virus clone using an Expand High Fidelity Plus PCR System (Roche Applied Sciences) and primers 5′-p2 (5′-AAGCAATGAGCCAGGTAACCAATTC-3′; positions 1883 to 1907) and 3′-prot-2 (5′-AATGCTTTTATTTTTTCTTCTGTCAATGGC-3′; positions 2650 to 2621) and a third mutagenesis primer which was either VPR1-out (5′-TTCCCGGCAAATTCATTTCTTCTAATATTGT-3′) or VPR3-out (5′-GGCAAATTTATTTCTTCTAATACTGTATCATCTGCT-3′). Subsequently, the PCR product was used to generate a recombinant virus clone, and sequence analysis was performed to confirm the generation of the correct recombinant virus clone.
Drug susceptibility analysis.
The infectious virus titer (50% tissue culture infective dose) was determined using end-point dilutions in MT-2 cells. The drug susceptibility of the viruses was determined in duplicate using a multiple cycle MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] assay (Antivirogram; Virco BVBA Mechelen, Belgium) (5).
Replication competition experiments.
To determine the relative replication capacity of the PR variants with and without insertions, competition experiments were performed in SupT1 cells. Replication competition experiments were performed by mixing two recombinant viruses based on the 50% tissue culture infective doses. In a total volume of 1 ml, 2 × 106 SupT1 cells were infected at a multiplicity of infection of 0.001. After 2 h of infection at 5% CO2 and 37°C, cells were washed and subsequently cultured in 10 ml of fresh culture medium. When full-blown syncytia were present in the culture, the viral supernatant was harvested, of which ca. 10 to 50 μl was used to infect 2 × 106 SupT1 cells until four serial passages were performed. RNA was extracted from the culture supernatant at several time points during the experiment, and the viral PR gene was sequenced as described previously. The relative amount of the insert in the population was determined by estimating the relative peak heights of the electrophoretograms.
Viral replication experiments.
The amount of p24 in each 293T cell recombinant virus batch was determined by enzyme-linked immunosorbent assay (Ampak; DAKO, Cambridgeshire, United Kingdom) (26). Viral replication experiments were performed by infecting 2.0 × 106 SupT1 cells with 100 ng of p24 from each recombinant virus batch. After 2 h of incubation, the cells were washed twice with RPMI 1640 medium with l-glutamine (Cambrex, Verviers, Belgium) and resuspended in 10 ml of culture medium (RPMI 1640 medium with l-glutamine supplemented with 10% fetal calf serum; Gibco, Breda, The Netherlands) and gentamicin (10 μg/ml; Invitrogen, Breda, The Netherlands). The cultures were maintained for 13 days, and each day two samples of 150 μl of cell-free viral supernatant was taken for p24 analysis.
DNA amplification and mutagenesis for recombinant protein expression.
The HIV-1 PR coding regions from the patients were amplified from the recombinant virus clones. The amino acid sequences (PR1 and PR3) are depicted in Fig. 1, and all the substitutions with respect to the HIV sequence database consensus B sequence (www.hiv.lanl.gov.) are summarized in Table 1. The amplifications were performed using the forward primer 5′-ATCCTTTCATATGCCTCAGATCACTCTTTGG-3′, which is specific to the 5′ end of the PR coding region and includes an NdeI site, and the reverse primer 5′-TTGAATTCGATATCATTAAAAATTTAAAGTGCAGCC-3′, which contains an EcoRI site. The PCR products were subsequently ligated into the expression vector pET24a (Novagen, Darmstadt, Germany). The E35EE insertion was removed from PR1 by ligation of two PCR products to obtain DNA encoding PR2. The primers used for generation of the first PCR product were 5′-GCGCTCCCGGCAAGTTCATTTCTTCTAATATTGTATCATC-3′, which includes a recognition site for BssKI endonuclease, and 5′-ATCCTTTCATATGCCTCAGATCACTCTTTGG-3′, which contains an NdeI site. The amplification of the second product was performed using primers 5′-TTGCCGGGAAGATGGACACCAAAAGTGGTAGGG-3′, which includes a BssKI site, and 5′-TTGAATTCGATATCATTAAAAATTTAAAGTGCAGCC-3′, which contains an EcoRI site. These products were cleaved by the above-mentioned endonucleases and ligated into pET24a. The same procedure was carried out for deletion of the L33LL insertion from PR3 to obtain PR4 using the following primers: 5′-GCGCTCCTGGCAAGTTAATCTCCTCTAATACTGTATCATCTGC-3′, 5′-ATCCTTTCATATGCCTCAGATCACTCTTTGG-3′, 5′-TTGCCAGGAAGATGGAAACCAAAAATAATAGGG-3′, and 5′-TTGAATTCGATATCATTAAAAATTTAAAGTGCAGCC-3′.
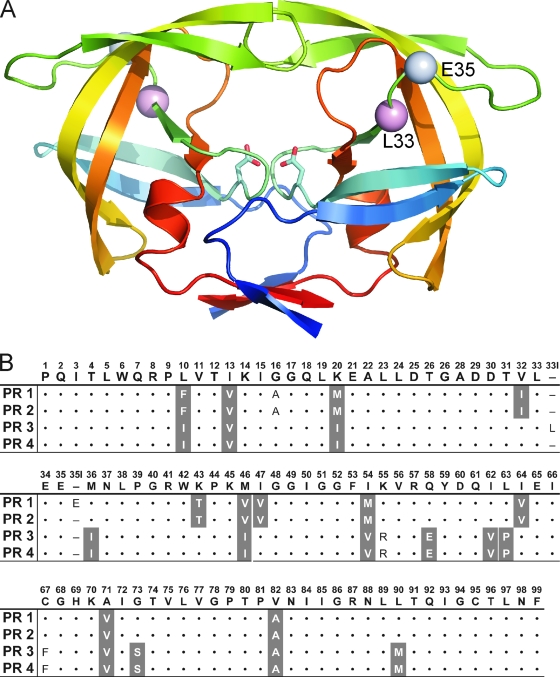
FIG. 1.
(A) Overall structure of HIV-1 PR showing the positions of the insertions (amino acid residues 33 and 35). The PR monomers are depicted in ribbon representation and are colored blue to red from the N terminus to the C terminus; Cα atoms of residues which are duplicated by insertion are shown as spheres and labeled. (B) HIV PR sequences of the corresponding mutant PRs with insertions (PR1 and PR3) and without insertions (PR2 and PR4) compared to the consensus B sequence. Primary resistance mutations (according to the International AIDS Society list, 2006 [current version available at http://www.iasusa.org/resistance_mutations/mutations_figures.pdf]) are shaded.
TABLE 1.
List of mutations and enzyme characteristics (Km, kcat, and catalytic efficiency) of PR variants analyzed in this study
| HIV-1 PR type | Mutationsa | Enzyme kineticsb
|
||
|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (mM−1·s−1) | ||
| Wild-type | 15.1 ± 1.3 | 30 ± 1.8 | 1,990 ± 210 | |
| PR1 | L10F, I13V, G16A, K20M, V32I, E35EE, K43T, M46V, I47V, I54M, I64V, A71V, V82A | 20.4 ± 1.6 | 5.5 ± 0.2 | 271 ± 23 |
| PR2 | L10F, I13V, G16A, K20M, V32I, K43T, M46V, I47V, I54M, I64V, A71V, V82A | 9.2 ± 1.0 | 6.5 ± 0.3 | 712 ± 80 |
| PR3 | L10I, I13V, K20I, L33LL, M36I, M46I, I54V, K55R, Q58E, I62V, L63P, C67F, A71V, G73S, V82A, L90M | 23 ± 1 | 9.1 ± 0.4 | 400 ± 25 |
| PR4 | L10I, I13V, K20I, M36I, M46I, I54V, K55R, Q58E, I62V, L63P, C67F, A71V, G73S, V82A, L90M | 11.3 ± 0.7 | 8.3 ± 0.4 | 735 ± 53 |
Amino acid insertions are underlined.
Values are means ± standard deviations. Catalytic efficiency is calculated as kcat/Km.
Protein expression and purification.
All of the PRs were overexpressed in Escherichia coli BL21(DE3)RIL (Novagen, Darmstadt, Germany), and insoluble recombinant protein, accumulated in the form of inclusion bodies, was isolated and solubilized in 67% (vol/vol) acetic acid. The recombinant PRs were refolded by dilution into a 25-fold excess of water and overnight dialysis against water at 4°C, followed by subsequent overnight dialysis against 50 mM morpholineethanesulfonic acid, pH 5.8, 10% (vol/vol) glycerol, 1 mM EDTA, and 0.05% (vol/vol) 2-mercaptoethanol. The PRs were purified by cation exchange chromatography using MonoS fast protein liquid chromatography (Amersham Bioscience, Uppsala, Sweden). The purified enzymes were stored at −70°C (34).
Activity and inhibition assay.
The inhibition constants (Ki values) were determined by spectrophotometric assay using the chromogenic peptide substrate KARVNle*NphEANle-NH2 (where Nle is norleucine and Nph is p-nitrophenylalanine and the asterisk signifies the PR cleavage site) as previously described (39). Typically, 8 pmol of PR was added to 1 ml of 0.1 M sodium acetate buffer, pH 4.7, 0.3 M NaCl, and 4 mM EDTA, containing substrate at a concentration near the Km of the enzyme and various concentrations of inhibitor dissolved in dimethyl sulfoxide (DMSO). The final concentrations of DMSO were kept below 2.5% (vol/vol). Substrate hydrolysis was followed as a decrease in absorbance at 305 nm using a Unicam UV500 UV-visible light spectrophotometer (ThermoSpectronic, Cambridge, United Kingdom). The data were analyzed using the equation for competitive inhibition according to Williams and Morrison (40).
Crystallization and data collection.
The enzyme-inhibitor complexes for crystallization were prepared by mixing the enzyme with a fivefold molar excess of lopinavir (in DMSO) and concentrated by ultrafiltration to a final protein concentration of 8 mg/ml. The crystals were grown using a hanging-drop vapor diffusion technique at 19°C. The crystallization drops contained 2 μl of protein-inhibitor complex solution and 1 μl of the reservoir solution. The optimized reservoir solutions for the PR1-lopinavir and PR2-lopinavir complexes contained 0.5 M ammonium sulfate and 0.1 M morpholineethanesulfonic acid, pH 5.4 or pH 5.5, respectively. Cross-seeding and microseeding techniques were used for crystal optimization (35). For diffraction measurement, the crystals were soaked in the reservoir buffer solution supplemented with 30% (vol/vol) glycerol and cryo-cooled in liquid nitrogen. Diffraction data were collected at 100 K at beam line 19-ID of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, at a wavelength of 0.97Å and were processed using the program HKL-3000 (25). Crystal parameters and data collection statistics are summarized in Table 2.
TABLE 2.
X-ray data collection and model refinement statistics
| Parameter | Value for the PRa
|
|
|---|---|---|
| PR1 (PDB 2RKG) | PR2 (PDB 2RKF) | |
| Data collection statistics | ||
| Wavelength (Å) | 0.97 | 0.97 |
| Temperature (K) | 100 | 100 |
| Space group | P61 | P61 |
| a = b, c (Å) | 62.0, 83.9 | 61.9, 84.1 |
| Resolution (Å) | 1.80 (1.83-1.80) | 1.80 (1.86-1.80) |
| Redundancy | 5.5 (4.1) | 6.0 (4.3) |
| Completeness (%) | 98.7 (94.1) | 98.6 (88.9) |
| Rmergeb | 0.04 (0.36) | 0.04 (0.32) |
| Average I/σ(I) | 34.8 (2.8) | 37.5 (2.4) |
| Wilson B (Å2) | 30.5 | 26.6 |
| Refinement statistics | ||
| Resolution range (Å) | 29.0-1.8 (1.85-1.80) | 25.0-1.8 (1.85-1.80) |
| R value (%)c | 19.14 (24.0) | 18.94 (22.7) |
| Rfree value (%)d | 24.84 (28.5) | 24.79 (25.1) |
| Average B factor (Å2) | 35.8 | 32.9 |
| B factor for LPV (Å2) | 28.1/28.1 | 29.3/25.3 |
| RMSD bond length (Å) | 0.011 | 0.012 |
| RMSD bond angle (°) | 1.39 | 1.45 |
| Ramachandran plot | ||
| Most favored regions (%) | 95.7 | 92.5 |
| Allowed regions (%) | 4.3 | 7.5 |
Statistics for the highest resolution shell are in parentheses.
Rmerge = (Ihkl − 〈I〉)/Ihkl, where the average intensity 〈I〉 is taken over all symmetry-equivalent measurements and Ihkl is the measured intensity for any given reflection.
R value = ‖Fo| − |Fc‖/|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree is equivalent to the R value but is calculated for 5% of the reflections chosen at random and omitted from the refinement process.
Structure refinement and analysis.
Since the hexagonal crystals we obtained appeared to be isomorphous with all other P61 crystals of HIV-1 PR complexes, the structure determination was performed by the difference-Fourier method using PR from Protein Data Bank (PDB) structure 1U8G (7) as the initial model. Initial rigid-body refinement and subsequent restrained refinement were performed with the program REFMAC, version 5.1.24 (27), from the CCP4 package (1). The program Coot (13) was used for manual model rebuilding and inhibitor building. Final TLS refinement (using the tensors translation [T], libration [L], and correlation of translation and libration [S]) (41) was done with TLS groups corresponding to HIV PR subdomains (33). The final refinement statistics are summarized in Table 2.
Protein structure accession numbers.
Atomic coordinates and structure factors have been deposited in the PDB under accession codes 2RKG and 2RKF for PR1 and PR2, respectively.
RESULTS
Prevalence of amino acid insertions in the PR sequence.
To analyze the prevalence of PR insertions in the clinical population, we surveyed a mutation database constructed from more than 208,000 clinical HIV-1 sequences submitted for genotypic analysis between 1999 and May 2007. The majority of PR insertions (313 out of 334, or 93.7%) occurred between residues 32 and 41, which include a portion of the substrate cleft and the loop region known as the PR flap elbow (residues 37 to 42). The overall prevalence of substrate cleft insertions was 0.15% of all sequences and 0.27% of sequences with predicted resistance to any PI or RT inhibitor. The prevalence of substrate cleft insertions significantly increased from 0.11% in 1999 to 2001 to 0.31% in the 3-year period from 2002 to 2004 (Fisher exact test, P < 0.0001; odds ratio, 2.8; 95% confidence interval, 1.9 to 4.1). The prevalence of the insertions remained at this higher level (0.34%) during the following 3-year period, 2005 to 2007. Eighty-one out of 313 sequences with PR insertions contained single amino acid insertions that appeared to be placed after codon 33. An additional 137 out of 314 insertions of 1 to 5 amino acids were placed after codon 35. To assess whether other PR mutations were associated with the insertions between amino acids 32 and 41 of the PR sequence, we compared the prevalence of PR mutations in 278 sequences with insertions identified between January 2002 and April 2007 to 34,777 PR sequences from the same period with predicted resistance to one or more PIs but with no insertions in the primary sequence. We found eight positions (11, 13, 16, 36, 47, 57, 89, and 91) that were mutated more frequently in the insertion sequences and two positions (30 and 88) that were mutated less frequently (see Table S2 in the supplemental material).
Enzymatic analysis of recombinant PRs with and without insertions.
We identified two patient-derived viral isolates representing the commonly observed amino acid insertions at positions 33 and 35 of HIV PR and set out to characterize the contribution of these insertions on the molecular and virological levels. The PR insertion variants identified in the HIV-positive patients were cloned, expressed in E. coli, purified, and characterized with respect to PI resistance. Since the insertions in the PRs studied occur on the background of many other resistance-conferring mutations (12 mutations in PR1 and 15 mutations in PR3), we decided to dissect the relative contribution of the insertions in positions 33 and 35. Therefore, we prepared the corresponding mutants lacking the insertions at positions 33 and 35 by site-directed mutagenesis (Table 1, PRs 2 and 4).
We investigated the impact of an insertion at position 33 or 35 on the catalytic activity of the viral PR with purified recombinant proteins using an in vitro activity assay. We analyzed the ability of PR variants with and without insertions to hydrolyze a chromogenic substrate in order to determine the impact of amino acid insertions on kcat and Km values. The data are summarized in Table 1; patient-derived PR1 (E35EE insertion) and PR3 (L33LL) show slightly increased Km values and more significantly decreased kcat values relative to the wild-type PR. However, if we compare the isolated influence of the PR insertion on the enzyme kinetics (i.e., if we directly compare PR 1 to 2 and PR 3 to 4), we conclude that the insertion itself leads to the increase in the Km values without affecting the substrate turnover. The specific activities (kcat/Km) of insertion mutants (PR1 and PR3) are approximately 7 or 5 times lower, respectively, than that of the wild-type HIV PR, and 2.6 or 1.8 times lower, respectively, than the control mutant proteases PR2 and PR4, which lack the insertions (Table 1).
The impact of PR insertions on viral replication capacity.
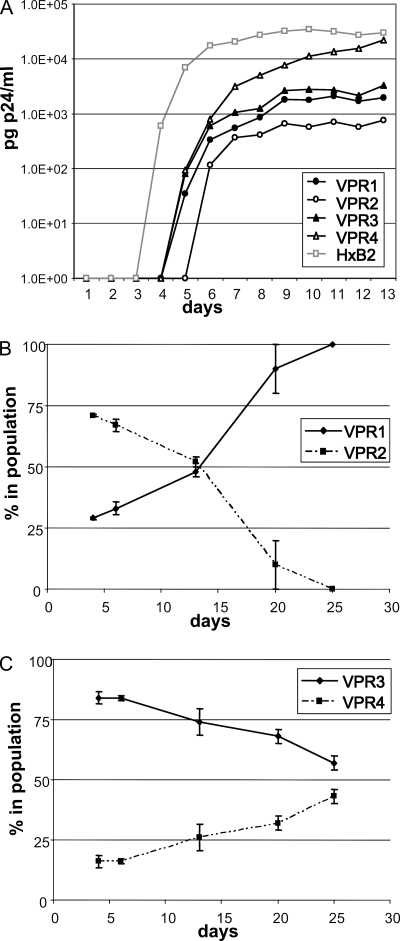
The impact of PR insertions on the replication capacity of the virus was investigated by performing viral replication and competition experiments in SupT1 cells (Fig. 2). To do so, the patient-derived viral PR and the C-terminal part of Gag (including the nucleocapsid/p1 cleavage site changes A431V in VPR1 and VPR2 and the A437V substitution in VPR3 and VPR4 (see Fig. S2 in the supplemental material) was cloned in an HIV-1 subtype B reference strain (HXB2). In line with the enzymatically determined catalytic efficiency of the PR variants, all viruses with PR mutants replicated less efficiently than wild-type HXB2 (Fig. 2A). A comparison of the replication capacity of the viral variants VPR3 and VPR4, with and without the L33LL insertion, respectively, showed in both the replication curves and the competition experiments that presence of the insertion (VPR3) results in a reduced viral replication capacity (Fig. 2C). This observation is in agreement with the reduced enzymatic catalytic efficiency of the PR mutant with the insertion (PR3) compared to PR4. Although the differences are much less pronounced in the replication curves, the competition experiments clearly show that VPR1 (with the E35EE insertion) replicates better than the variant without the insertion (VPR2) (Fig. 2B).
FIG. 2.
(A) Replication kinetics of the viral PR mutants. Recombinant viruses containing patient-derived viral PRs with multiple amino acid substitutions were prepared. VPR1 contains an E35EE insertion and VPR3 contains an L33LL insertion. VPR2 and VPR4 are derivatives of VPR1 and VPR3, respectively, lacking the corresponding insertions. Viral replication experiments were performed with these recombinant viruses in SupT1 cells. Virus replication was monitored by p24 production in the culture supernatant. (B and C) Replication competition experiments, comparing the replication capacity of VPR1 relative to VPR2 (B) and VPR3 relative to VPR4 (C). Replication competition assays were performed in SupT1 cells in two independent experiments. At several time points during viral culture, the relative presence of both viruses in the population was determined by sequence analysis.
Drug susceptibility analysis: relative Ki and vitality values for the recombinant PRs.
The recombinant PRs with or without insertions were assayed in vitro using a chromogenic peptide substrate and a panel of clinically available PIs (Table 3). Both insert-containing PR variants proved highly resistant to all PIs tested. Interestingly, comparison of the variants with and without insertions revealed that the insert-containing variants had higher Ki values than their counterparts without the insert. This was the case for all of the inhibitors tested, with the exception of PR1 (E35EE) and nelfinavir. Strikingly, a greater than 10-fold increase in relative Ki values was observed for saquinavir, lopinavir, and ritonavir.
TABLE 3.
Ki values for the inhibition of PR mutants by clinically available inhibitors
| HIV-1 PR type (recombinant virus)a |
Ki (nM)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SQV | RTV | IDV | NFV | LPV | APV | ATV | DRV | BCV | |
| Wild type | 0.04 ± 0.01 | 0.015 ± 0.003 | 0.12 ± 0.02 | 0.07 ± 0.01 | 0.018 ± 0.009 | 0.18 ± 0.02 | 0.024 ± 0.005 | 0.005 ± 0.003 | 0.0005 ± 0.002 |
| PR1 (VPR1) | 10.6 ± 0.9 (1.7) | 4.3 ± 0.3 (ND) | 87 ± 7 (23.9) | 13 ± 1 (52.3) | 8.2 ± 0.7 (2360) | 48.7 ± 2.9 (45.2) | 1.9 ± 0.2 (5) | 1.9 ± 0.3 (19.6) | 0.29 ± 0.04 (ND) |
| PR2 (VPR2) | 0.93 ± 0.16 (0.6) | 0.46 ± 0.03 (ND) | 30 ± 4.5 (9.7) | 13 ± 1.3 (11.8) | 0.83 ± 0.11 (688) | 10.4 ± 1.1 (26.2) | 0.43 ± 0.05 (2.3) | 0.68 ± 0.17 (2.1) | 0.11 ± 0.04 (ND) |
| PR3 (VPR3) | 84 ± 4 (20.8) | 67 ± 5 (ND) | 550 ± 30 (>55.5) | 62 ± 5 (>68) | 4.9 ± 0.4 (534) | 1.7 ± 0.05 (5.3) | 6.61 ± 0.56 (104) | 0.025 ± 0.011 (0.8) | 0.027 ± 0.015 (ND) |
| PR4 (VPR4) | 9.4 ± 1.4 (18) | 4.9 ± 0.4 (ND) | 102 ± 14 (>71.5) | 7.9 ± 0.6 (>85.3) | 0.47 ± 0.01 (72.3) | 0.5 ± 0.05 (5.1) | 1.6 ± 0.2 (82.4) | 0.027 ± 0.001 (1.1) | 0.012 ± 0.009 (ND) |
PR1 has the E35EE insertion, and PR3 carries the L33LL insertion.
Values are means ± standard deviations. The inhibition constants were determined by spectrophotometric assay at the pH optimum of HIV PR (pH 4.7). Numbers in parentheses show relative increases in IC50 values for the corresponding recombinant viruses VPR1 to VPR4 compared to the wild-type virus. These values were obtained by a multiple-cycle MTT recombinant virus assays. SQV, saquinavir; RTV, ritonavir; IDV, indinavir; NFV, nelfinavir; LPV, lopinavir; APV, amprenavir; ATV, atazanavir; DRV, darunavir; BCV, brecanavir; ND, not done.
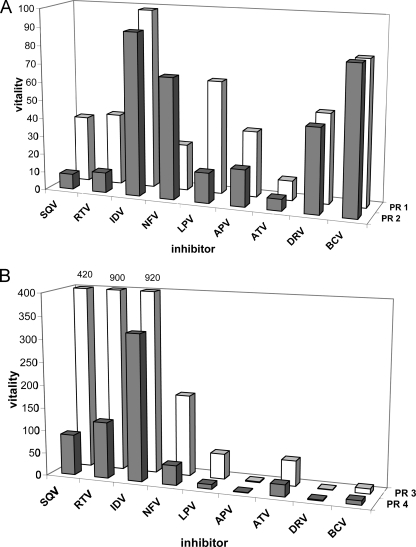
In order to correlate relative Ki values with changes in catalytic efficiency caused by PR mutations, Gulnik et al. (14) introduced vitality as a measure of the relative capability of the mutated enzyme to cleave its substrate in the presence of an inhibitor. The vitalities of individual mutants with nine clinically available inhibitors are shown in Fig. 3. A direct comparison of PR1 (E35EE) and PR2 (without the mutation) reveals that the E35EE mutation increases the vitality of the PR in the presence of saquinavir, ritonavir, and lopinavir by an order of magnitude, does not significantly change the vitalities in the presence of other PIs, and actually decreases the vitality of the PR mutant in the presence of nelfinavir (Fig. 3A). Along similar lines, the L33LL insertion significantly increases the vitality in the presence of saquinavir, ritonavir, lopinavir, and nelfinavir. Interestingly, the L33LL insertion has a different effect on nelfinavir resistance than the E35EE insertion. While the affinity of nelfinavir to the PR is almost unaffected by the E35EE mutation (and the overall vitality is, in fact, decreased due to the lower catalytic efficiency of the mutant), the L33LL insertion decreases the binding affinity of nelfinavir to the PR mutant by a factor of 10 (compare Table 3 and Fig. 3).
FIG. 3.
(A) Relative vitality values for recombinant PR with (PR1) and without (PR2) the E35EE insertion in the presence of various HIV PIs. (B) Relative vitality values for recombinant PR with (PR3) and without (PR4) the L33LL insertion in the presence of various HIV PIs. APV, amprenavir; ATV, atazanavir; BCV, brecanavir; DRV, darunavir; IDV, indinavir; NFV, nelfinavir; RTV, ritonavir; SQV, saquinavir; LPV, lopinavir.
Phenotypic protease susceptibility to PIs.
The impact of the inserts in the viral PR on the phenotypic PR susceptibility was analyzed by a recombinant virus assay in which the PI susceptibility of the viral protease and the C-terminal part of Gag was assayed in the background of a subtype B reference virus. In parallel, recombinant viruses harboring the mutated region without the inserts were prepared by site-directed mutagenesis as controls. In line with the enzymatic data, PR insertion variant VPR1 (corresponding to PR1) has clinically significant levels of resistance to all PIs except for saquinavir (Table 3). PR insertion variant VPR3 shows clinically significant levels of resistance to all PIs but darunavir. Comparison of the PR variants with and without the insertion (i.e., VPR1 and VPR2) confirmed that the E35EE insertion is associated with an increase in viral resistance, although the difference is smaller in comparison to the Ki data. For the other PR insertion L33LL (VPR3), the effect on PI susceptibility is less pronounced. While we see a significant increase in resistance to lopinavir in the presence of the insertions, the effect on other PIs is limited or cannot be investigated because both variants have a 50% inhibitory concentration (IC50) value above the cutoff of the assay.
Crystal structures of recombinant PRs with and without the E35EE mutation.
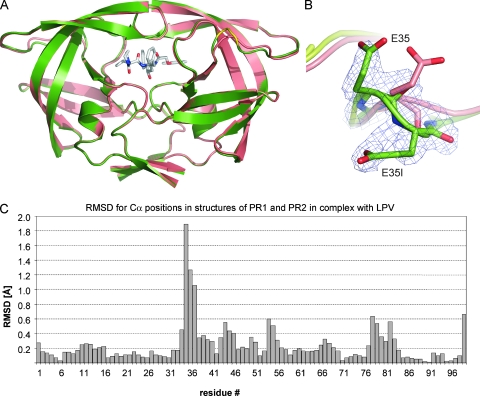
X-ray crystal structures of PR1 and PR2 in complex with lopinavir were determined using data to a resolution of 1.8 Å (Fig. 4A; see also Table S1 in the supplemental material). Both complexes crystallized in the same hexagonal form with one PR dimer in the asymmetric unit, and structures were refined with two inhibitor molecules bound in alternative orientations with 50% relative occupancy. Overall, the two crystal structures are very similar with a root mean square deviation (RMSD) of 0.39 Å for all main-chain atoms, a value within the range observed for different crystal structures of identical proteins (2). However, a detailed analysis of the RMSDs of the individual Cα positions after superimposition of the two structures revealed several local structural changes caused by the E35EE mutation (Fig. 4C). The insertion causes direct rearrangement primarily in the vicinity of position 35, as documented by the large RMSDs for the Cα atoms of residues 34 to 37. The quality of the electron density maps and higher atomic displacement parameters (B factors) also suggested increased disorder and mobility of the two glutamic acid residues in PR1 (Fig. 4B). Other structural differences can be found in the flap region (residues 44 to 46 and 54 to 55) and in the vicinity of residues 78, 79, and 82, which are part of the substrate binding subsites (Fig. 4C).
FIG. 4.
(A) Superposition of the crystal structures of PR1 and PR2 in complex with lopinavir. The protease is shown in ribbon representation: PR1 in green with insertion 35EE is shown in yellow, and PR2 is shown in salmon. The active-site-bound inhibitor is shown in sticks with carbon atoms shown in gray and nitrogen and oxygen atoms shown in blue and red, respectively. For the sake of clarity only one of the two alternative conformations is shown. (B) Detail of insertion region shown on superimposed structures of PR1 (green) and PR2 (salmon). The 2Fo − Fc (the observed and calculated structure factors, respectively) electron density map contoured at the 1.0 σ level is shown for the E35EE insertion in PR1. (C) Plot showing the RMSDs for positions of individual Cα atoms after superposition of the PR1 and PR2 structures.
The position and conformation of the inhibitor within the enzyme catalytic site remained the same in both structures (the RMSD for all lopinavir atoms is bellow 0.18 Å). Also, all polar interactions, direct and water-mediated hydrogen bonds, between lopinavir and PR1 or PR2 are preserved, and parameters describing PI binding (15, 20) are not significantly altered (see Table S1 in the supplemental material).
DISCUSSION
Insertions in various codons of PR have been described as rare events, and they always appear in combination with well-described PI resistance mutations. We show that PR insertions, particularly those between residues 32 and 42, have become more prevalent since 1999. These insertions are positively correlated with PR codons associated with resistance to PIs whose usage has increased in recent years, including atazanavir (position 16) (38), lopinavir and amprenavir (position 47) (21, 17), and tipranavir (positions 13, 36, and 47) (11). Additionally, a mutation at position 57 is a predictor of early virologic failure (23), and codon 91 mutations are more common in patients on long-term nonsupressive PI therapy (16). Mutations at position 11 are also associated with PI treatment (44, 32).
While the insertions lead to a decrease in PI susceptibility and modestly improve viral fitness (18), they seem to contribute to PI resistance only in combination with other mutations either in the PR or in Gag. To our knowledge, however, no structural or enzymological analysis of HIV PR variants with insertions has been provided. We investigated the impact of PR insertions on PI susceptibility both in vitro and in vivo using recombinant enzymes and viruses. We aimed to elucidate the effect of insertions in the PR on viral replication capacity as well as on the activity, vitality, and three-dimensional structure of the mutated enzyme.
Analysis of the recombinant patient-derived resistant HIV PRs (with or without insertions in positions 35 and 33) shows a severalfold decrease in the relative enzymatic activities compared to the wild-type HIV-1 PR. The drop is mainly due to the decrease in kcat values for hydrolysis of the chromogenic peptide substrate. Control PRs lacking the insertions (PR2 and PR4) show a two- to threefold increase in relative activity compared to PR1 and PR3, respectively, mostly due to lower Km values, which suggest improved substrate binding. This observation is in line with the reduced viral replication kinetics of all mutant viruses, with or without insertions, compared to the wild-type reference virus. However, it should be noted that a direct comparison of the enzymatic data to the virological data is intrinsically complicated because of all the differences between the assays, including the presence of patient-derived Gag sequences in the recombinant virus. Gag cleavage site mutations have been shown to partially restore the PI resistance mutation-associated loss of viral fitness seen in PI-resistant viruses (21, 22). Even though the recombinant viruses containing E35EE (VPR1 and 2) and L33LL (VPR3 and 4) insertions are thus not directly comparable, the trends between the enzyme or the virus with and without insertion can be reliably investigated.
As indicated above, both PR insertions increased the resistance to most clinically available PIs. Investigation of the replication capacity revealed that all PR mutants, with and without insertions, had to pay a price for PR resistance and replicated less well than wild-type HXB2. Furthermore, the impact of the insertions on the relative replication capacity was investigated and revealed that selection of the L33LL insertion in the background of 15 known PR resistance mutations decreased the replication capacity of the virus. Analysis of the relative replication capacity of the other PR insertion (E35EE) demonstrated that this variant replicated better that the one without the insertion. This observation is in line with another position 35 insertion mutant described by Kim et al. (18). They demonstrated that the replication capacity of virus containing PR with an insertion at position 35 is higher than that of the corresponding insertion-lacking mutant, both in viral replication experiments and in direct competition experiments (18).
The presence of the E35EE insertion together with 12 known PR resistance mutations clearly increases the resistance to PIs, as shown by both vitality values obtained from recombinant PRs and by phenotypic susceptibility assays with recombinant viruses. It is important that the effect of insertions at positions 33 and 35 was studied in the context of a background of other mutations found in two particular viral samples. Further extensive experimental work would be necessary to completely describe the role of background mutations for HIV resistance by amino acid insertions in the PR region. Remarkably, the presence of the E35EE insertion in the background of other PR resistance-associated mutations was associated with an almost 10-fold increase in the virological resistance to the novel high genetic barrier PI darunavir. A severalfold increase in virological resistance was also observed for saquinavir, lopinavir, indinavir, amprenavir, and atazanavir. Vitality values for the corresponding recombinant PRs with and without the mutations reveal a similar pattern. As far as we know, there are very few studies in which the impact of an insertion in the viral PR on the virological susceptibility has been investigated (18, 6, 30). These studies showed that, in general, no large differences in susceptibility to PIs as a result of insertions could be identified. However, the presence of a 5-amino-acid insertion at position 35 in combination with two known PR resistance mutations did confer a five- to sixfold increase in resistance to indinavir, which agrees with the observation that the insertion variant replicated at a higher rate than the variant without insertion in the presence of low levels of indinavir (18).
It is interesting that the prevalence of the insertions started to increase after 2002. Lopinavir was approved for clinical use by the Food and Drug Administration in September 2000 and has been used extensively ever since. It could be speculated that lopinavir selects for these insertions.
With respect to the other PR insertion studied here, L33LL, the effect on PI susceptibility was much less clear. In in vitro assays using recombinant enzymes, an increase in Ki for all PIs tested except darunavir was observed, whereas the virological assay indicates an increase in resistance to lopinavir only. The virological resistance to the other PIs is limited or cannot be investigated because both variants have IC50 values above the cutoff of the assay. In general, the differences in resistance are more pronounced on an enzymatic level than in the virus, which corresponds to our earlier finding when replicative capacity of a series of drug-resistant HIV mutants was correlated with the kinetic parameters of corresponding recombinant mutated PRs (29).
In order to explain the phenotypic changes brought about by PR insertions in structural terms, we solved the structures of PRs with and without the E35EE insertion (PR1 and PR2, respectively) in complex with the PI lopinavir. The comparison of these structures shows that the E35EE insertion does not significantly change the overall inhibitor binding to the substrate binding sites (see Table S1 in the supplemental material). Detailed comparison of van der Waals distances between inhibitor and residues within individual substrate binding subsites revealed a slight enlargement of the P1 and P1′ pockets in the PR1 complex structure compared to the PR2-lopinavir structure. This enlargement is caused by a minor rearrangement of residues 78, 79, and 82, and it results specifically in the loss of van der Waals contacts between the S1 and S1′ benzyl rings of lopinavir and the side chain of Ala 82 (see Fig. S1 in the supplemental material). This subtle change in inhibitor binding could account for the weaker binding of lopinavir to PR1. However, it is more likely that additional factors are involved in the PI resistance caused by the E35EE insertion. The increased disorder of two glutamic acid residues observed in the PR1 structure, together with the slight structural differences of several flap resides (44 to 46 and 54 to 55) relative to the PR2 structure, might indicate that the insertion also influences the dynamics of flap movements during inhibitor binding. Impaired dynamics of flap movement could explain the overall trend of weaker inhibitor binding to insertion mutants.
In conclusion, we confirmed by enzymological characterization using recombinant PRs and by phenotypic analysis with recombinant viruses that the amino acid insertions in positions 33 and 35 contribute to viral resistance to many PIs currently in clinical use. X-ray analysis of the complexes of PRs with lopinavir revealed minor secondary structural changes in the flap region and the P1 and P1′ subsites, leading to the enlargement of the PR binding site. This structural difference together with possibly impaired flap dynamics could account for the weaker binding of the inhibitor by insertion mutants. Amino acid insertions into the PR sequence in the vicinity of the binding cleft therefore represent a novel mechanism of HIV resistance development.
Supplementary Material
Acknowledgments
This work was supported by grant NR-8571-3 from the Ministry of Health Care of the Czech Republic, a grant from the Ministry of Education (MSMT) of the Czech Republic within Programme 1M0508, Research Centre for New Antivirals and Antineoplastics, and by Research Plans AVZ40550506 and AVO250520514 from the Academy of Sciences of the Czech Republic.
We thank Virco (Mechelen, Belgium) for phenotyping analysis and the APS synchrotron facility in Argonne, IL, for access to their beam line; we also thank Hillary Hoffman for critical proofreading of the manuscript and many valuable comments.
Footnotes
Published ahead of print on 9 April 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bailey, S. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 2.Betts, M. J., and M. J. Sternberg. 1999. An analysis of conformational changes on protein-protein association: implications for predictive docking. Protein Eng. 12271-283. [DOI] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A. M., S. Paulos, and F. Clavel. 1996. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J. Gen. Virol. 77419-426. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, C. A., W. Keulen, T. van Bommel, M. Nijhuis, D. de Jong, M. D. de Jong, P. Schipper, and N. K. Back. 1996. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob. Agents Chemother. 402404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brann, T. W., R. L. Dewar, M. K. Jiang, A. Shah, K. Nagashima, J. A. Metcalf, J. Falloon, H. C. Lane, and T. Imamichi. 2006. Functional correlation between a novel amino acid insertion at codon 19 in the protease of human immunodeficiency virus type 1 and polymorphism in the p1/p6 Gag cleavage site in drug resistance and replication fitness. J. Virol. 806136-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brynda, J., P. Rezacova, M. Fabry, M. Horejsi, R. Stouracova, M. Soucek, M. Hradilek, J. Konvalinka, and J. Sedlacek. 2004. Inhibitor binding at the protein interface in crystals of a HIV-1 protease complex. Acta Crystallogr. D 601943-1948. [DOI] [PubMed] [Google Scholar]
- 8.Cigler, P., M. Kozisek, P. Rezacova, J. Brynda, Z. Otwinowski, J. Pokorna, J. Plesek, B. Grüner, L. Doleckova-Maresova, M. Masa, J. Sedlacek, J. Bodem, H. G. Kräusslich, V. Kral, and J. Konvalinka. 2005. From nonpeptide toward noncarbon protease inhibitors: metallacarboranes as specific and potent inhibitors of HIV protease. Proc. Natl. Acad. Sci. USA 10215394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Tepplert, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374569-571. [DOI] [PubMed] [Google Scholar]
- 10.De Clercq, E. 2007. Anti-HIV drugs. Verh. K. Acad. Geneeskd. Belg. 6981-104. [PubMed] [Google Scholar]
- 11.de Mendoza, C., J. Morello, P. Garcia-Gasco, S. Rodriguez-Novoa, and V. Soriano. 2007. Tipranavir: a new protease inhibitor for the treatment of antiretroviral-experienced HIV-infected patients. Expert Opin. Pharmacother. 8839-850. [DOI] [PubMed] [Google Scholar]
- 12.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 703763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 14.Gulnik, S. V., L. I. Suyorov, B. Lju, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 349282-9287. [DOI] [PubMed] [Google Scholar]
- 15.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 9313-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kagan, R. M., P. K. Cheung, T. K. Huard, and M. A. Lewinski. 2006. Increasing prevalence of HIV-1 protease inhibitor-associated mutations correlates with long-term non-suppressive protease inhibitor treatment. Antivir. Res. 7142-52. [DOI] [PubMed] [Google Scholar]
- 17.Kagan, R. M., M. D. Shenderovich, P. N. Heseltine, and K. Ramnarayan. 2005. Structural analysis of an HIV-1 protease I47A mutant resistant to the protease inhibitor lopinavir. Protein Sci. 141870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, E. Y., M. A. Winters, R. M. Kagan, and T. C. Merigan. 2001. Functional correlates of insertion mutations in the protease gene of human immunodeficiency virus type 1 isolates from patients. J. Virol. 7511227-11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 854686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowski, R. A. 1995. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph 13323-328. [DOI] [PubMed] [Google Scholar]
- 21.Maguire, M., D. Shortino, A. Klein, W. Harris, V. Manohitharajah, M. Tisdale, R. Elston, J. Yeo, S. Randall, F. Xu, H. Parker, J. May, and W. Snowden. 2002. Emergence of resistance to protease inhibitor amprenavir in human immunodeficiency virus type 1-infected patients: selection of four alternative viral protease genotypes and influence of viral susceptibility to coadministered reverse transcriptase nucleoside inhibitors. Antimicrob. Agents Chemother. 46731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 727632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masquelier, B., C. Droz, M. Dary, C. Perronne, V. Ferré, B. Spire, D. Descamps, F. Raffi, F. Brun-Vézinet, G. Chêne, and the APROCO/COPILOTE Study Group. 2003. R57K polymorphism in the human immunodeficiency virus type 1 protease as predictor of early virologic failure in a cohort of antiretroviral-naive patients treated mostly with a nelfinavir-containing regimen. Antimicrob. Agents Chemother. 473623-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masquelier, B., E. Race, C. Tamalet, D. Descamps, J. Izopet, C. Buffet-Janvresse, A. Ruffault, A. S. Mohammed, J. Cottalorda, A. Schmuck, V. Calvez, E. Dam, H. Fleury, and F. Brun-Vezinet. 2001. Genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 variants with insertions or deletions in the reverse transcriptase (RT): multicenter study of patients treated with RT inhibitors. Antimicrob. Agents Chemother. 451836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minor, W., M. Cymborowski, Z. Otwinowski, and M. Chruszcz. 2006. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D 62859-866. [DOI] [PubMed] [Google Scholar]
- 26.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 2501139-1142. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 28.Nijhuis, M., N. M. van Maarseveen, S. Lastere, P. Schipper, E. Coakley, B. Glass, M. Rovenska, D. de Jong, C. Chappey, I. W. Goedegebuure, G. Heilek-Snyder, D. Dulude, N. Cammack, L. Brakier-Gingra, J. Konvalinka, N. Parkin, H.-G. Kräusslich, F. Brun-Vezinet, and C. A. Boucher. 2007. A novel substrate based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 4152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 132349-2359. [DOI] [PubMed] [Google Scholar]
- 30.Paolucci, S., F. Baldanti, L. Dossena, and G. Gerna. 2006. Amino acid insertions at position 35 of HIV-1 protease interfere with virus replication without modifying antiviral drug susceptibility. Antivir. Res. 69181-185. [DOI] [PubMed] [Google Scholar]
- 31.Prejdova, J., M. Soucek, and J. Konvalinka. 2004. Determining and overcoming resistance to HIV protease inhibitors. Curr. Drug Targets Infect. Disord. 4137-152. [DOI] [PubMed] [Google Scholar]
- 32.Rhee, S. Y., W. J. Fessel, A. R. Zolopa, L. Hurley, T. Liu, J. Taylor, D. P. Nguyen, S. Slome, D. Klein, M. Horberg, J. Flamm, S. Follansbee, J. M. Schapiro, and R. W. Shafer. 2005. HIV-1 Protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug-resistance surveillance. J. Infect. Dis. 192456-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose, R. B., C. S. Craik, and R. M. Stroud. 1998. Domain flexibility in retroviral proteases: structural implications for drug resistant mutations. Biochemistry 372607-2621. [DOI] [PubMed] [Google Scholar]
- 34.Strisovsky, K., U. Tessmer, J. Langner, J. Konvalinka, and H. G. Kräusslich. 2000. Systematic mutational analysis of the active-site threonine of HIV-1 proteinase: re-thinking the “fireman's grip” hypothesis. Protein Sci. 91631-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stura, E. A., and I. A. Wilson. 1991. Applications of the streak seeding technique in protein crystallization. J. Cryst. Growth 110270-282. [Google Scholar]
- 36.van Maarseveen, N. M., D. de Jong, C. A. Boucher, and M. Nijhuis. 2006. An increase in viral replicative capacity drives the evolution of protease inhibitor-resistant human immunodeficiency virus type 1 in the absence of drugs. J. Acquir. Immune Defic. Syndr. 42162-168. [DOI] [PubMed] [Google Scholar]
- 37.van Maarseveen, N. M., M. C. Huigen, D. de Jong, A. M. Smits, C. A. Boucher, and M. Nijhuis. 2006. A novel real-time PCR assay to determine relative replication capacity for HIV-1 protease variants and/or reverse transcriptase variants. J. Virol. Methods 133185-194. [DOI] [PubMed] [Google Scholar]
- 38.Vora, S., A. G. Marcelin, H. F. Gunthard, P. Flandre, H. H. Hirsch, B. Masquelier, A. Zinkernagel, G. Peytavin, V. Calvez, L. Perrin, and S. Yerly. 2006. Clinical validation of atazanavir/ritonavir genotypic resistance score in protease inhibitor-experienced patients. AIDS 2035-40. [DOI] [PubMed] [Google Scholar]
- 39.Weber, J., J. R. Mesters, M. Lepsik, J. Prejdova, M. Svec, J. Sponarova, P. Mlcochova, K. Skalicka, K. Strisovsky, T. Uhlikova, M. Soucek, L. Machala, M. Stankova, J. Vondrasek, T. Klimkait, H.-G. Kräusslich, R. Hilgenfeld, and J. Konvalinka. 2002. Unusual binding mode of an HIV-1 protease inhibitor explains its potency against multi-drug-resistant virus strains. J. Mol. Biol. 324739-754. [DOI] [PubMed] [Google Scholar]
- 40.Williams, J. W., and J. F. Morrison. 1979. The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63437-467. [DOI] [PubMed] [Google Scholar]
- 41.Winn, M. D., G. N. Murshudov, and M. Z. Papiz. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374300-321. [DOI] [PubMed] [Google Scholar]
- 42.Winters, M. A., R. M. Kagan, L. Kovari, P. N. Heseltine, and T. C. Merigan. 2005. Rare one and two amino acid inserts adjacent to codon 103 of the HIV-1 reverse transcriptase (RT) affect susceptibility to non-nucleoside RT inhibitors. Antivir. Ther. 10363-366. [PubMed] [Google Scholar]
- 43.Winters, M. A., and T. C. Merigan. 2005. Insertions in the human immunodeficiency virus type 1 protease and reverse transcriptase genes: clinical impact and molecular mechanisms. Antimicrob. Agents Chemother. 492575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, T. D., C. A. Schiffer, M. J. Gonzales, J. Taylor, R. Kantor, S. Chou, D. Israelski, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 774836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin, P. D., D. Das, and H. Mitsuya. 2006. Overcoming HIV drug resistance through rational drug design based on molecular, biochemical, and structural profiles of HIV resistance. Cell Mol. Life Sci. 631706-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 716662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.