Abstract
Epstein-Barr virus (EBV) replicates its genome as a licensed plasmid in latently infected cells. Although replication of this plasmid is essential for EBV latent infection, its synthesis still fails for 16% of the templates in S phase. In order to understand these failures, we sought to determine whether the affinity of the initiator protein (EBNA1) for its binding sites in the origin affects the efficiency of plasmid replication. We have answered this question by using several engineered origins modeled upon the arrangement of EBNA1-binding sites found in DS, the major plasmid origin of EBV. The human TRF2 protein also binds to half-sites in DS and increases EBNA1's affinity for its own sites; we therefore also tested origin efficiency in the presence or absence of these sites. We have found that if TRF2-half-binding sites are present, the efficiency of supporting the initiation of DNA synthesis and of establishing a plasmid bearing that origin directly correlates with the affinity of EBNA1 for that origin. Moreover, the presence of TRF2-half-binding sites also increases the average level of EBNA1 and ORC2 bound to those origins in vivo, as measured by chromatin immunoprecipitation. Lastly, we have created an origin of DNA synthesis from high-affinity EBNA1-binding sites and TRF2-half-binding sites that functions severalfold more efficiently than does DS. This finding indicates that EBV has selected a submaximally efficient origin of DNA synthesis for the latent phase of its life cycle. This enhanced origin could be used practically in human gene vectors to improve their efficiency in therapy and basic research.
For Epstein-Barr virus (EBV) the origin of DNA synthesis within oriP is the dyad symmetry (DS) element, which has been genetically dissected to identify several of its key features. DS is composed of two pairs of binding sites for its only required viral protein EBNA1, three 9-bp elements that resemble the telomeric repeats of the ends of human chromosomes and are half-binding sites for TTAGGG-repeat binding factor 2 (TRF2), a dyad element from which the name was derived, and a region upstream of DS that was found to be helically unstable (4, 6, 9, 19, 21, 23, 29). Of these elements, a pair of appropriately spaced EBNA1-binding sites was found to be the minimal cis-acting element required for the replicative function of DS (6).
The DS element recruits multiple licensing factors for DNA replication (e.g., ORC1 to -6, MCM2 to -7, Cdt1, and geminin), which appear to function indistinguishably from the roles assigned to them with human chromosomes (7, 10, 13, 24, 25). The DS element recruits these proteins either directly by serving as a substrate for sequence-specific binding as in the case of TRF2 or indirectly through proteins that bind it site specifically, also binding additional proteins. TRF2, for example, can bind ORC1 (3). EBNA1 has an N-terminal domain that is required for the synthesis of oriP and mimics the human HMGA1a protein in both its overall amino acid composition and its AT-hook sequence motifs (11, 26). This molecular mimicry has been demonstrated by the finding that HMGA1a fused to the DNA-binding and dimerization domain of EBNA1 restores the ability to support DNA synthesis to this derivative of EBNA1 (1, 11, 26, 27). These experiments show that the N-terminal domains of EBNA1 and HMGA1a both confer origin function to DS when bound by it site specifically.
EBV has a second plasmid origin of DNA synthesis, Rep*, which has been experimentally compared to DS (28). Rep* contains two EBNA1-binding sites with 21-bp center-to-center spacing, which, as with DS, is critical for pre-RC recruitment and replicative function (28). However, Rep* does not contain a dyad, TRF2-half-binding sites, or a helically unstable region upstream of it, indicating that these elements are not essential for the initiation of DNA synthesis and most likely play auxiliary roles in these processes (28).
The surprising finding that DS supports the initiation of DNA synthesis in only 84% of S phases led us to identify the features of DS that contribute to its efficiency in supporting the initiation of DNA synthesis (18). We engineered several candidates for EBNA1-dependent origins of DNA synthesis, modeled upon the evolutionarily selected arrangement of the cis elements of DS. Like DS, these engineered candidates contain four natural EBNA1-binding sites but have a wide range of different affinities for EBNA1 (Fig. 1) (2, 12, 28). We also constructed derivatives of the engineered candidate origins that include TRF2-half-binding sites in the arrangement present in DS because the interaction between TRF2 and EBNA1 increases the apparent affinity of EBNA1 for its binding site (Fig. 1). The sequences of each origin tested are listed in Fig. S1 in the supplemental material.
FIG. 1.
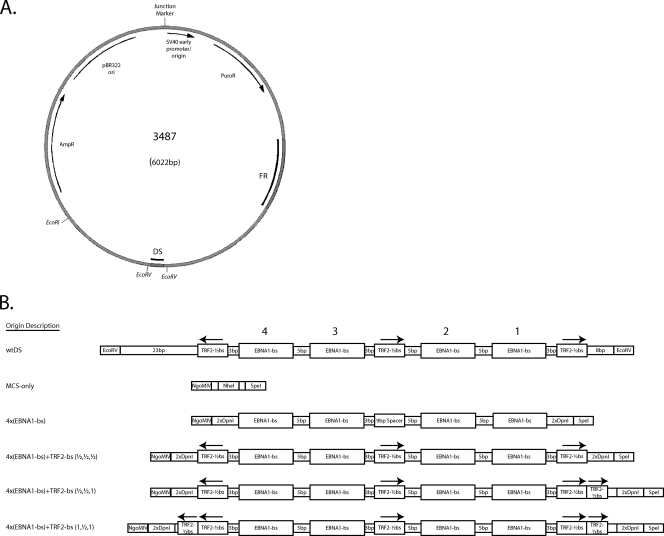
oriP plasmid backbone used to introduce the MCS and engineered origins of DNA synthesis tested in the present study. (A) The plasmid p3487 is derived from the pPUR vector (Clontech), which contains a pBR322 origin for plasmid replication in Escherichia coli and resistance to both ampicillin and puromycin. The wild-type oriP sequence has been inserted into a unique BamHI restriction site, which was destroyed near FR but was retained near DS. The MCS of plasmid p3488 was inserted between the EcoRV sites flanking DS. Subsequent insertions of the engineered origins, or the reintroduction of wtDS, were made into unique NgoMIV and SpeI sites present within this MCS. (B) Graphical representation of each class of origin tested. Within each class, only the sequences corresponding to the EBNA1-binding sites differ. Plasmids p3487 and p3567, which includes 1.2 kb of lambda phage DNA inserted in the EcoRI site of p3487, contain the wtDS element between the EcoRV sites as found in the B95-8 strain of EBV (4) and serve as positive-control origins. Plasmid p3488 has a 22-bp MCS inserted between the EcoRV sites of p3487 to remove and replace wtDS (MCS-only) and serves as an origin-less negative control. Test plasmids contain four EBNA1-binding sites of a range of different affinities in the same arrangement observed in wtDS, in the absence or presence of TRF2-half-binding sites. Two origins (wtDS and 4×FRbs) were further modified to include one or two complete TRF2-binding sites as well. The arrows above the TRF2-binding sites denote the N-to-C-terminal orientation that TRF2 would use to bind to these sites, as are found in wtDS. All origins were inserted between the NgoMIV and SpeI sites of p3488 to ensure that the only differences between the test plasmids were the origin sequences. The sequences of all origins tested are given in Fig. S1 in the supplemental material.
Using these engineered candidate origins, we found a direct correlation between the affinity of EBNA1's binding to an origin if the TRF2-half-binding sites are present and (i) the efficiency of initiating DNA synthesis and (ii) the efficiency of supporting establishment with that candidate origin. In addition, the presence of half-binding sites for TRF2 flanking the pairs of EBNA1-binding sites enhanced the synthesis of these candidate origins to a greater extent than could be expected from previous analyses (3, 8, 9). We also constructed origins of DNA synthesis that are severalfold more efficient than wild-type DS (wtDS) in their abilities to initiate DNA synthesis and to promote their extrachromosomal establishment. These findings indicate that the efficiency of plasmid establishment in part reflects the efficiency of the initiation of DNA synthesis. We propose a model incorporating these findings in which the affinity of EBNA1 for its origin binding sites differentially affects the structure of the bound DNA and/or EBNA1 to promote the recruitment of TRF2 and the cellular replicative machinery.
MATERIALS AND METHODS
Plasmid construction.
All origins tested in the present study are depicted graphically in Fig. 1. Complete representations of these origins, including their sequences, are given in Fig. S1 in the supplemental material. A positive-control oriP plasmid (p3487) was digested with EcoRV to remove the DS element, and an artificial multiple cloning site (MCS) was inserted in its place to create a plasmid lacking origin activity (p3488, “MCS-only” plasmid). All origins were designed to position four naturally occurring EBNA1-binding sites of differing affinities [e.g., high affinity, 4×FRbs; medium affinity, 2×(DS1+2) and 4×SiteIIIbs; and low affinity, 4×Rep*bs] in an arrangement identical to that found in the DS element. This arrangement was chosen to allow comparisons with the natural element that EBV has evolved to use. The interaction between TRF2 and EBNA1 increases the apparent affinity of EBNA1 for its binding site when a TRF2-half-binding site is positioned 3 bp away from it (9). Therefore, we also constructed derivatives of the engineered origins that would include three 9-bp TRF2-half-binding sites in the arrangement present in DS to determine whether any further effect on DNA synthesis could be attributed to the interaction of TRF2 with ORC1 and/or EBNA1. Lastly, because DS contains two EBNA1-binding sites of higher affinity that are paired with two EBNA1-binding sites of lower affinity, we created an origin from EBNA1-binding sites derived from the high-affinity family of repeats (FR) binding sites and the medium-affinity binding sites from site III (23). This produced an origin with EBNA1-binding sites in the same “strong-weak-weak-strong” (S-W-W-S) arrangement that is observed in DS, but with higher affinities than those found in wtDS. All engineered origins of DNA synthesis were inserted between the NgoMIV and SpeI sites of the MCS (p3508 to p3516, p3591, p3592, p3603, and p3604). This strategy also preserved the sequences present in EBV between FR and DS. Moreover, the plasmids produced differ only in the sequence of the origin of DNA synthesis, thus avoiding the potential effects that any other sequence in cis might have on replicative function. In addition, wtDS was reintroduced into the MCS to control for effects caused by the residual bases introduced by the MCS (p3499). Finally, an additional positive-control plasmid (p3567) was created by inserting 1.2 kb of lambda phage DNA known not to support initiation of DNA synthesis into a unique EcoRI site present in the vector backbone of the parental oriP plasmid.
Cell culture.
Raji, an EBV-positive cell line, and 293/EBNA1, a human embryonic kidney cell line that stably expresses EBNA-1 and neomycin phosphotransferase (ATCC CRL 10852), have been previously described (16, 17, 22). Raji cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 200 U of penicillin/ml, and 200 μg of streptomycin sulfate (Gibco-BRL, Rockville, MD)/ml (R10F) at 37°C in a 5% CO2 humidified atmosphere. 293/EBNA1 cells were similarly maintained, but in Dulbecco modified Eagle medium (DMEM) with high glucose and supplemented with 10% FBS, 200 U of penicillin/ml, 200 μg of streptomycin sulfate/ml, and 220 μg of G418 sulfate/ml (D10F). Clones of Raji cells stably transfected with the test plasmids were cultured under selection with 1 μg of puromycin/ml.
Transfection.
Electroporation of 5 × 106 Raji cells was performed in 500 μl of R10F with a custom-built electroporator set at 1,100 V, three capacitor banks with 1,540 μF of capacitance, the R-adjust set at 6 o'clock, and a rise time set at 10 o'clock as described previously (14, 28). Transfection efficiencies were determined by introducing 2 μg of an enhanced green fluorescent protein (eGFP) expression plasmid per 107 cells along with the test plasmids. The percentage of eGFP-positive cells was determined by counting green cells using a UV microscope.
Transfection of 293/EBNA1 cells was performed by plating 3 × 105 293/EBNA1 cells/well in a six-well plate 1 day prior to transfection. For each well, 2 μg of each plasmid were mixed in 100 μl of DMEM without FBS and, in parallel, 5 μl of Polyfect was also mixed in 100 μl of medium without FBS. Both solutions were combined and incubated at room temperature for 15 min. Each plasmid was transfected in duplicate. The cell growth medium was replaced with 800 μl of DMEM without FBS, and 200 μl of the DNA-Polyfect mixture was added to each well for 4 to 5 h in a humidified incubator at 37°C. This transfection medium was then aspirated and replaced with 2 ml of D10F per well. The next day, two identically transfected wells were transferred onto a 15-cm plate and grown under selection for at least 2 weeks with D10F with 220 μg of G418 sulfate/ml.
Short-term replication assay and Southern blotting.
Extrachromosomal DNA was extracted and assayed by Southern blotting as previously described (28). Briefly, low-molecular-weight DNA was extracted from 107 Raji cells by alkaline lysis and subsequently treated with phenol-chloroform and precipitated with ethanol. The precipitated DNA was resuspended in 1× Tris-EDTA and further treated with RNase A and proteinase K to digest contaminating RNA and protein, respectively. DNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in distilled and/or deionized water. A total of 90% of the DNA was digested with a single-cut restriction endonuclease (e.g., MluI) to linearize the test and control plasmids and with DpnI to remove the unreplicated, bacterially methylated input DNA. The remaining 10% of the DNA was digested only with the linearizing restriction endonuclease. The DNA was subsequently concentrated by precipitation with ethanol and electrophoresed through a 0.8% agarose gel in 1× Tris-acetate-EDTA buffer containing 500 ng of ethidium bromide/ml at 30 V (2 V/cm) for 14 to 18 h. The separated DNA fragments were nicked and denatured in situ, neutralized, and then transferred by capillary action to GeneScreen Plus membrane (NEN Life Sciences). A total of 25 ng of the parent vector backbone DNA was labeled by using a RediPrime II kit (Amersham Biosciences) and purified by using QuickSpin columns (Roche). The blots were hybridized with the labeled probe in Ultrahyb (Ambion) at 42°C for 16 h or longer, extensively washed, and then exposed to a storage phosphor screen (Molecular Dynamics). Signals were visualized by a Storm 860 PhosphorImager, and band intensities were quantified by using ImageQuant 5.2 (Molecular Dynamics).
Long-term replication assay.
In order to measure the number of Raji cells in which transfected DNAs became established as plasmids, the percentage of GFP-positive cells was determined at 3 days posttransfection. These cell populations harboring the test plasmids were serially diluted and plated in 96-well plates to contain a known number of green, transfected cells per well in 200 μl of R10F plus 1 μg of puromycin/ml. The number of puromycin-resistant colonies was counted after 3 weeks and verified for continued colony viability after 4 weeks. The number of colony-free wells was used to calculate the colony formation efficiency by the Poisson distribution as follows: colony formation efficiency = [−ln(fraction of negative wells)]/number of transfected cells per well.
Competitive EMSA.
A competitive electrophoretic mobility shift assay (EMSA) was conducted essentially as described previously (28). Briefly, 10 fmol of a double-stranded DNA probe containing one copy of a high-affinity, palindromic EBNA1-binding site was end labeled and incubated either in the absence or in the presence of 20 mM dnEBNA1/Softag1 and excess poly(dI-dC). Additional unlabeled competitor DNA containing wtDS or one of the complete engineered origins [e.g., 2×(DS1+2), 4×FRbs, 4×SiteIIIbs, 4×Rep*bs, or S-W-W-S] was included in increasing concentrations from 1- to 14-fold EBNA1-binding site excess. Competition for binding EBNA1 by the unlabeled DNAs was analyzed by ImageQuant analysis of the bound and free probe bands as described for Southern blotting.
ChIP assay and real-time PCR analysis.
For chromatin immunoprecipitation (ChIP) experiments, nuclei were prepared as described previously (24). For each sample 107 cells were harvested, washed with phosphate-buffered saline, and resuspended in 250 μl of hypotonic buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, Complete protease inhibitor mixture [Roche]). Cells were lysed by adding 0.04% Triton X-100 and incubated for 10 min on ice. Samples were centrifuged (4 min, 1,300 × g, 4°C) to separate soluble cytosolic and nucleosolic proteins from the chromatin. Nuclei were washed at a concentration of 108 nuclei/ml in ice-cold buffer A supplemented with 200 mM NaCl. After centrifugation (1,300 × g, 5 min, 4°C) nuclei were carefully resuspended in 1 ml of buffer A. Then, 9 ml of prewarmed buffer A supplemented with formaldehyde to a final concentration of 1.1% was added, and the nuclei were cross-linked for 5 min at 37°C. Fixed nuclei were washed twice with phosphate-buffered saline-0.5% NP-40, resolved in 2.7 ml of LSB (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2), and lysed by adding 300 μl of 20% Sarkosyl. The chromatin was transferred onto a 40-ml sucrose cushion (LSB plus 100 mM sucrose) and centrifuged (10 min, 4°C, 4,000 × g). The supernatant was removed, and the chromatin was resuspended in 2 ml of Tris-EDTA and sonicated (Branson Sonifier 250-D, 35% amplitude) for 2 min in 1-s intervals. For each immunoprecipitation, 500 μg of the nucleoprotein was adjusted with a 1/10 volume of 11× NET (50 mM Tris, 150 mM NaCl, 0.5 M EDTA, 0.5% NP-40). Then, 10 μg of polyclonal rabbit antibodies directed against the human ORC2 protein or 50 μl of hybridoma supernatant of the monoclonal EBNA1-specific antibody 1H4 was added. Immunoprecipitation and purification of coprecipitated DNA was performed as described previously (24). Real-time PCR analysis was performed according to the manufacturer's instructions using the same parameters and the primer pairs described previously (24). The primer pairs used were as follows: origin for, TCTTCAGCCACTGCCCTTGTG; origin back, CAGATATCAAGCTTGTTAACCCT; control for, CACGACGGGGAGTCAGGC; and control back, GTAGCGGTGGTTTTTTTGTTTGC. The copy number of each test plasmid per cell for the ChIP experiments was determined by real-time PCR with a serial dilution of p3508 plasmid DNA as the standard. Input DNA from two independent experiments was digested with DpnI, and 1, 10, or 100 ng of this total input DNA was used as a template for real-time PCR analysis.
RESULTS
Engineering of EBNA1-dependent origins of DNA synthesis.
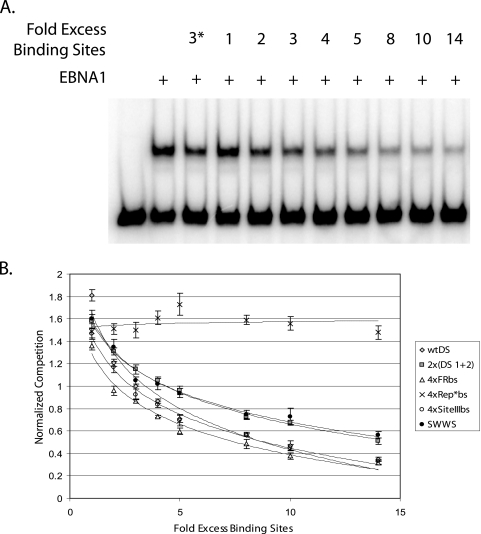
We engineered 13 candidate origins of DNA synthesis modeled upon DS, with a range of different affinities for the initiator protein, EBNA1 (Fig. 1). The rank order of the affinity of EBNA1 for these engineered origins was compared directly by a competitive EMSA, in which each origin was used as unlabeled competitor DNA (Fig. 2). A comparison of the level of competition that each origin produced yielded the following rank order of affinity for EBNA1: 4×FRbs > wtDS ≈ 4×SiteIIIbs > 2×(DS1+2) ≈ S-W-W-S ≫ 4×Rep*bs. The fold excess of EBNA1-binding sites for each origin type that was required to produce a comparable competition of dnEBNA1 from the labeled probe was quantified as follows: 2.1-fold (4×FRbs), 2.8-fold (4×SiteIIIbs), 3.0-fold (wtDS), 4.5-fold [2×(DS1+2) and S-W-W-S], and 25-fold (4×Rep*bs). This ranking of affinities is consistent with the relative affinities of their individual EBNA1-binding sites previously reported (2, 12, 28). The binding of the human TRF2 protein to DS increases the apparent affinity of EBNA1 for this origin (9); we therefore incorporated half-binding sites for TRF2 into a subset of these engineered origins with the same arrangement for these half-binding sites as found in DS (Fig. 1). We also hypothesized that stabilizing the binding of TRF2 to the EBNA1-dependent origins would enhance an origin's efficiency. To test this hypothesis, two origins—wtDS and 4×FRbs+TRF2hbs—were modified to contain one or two complete TRF2-binding sites in place of half-binding site(s) (Fig. 1).
FIG. 2.
Rank order of the affinity of EBNA1 for its binding sites in origins of DNA synthesis as determined by competitive EMSA. (A) A total of 5 fmol of end-labeled probe containing one EBNA1-binding site from FR was incubated either in the absence or in the presence of 1.6 nM dnEBNA1/Softag1 in a reaction volume of 25 μl at room temperature for 30 min. Reactions contained increasing amounts (1- to 14-fold excess of binding sites) of unlabeled DNA fragments containing either wtDS or one of the engineered origins of DNA synthesis. A threefold excess of wtDS EBNA1-binding sites was included in one lane on all gels, marked with an asterisk, as an unlabeled competitor control and was used for sample normalization across experiments. Samples were electrophoresed through a 4% polyacrylamide gel at 300 V at 4°C for 1.5 to 2 h, dried onto Whatman paper, exposed to a storage phosphor screen, and visualized by scanning on a Storm 640 PhosphorImager. (B) The amount of competition for binding by EBNA1 produced from each origin tested was normalized to the amount of competition for binding by EBNA1 produced from a threefold excess of wtDS EBNA1-binding sites. These values were plotted on the y axis against the fold excess of unlabeled EBNA1-binding sites added to each reaction in order to determine the rank order of the affinity of EBNA1 for all origins tested. A 25-fold excess in the binding sites of the 4×Rep*bs containing DNA fragment was required to produce an amount of competition similar to that of 3-fold excess wtDS EBNA1-binding sites (data not shown). The rank order is 4×FRbs > wtDS ≈ 4×SiteIIIbs > 2×(DS1+2) ≈ S-W-W-S ≫ 4×Rep*bs, as determined by experiments conducted in triplicate.
The affinity of EBNA1 for its binding to candidate origins with binding sites for TRF2 correlates with their efficiency for initiating DNA synthesis.
In order to determine whether the replicative efficiency of candidate origins of DNA synthesis correlates with the affinity of EBNA1 for its origin binding sites, the 13 plasmids described above with 2 control plasmids were scored for their ability to replicate shortly after introduction into EBV-positive B cells. Raji cells (5 × 106 cells) were electroporated with 5 μg of an internal positive control plasmid (p3567), 5 μg of a test plasmid, and 1 μg of an eGFP expression plasmid (p2134) and were cultured at appropriate cell densities without selection for 4 days. Extrachromosomal DNA from 107 cells was then harvested by alkaline lysis, purified, and subsequently digested with either MluI alone or MluI plus DpnI. The digestion of this DNA was accomplished by two rounds of enzyme addition and was allowed to progress for at least 24 h to ensure its completion. These DNAs were concentrated by precipitation with ethanol, and electrophoresed through a 0.8% Tris-acetate-EDTA agarose gel, and subsequently detected by Southern blotting. This assay measures the accumulation of DNAs synthesized during the 4 days after their introduction into the cells and reflects their homing to the compartments that support DNA synthesis, their synthesis, and their maintenance in the proliferating cells.
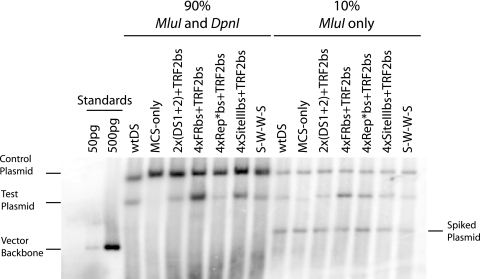
The signal from the replicated, larger, positive-control plasmid (p3567) was present in all lanes and was used as a control for plasmid recovery, to normalize the signal of the replicated test plasmids, and to facilitate comparison between samples (Fig. 3). A total of 5 ng of the pPUR vector backbone plasmid (Clontech) was also introduced prior to cell lysis to serve as an additional control for plasmid recovery and as a control for the completion of digestion with DpnI. The signal corresponding to this bacterially methylated DNA is only present in lanes containing 10% of the total sample that has only been linearized by digestion with MluI so long as the digestion with DpnI in the other samples was complete. Both the larger (p3567) and the smaller (p3487) positive-control oriP plasmids replicated efficiently and consistently produced 40 to 100 replicated plasmids per transfected cell detected 4 days postelectroporation. In addition, a plasmid with wtDS reintroduced into the MCS of p3488 replicated indistinguishably from the other positive control plasmids (data not shown). The MCS-only (p3488) plasmid produced signals indistinguishable from the background when quantified with ImageQuant 5.2 (Molecular Dynamics). In all cases, the candidate origins of DNA synthesis engineered to contain half-binding sites for TRF2 (p3512 to p3515) replicated more efficiently than did the comparable origins lacking them (p3508 to p3511) (compared in Table 1). Signals arising from the replicated test plasmids were normalized to the signal from the positive control oriP plasmid, which was set at 100%. Unexpectedly, the origin composed of four high-affinity EBNA1-binding sites from FR with flanking TRF2-half-binding sites (p3513) supported DNA replication 220% as efficiently as did wtDS. In comparison, the origins composed of two pairs of medium-affinity EBNA1-binding sites from DS (p3512) or site III (p3515) with TRF2-half-binding sites flanking them replicated 57 and 86%, respectively, as efficiently as did wtDS. The origin composed of four pairs of the slightly lower affinity EBNA1-binding site from Rep* with TRF2-half-binding sites flanking them (p3514) replicated only 28% as efficiently as did wtDS. In contrast to these origins that are flanked by TRF2-half-binding sites, the same origins lacking them replicated much less efficiently in all cases. The origin composed of four EBNA1-binding sites from FR (p3509) replicated 12% as efficiently as did wtDS and was not significantly different from the origin composed of two pairs of EBNA1-binding sites from DS, which replicated 15% as efficiently as wtDS. However, the candidate origin composed of four EBNA1-binding sites from Rep* did not produce a detectable signal. Among the derivatives without half-sites for TRF2, two replicated better than the FR- and DS-derived origins lacking those half-sites. The origin composed of four medium-affinity EBNA1-binding sites derived from site III replicated at 27% the efficiency of wtDS. Finally, the hybrid origin created from EBNA1-binding sites derived from both FR and site III (S-W-W-S, p3516) replicated at 61% the efficiency of wtDS. This result is interesting because the arrangement of “strong” and “weak” EBNA1-binding sites, as found in wtDS, replicated more efficiently than did the related candidate origins composed of four of the “strong” (p3509, 4×FRbs) or “weak” (p3511, 4×SiteIII) EBNA1-binding sites. This result indicates that not only the affinities of binding sites for a replicator but also their arrangements can be determinants of the efficiency of that origin.
FIG. 3.
The efficiency of the initiation of DNA synthesis from engineered origins containing TRF2-half-binding sites correlates with the affinity of EBNA1 for its origin-binding sites. Equal masses of the control plasmid (p3567) and of one of the test plasmids containing wtDS (p3487), the MCS-only (p3488), an origin with the flanking TRF2-half-binding sites (p3512 to p3515), or the S-W-W-S origin (p3516) were electroporated into Raji cells. After 4 days, the extrachromosomal DNA was extracted from 107 cells by alkaline lysis. A total of 90% of the samples was digested with both MluI and DpnI, and 10% was digested with MluI only to linearize the DNA. To test the completeness of DpnI digestion, 5 ng of pPur plasmid was added as “spiked DNA” during extraction. For the analysis of this DNA, a Southern blot using a radiolabeled probe produced by random priming of the pPUR vector backbone was hybridized to the electrophoresed and transferred DNAs. The identity of the test plasmid in each lane is given above it. As standards, 50 and 500 pg of linearized vector backbone were loaded on the left of the blot. This is a representative blot from an experiment conducted in triplicate.
TABLE 1.
Comparison of the short-term replication efficiencies of the natural and artificial origins of DNA synthesis in Raji cells
| Plasmid | Origin description | Normalized replicative efficiency | SD |
|---|---|---|---|
| 3487 | wtDS | 1 | NA |
| 3488 | MCS-only | 0.03 | 0.04 |
| 3508 | 2x(DS1+2) | 0.15 | 0.06 |
| 3509 | 4xFRbs | 0.12 | 0.08 |
| 3510 | 4xRep*bs | 0.001 | 0.01 |
| 3511 | 4xSiteIIIbs | 0.27 | 0.01 |
| 3516 | S-W-W-S | 0.61 | 0.19 |
| 3512 | 2x(DS1+2) + TRF2(1/2, 1/2, 1/2) | 0.57 | 0.28 |
| 3513 | 4xFRbs + TRF2(1/2, 1/2, 1/2) | 2.20 | 0.60 |
| 3514 | 4xRep*bs + TRF2(1/2, 1/2, 1/2) | 0.28 | 0.09 |
| 3515 | 4xSiteIIIbs + TRF2(1/2, 1/2, 1/2) | 0.86 | 0.10 |
| 3591 | wtDS + TRF2(1/2, 1/2, 1) | 1.7 | 0.65 |
| 3592 | 4xFRbs + TRF2(1/2, 1/2, 1) | 0.73 | 0.03 |
| 3603 | wtDS + TRF2(1, 1/2, 1) | 1.77 | 0.92 |
| 3604 | 4xFRbs + TRF2(1, 1/2, 1) | 0.92 | 0.33 |
In comparing these two classes of engineered origins, we found that the presence of TRF2-half-binding sites flanking the EBNA1-binding sites increased the efficiency of the initiation of DNA synthesis differently for different origins. Moderate increases in replicative efficiency were observed for the DS and site III-derived origins (3.8- and 3.2-fold increases, respectively). However, more dramatic increases were observed for the Rep*-derived origin, which supported detectable replicative signals in the presence of TRF2-half-binding sites but not in their absence, and for the FR-derived origin (a 18.4-fold increase). Surprisingly, the 4×FRbs+TRF2hbs origin (p3513) initiated DNA synthesis 2.2-fold more efficiently than did wtDS. This finding indicates that EBV has evolved to use a submaximally efficient origin of DNA synthesis during the latent phase of its life cycle.
We tested whether complete TRF2-binding sites would further enhance the efficiency of DNA replication with the notion that increasing the affinity of TRF2 dimers for candidate origins would increase the apparent affinity of EBNA1 for them. We modified the wtDS and 4×FRbs+TRF2hbs origins to introduce one or two complete TRF2-binding sites in the place of the flanking half-binding sites and found modest differences in their effect. Including one complete TRF2-binding site to wtDS (p3591) enhanced its replicative efficiency (170% of wtDS), while the inclusion of two such binding sites (p3592) inhibited origin function (73% of wtDS). Similar constructions with 4×FRbs+TRF2hbs (p3603, p3604) increasingly inhibited origin function (80 and 42%, respectively, of 4×FRbs+TRF2hbs).
The affinity of EBNA1 for binding to candidate origins with binding sites for TRF2 is a major determinant for the establishment of extrachromosomal replicons.
EBV-derived plasmid replicons that are introduced into mammalian cells must proceed successfully through a process termed “establishment” in order to be stably replicated and maintained (15). Newly introduced plasmids are rapidly lost from the cell population at ∼15 to 25% per cell generation for approximately 10 to 15 generations. Subsequently, the plasmids that remain are lost from the population at a rate of 3 to 4% per generation and are said to be “established” in the cell population. Little is known mechanistically of how establishment occurs. In order to determine whether the affinity of EBNA1 for its origin-binding sites has an effect upon the establishment of plasmids, the plasmids bearing either a natural or an engineered origin of DNA synthesis were scored for their ability to form colonies in a long-term replication assay. Briefly, 5 × 106 Raji cells were electroporated as described above and were cultured at appropriate cell densities for 3 days. At this time, the efficiency of electroporation for each introduced plasmid was determined by measuring the percentage of GFP-positive cells present in the population. These cell populations harboring the test plasmids were serially diluted and divided into aliquots into 96-well plates so that a known number (between 1 and 10,000) of GFP-positive cells were present per well. This range allowed the growth of cells for all plasmids bearing an origin of DNA synthesis at 3 weeks postelectroporation, enabling an accurate comparison of the efficiencies of colony formation of each plasmid after growth under selection with puromycin.
The efficiency of long-term colony formation depends on the efficiency of establishment; 1 to 10% of cells transfected or electroporated with plasmids bearing wtDS can give rise to colonies (15, 28; C. Wang, unpublished results). The efficiency of colony formation of cells with the plasmid bearing wtDS (p3487) in these experiments (8.2%) is consistent with that in previous reports. The efficiencies of colony formation of cells with the plasmids bearing no origin or one of the engineered, candidate origins of DNA synthesis were normalized to the efficiency of cells with wtDS and are compared in Table 2. The cells bearing the MCS-only (p3488) plasmid yielded no colonies, and hence the background efficiency of colony formation was set to be less than 1 in 10,000, or <0.056% overall and <0.68% of the efficiency of wtDS. The origins lacking TRF2-binding sites generally were established quite inefficiently (e.g., <1.7 to 5% of wtDS). In contrast, the one origin constructed with binding sites from both FR and site III and having the same arrangement of sites with different affinities for binding EBNA1 as found in DS (S-W-W-S, p3516) was more efficient at supporting the formation of colonies (17.4% that of cells with wtDS) than either of the related candidate origins composed solely of binding sites from FR or site III (p3509, p3511).
TABLE 2.
Comparison of the efficiencies of colony formation of the natural or artificial origins of DNA synthesis in Raji cells
| Plasmid | Origin description | Normalized replicative efficiency |
|---|---|---|
| 3487 | wtDS | 1 |
| 3488 | MCS-only | <0.007 |
| 3508 | 2x(DS1+2) | 0.051 |
| 3509 | 4xFRbs | <0.017 |
| 3510 | 4xRep*bs | <0.017 |
| 3511 | 4xSiteIIIbs | 0.051 |
| 3516 | S-W-W-S | 0.174 |
| 3512 | 2x(DS1+2) + TRF2(1/2, 1/2, 1/2) | 0.406 |
| 3513 | 4xFRbs + TRF2(1/2, 1/2, 1/2) | 6.21 |
| 3514 | 4xRep*bs + TRF2(1/2, 1/2, 1/2) | 0.014 |
| 3515 | 4xSiteIIIbs + TRF2(1/2, 1/2, 1/2) | 1.88 |
| 3591 | wtDS + TRF2(1/2, 1/2, 1) | 1.51 |
| 3592 | 4xFRbs + TRF2(1/2, 1/2, 1) | 0.370 |
| 3603 | wtDS + TRF2(1, 1/2, 1) | 4.68 |
| 3604 | 4xFRbs + TRF2(1, 1/2, 1) | 3.13 |
The addition of the TRF2-half-binding sites to these origins had a significant effect on the efficiencies of colony formation for all origins tested. The 2×(DS1+2)+TRF2hbs (p3512) origin formed colonies 40.6% as efficiently as wtDS, resulting in a ∼8-fold increase over the identical origin that lacked the TRF2-half-binding sites. The inclusion of TRF2-half-binding sites to 4×FRbs (p3513), 4×Rep*bs (p3514), and 4×SiteIIIbs (p3515) resulted in even more striking increases in the efficiency of colony formation relative to these mediated by the related candidate origins lacking those sites. The 4×SiteIIIbs+TRF2hbs origin supported colony formation 1.88-fold better than did wtDS, or ∼37-fold better than the same origin lacking TRF2-half-binding sites (p3511). Moreover, introduction of TRF2-half-binding sites to the 4×FRbs and 4×Rep*bs origins supported detectable levels of colony formation, whereas in their absence no colonies were observed above the background. The 4×FRbs+TRF2hbs origin functioned significantly better than wtDS in forming colonies (e.g., a 6.2-fold increase). Thus, EBV has been selected to have a submaximally efficient origin for the establishment of its plasmids, which is consistent with the findings of the short-term replication assays.
The modification of the half-binding sites for TRF2 in wtDS and 4×FRbs+TRF2hbs to create one or two complete TRF2-binding sites flanking the EBNA1-binding sites also affected the efficiency of colony formation of these origins. Similar to what was observed in the short-term assay, the wtDS origin with one complete TRF2-binding site supported colony formation slightly better than wtDS (150% of wtDS), whereas the inclusion of two such binding sites inhibited the efficiency of colony formation (37% of wtDS). The inclusion of complete TRF2-binding sites to the 4×FRbs+TRF2hbs origin increasingly inhibited colony formation (75 and 50%, respectively, of 4×FRbs+TRF2hbs).
The presence of binding sites for TRF2 correlates with increased binding of EBNA1 and ORC2 in vivo at candidate origins.
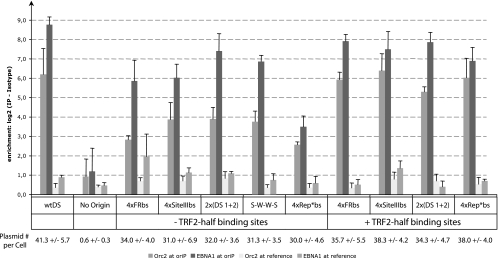
Having found that candidate origins with binding sites for TRF2 supported replication in proportion to their affinities for binding of EBNA1 as measured in vitro, we sought to determine whether their binding of EBNA1 and ORC2 in vivo was also in proportion to their support of replication. We assayed the levels of binding EBNA1 and ORC2 by using ChIP with antibodies to EBNA1 and ORC2 and 293/EBNA1 cells carrying the introduced plasmids with the candidate origins. These cells do not have endogenous EBV as do Raji cells that might confound the ChIP assays. The levels of the plasmid DNAs in all of the studied samples were measured by real-time PCR and found to be similar (Fig. 4). The levels of EBNA1 and ORC2 bound to the candidate origins and to a distal DNA sequence common to all of the plasmids were determined by measuring the differences between the amount of those DNAs precipitated by an antibody specific to EBNA1 or ORC2 and that precipitated by an isotype control antibody. The five origins with binding sites for TRF2 bound on average two times the levels of EBNA1 and four times the level of ORC2 as did the three lacking those binding sites, as measured in vivo (Fig. 4).
FIG. 4.
Histogram comparing the association of ORC2 and EBNA1 with the engineered origins of DNA synthesis after establishment in 293/EBNA1 cells. The y axis gives the log2 values that denote the difference in crossing point values between DNA fragments containing the test origin or a distal control region on the plasmid when immunoprecipitated with a specific antibody to ORC2 or EBNA1 and the observed signal when immunoprecipitated with an isotype control antibody (25). The wtDS origin, a no-origin control, and the engineered origins of DNA synthesis are listed on the x axis.
DISCUSSION
Previous studies of DS and Rep*, two latent origins of DNA synthesis of EBV, have uncovered shared properties that contribute significantly to their replicative function. We sought to determine whether one of these features, EBNA1's affinity for its binding sites in the origin of DNA synthesis, affected the efficiency of the replication of plasmids bearing them. To this end, we constructed several plasmids containing candidate origins of DNA synthesis composed of EBNA1-binding sites with a range of different affinities and introduced them into EBV-positive, human B cells. These EBNA1-dependent origins of DNA synthesis were designed to mimic the arrangement of the EBNA1 and TRF2-half-binding sites found in DS. Some of these constructed plasmids produced robust replicative signals in both short and long-term replication experiments and permitted the comparison of the replicative efficiencies of the various candidate origins. From this study, we have arrived at three general findings: (i) there is a direct correlation between EBNA1's affinity for an origin containing TRF2-half-binding sites and its frequency of supporting the initiation of DNA synthesis; (ii) there is a direct correlation between EBNA1's affinity for an origin containing TRF2-half-binding sites and the frequency of a plasmid's being established; and (iii) half-binding sites for TRF2 increase on average the levels of EBNA1 and ORC2 bound to origins in vivo.
A comparison of the efficiencies by which these engineered origins initiate DNA synthesis to that of wtDS at 4 days postelectroporation yielded several unexpected findings (Table 1). First, EBNA1's affinity for its binding different sites in engineered origins correlated with their efficiency in supporting the initiation of DNA synthesis in the presence of TRF2-half-binding sites but not in their absence (Kendall's rank correlation test, P = 0.021) (depicted graphically in Fig. 5). Second, the origin consisting of 4×FRbs+TRF2hbs (p3513) initiated DNA synthesis 2.2-fold more efficiently than did wtDS. The EBV genome therefore uses a less than maximally efficient origin of DNA synthesis during the latent phase of its life cycle. It is known that imperfections in the synthesis and partitioning of EBV's genome are balanced to generate a wide distribution in the numbers of viral plasmids per cell (18). This balance is achieved in part by synthesis and partitioning being mechanistically coupled (18) and allows the viral replicon with minimal requirements to be maintained stably in cells to which it provides a selective advantage. Thus, DS is likely tailored to foster EBV's successful lifestyle.
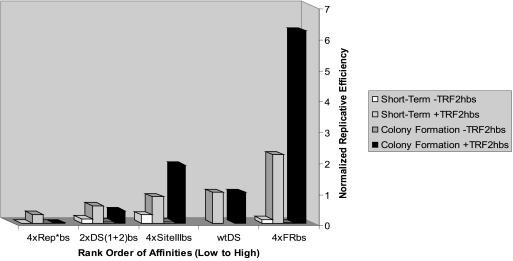
FIG. 5.
Graphical representation of the correlation of EBNA1-binding affinity for its origin binding sites in the presence or absence of TRF2-half-binding sites with measured replicative efficiencies. The rank order of affinities of EBNA1-binding sites are placed on the x axis, from weakest to strongest (left to right); the replicative efficiencies in the short-term assay (from Table 1) and the colony formation assay (from Table 2) are placed on the y axis. A positive correlation between the affinity of EBNA1 for its origin-binding sites with both replicative efficiencies is observed in the presence of TRF2-half-binding sites but not in their absence. Derivatives of wtDS lacking the TRF2-half-binding sites were not constructed for the present study, and hence the replicative efficiencies were not determined in these assays.
A third finding from the short-term replication assay was the magnitude of the effect produced by TRF2-half-binding sites flanking the pairs of EBNA1-binding sites. When the TRF2-half-binding sites were present in the origins consisting of 2×(DS 1+2) (p3512) and of 4×SiteIIIbs (p3514), they supported 3.6- and 3.2-fold increases, respectively, in the efficiency of initiating DNA synthesis relative to the same origins lacking them (p3508 and p3511, respectively). Even more strikingly, however, was the 18.4-fold effect observed with the origin consisting of 4×FRbs+TRF2hbs (p3513) compared to the same origin lacking the TRF2-half-binding sites (p3509). Our findings indicate that TRF2 contributes more to the initiation of DNA synthesis at EBNA1-dependent origins than previously appreciated. Deng et al. have shown by using EMSAs that the binding of TRF2 to sites identical in sequence and arrangement to those in 4×FRbs+TRF2hbs (p3513) increases the binding of EBNA1 cooperatively (9). This cooperative binding likely underlies some of the efficiency with which this plasmid is replicated and indicates that further characterization of the mechanism of TRF2's stimulation of the initiation of DNA synthesis is warranted.
When these engineered origins were introduced into Raji cells and selected with puromycin for 3 weeks, a similar trend (depicted graphically in Fig. 5) in the efficiencies of colony formation of these origins was found, as determined by Kendall's rank correlation test (P = 0.035). All plasmids bearing origins of DNA synthesis that contained TRF2-half-binding sites formed colonies more efficiently than the same plasmids lacking them. The inclusion of flanking TRF2-binding sites had a similar effect on both the initiation of DNA synthesis and the formation of colonies, as did the affinity of EBNA1 for its origin binding sites. However, when TRF2-half-binding sites were absent from the candidate origins, the affinity of EBNA1 for its origin binding sites alone was not a determinant of the replicative efficiency. The hybrid S-W-W-S origin (p3516) replicated better in both the short and the long term than either candidate origin composed of the same EBNA1-binding sites (4×FRbs, p3509; 4×SiteIIIbs, p3511). This finding is in agreement with previous studies of 8×Rep*, which replicates as well as wtDS with eight pairs of low-affinity EBNA1-binding sites in the absence of TRF2-half-binding sites (28). It indicates that the relative positioning of distinct binding sites for EBNA1 can also be a determinant of the efficiency of engineered origins.
These data lead us to two new interpretations of this well-studied model replicon. First, we have found that EBNA1's affinity for an origin may affect some function(s) in addition to the initiation of DNA synthesis. Potentially, a higher affinity of EBNA1 for its origin-binding sites could stimulate the homing of the introduced plasmids to the replication-competent compartments of the cell, thus allowing a higher percentage of the plasmids to begin replicating in the initial S phases. Alternatively, higher-affinity EBNA1-binding sites present in the origin of DNA synthesis may act cooperatively with these FR sites to stimulate FR's beneficial effects upon plasmid duplication. Second, our data indicate that EBNA1's affinity for an origin is one major determinant of the efficiency of establishment of that origin. In addition, it is clear that the association of TRF2 with an origin of DNA synthesis plays a substantial role in this process as well. However, the mechanism(s) by which TRF2 stimulates the initiation of DNA synthesis and the establishment of plasmids is unclear. If TRF2 were only stimulating the initiation of DNA synthesis by increasing the apparent affinity of EBNA1 for its binding sites in an origin, then sites bound by EBNA1 with the highest affinity should be the least affected. However, this inverse correlation was not observed in short- or long-term replication assays or in the ChIP measurements in vivo. Thus, the activities of TRF2 other than its cooperative binding with EBNA1 likely stimulate the replication of the test plasmids. TRF2 has also been shown to interact via its N-terminal basic domain with a region directly adjacent to the bromo adjacent homology (BAH) domain of ORC1 (3). The BAH domain of ORC1 has been implicated as being important for the replication of oriP plasmids (20). Derivatives of ORC1's BAH domain or with a point mutation within it (E111K), for example, inhibit the recruitment of ORC1 to DS and the replication of oriP plasmids (ibid). The contribution of TRF2-binding sites to replication may therefore reflect the synergistic effects of an increased affinity of EBNA1 for its origin binding sites and the enhanced recruitment of ORC to the origin. This explanation is supported by our finding that in the presence of half-binding sites for TRF2 at test origins correlates with increased binding by both EBNA1 and ORC2 in vivo.
A model consistent with our findings posits that structural differences in either the DNA or the EBNA1 are caused by differences in EBNA1's interaction with the different bases of the binding sites it binds with different affinities. This type of differential interaction has been demonstrated with the trp repressor binding to sites where the central 2 bp of the full binding site differ. An alteration of the dinucleotide between the half-sites for binding did not significantly affect the stability of the trp repressor binding to the primary, full binding site (5). However, the assembly of additional trp repressors at two half-sites flanking the full binding site was greatly affected (5). This effect of differential binding of a primary binding site, which affects the recruitment and loading of additional repressor molecules to flanking half-sites, has been implicated as the mechanism for differential repression of the various trp operators. (5). Based upon this model, we hypothesize that the differences in the overall EBNA1/DNA structure affects the positioning, recruitment, and/or loading of TRF2 to its flanking half-sites, as well as other cellular replicative factors such as the members of the prereplicative complex. This EBV-derived model system will permit further analysis of what trans-acting factor(s) are necessary and/or sufficient for affecting the efficiency of initiation of DNA synthesis in concert with the cis features identified in the present study.
Supplementary Material
Acknowledgments
This study was supported by a grant from the NIH to Bill Sugden (CA022443 and T32-CA09135), by grants from the Deutsche Forschungsgemeinschaft (SPP1230 and SFB646), and by institutional grants to Aloys Schepers. Bill Sugden is an American Cancer Society Research Professor.
Footnotes
Published ahead of print on 2 April 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altmann, M., D. Pich, R. Ruiss, J. Wang, B. Sugden, and W. Hammerschmidt. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. USA 10314188-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder, R. F., W. A. Shah, D. R. Rawlins, G. S. Hayward, and S. D. Hayward. 1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 642369-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atanasiu, C., Z. Deng, A. Wiedmer, J. Norseen, and P. M. Lieberman. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310207-211. [DOI] [PubMed] [Google Scholar]
- 5.Bareket-Samish, A., I. Cohen, and T. E. Haran. 1997. Repressor assembly at trp binding sites is dependent on the identity of the intervening dinucleotide between the binding half sites. J. Mol. Biol. 267103-117. [DOI] [PubMed] [Google Scholar]
- 6.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 7510603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 9810085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, Z., C. Atanasiu, J. S. Burg, D. Broccoli, and P. M. Lieberman. 2003. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 7711992-12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9493-503. [DOI] [PubMed] [Google Scholar]
- 10.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106287-296. [DOI] [PubMed] [Google Scholar]
- 11.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 981865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, C. H., S. D. Hayward, and D. R. Rawlins. 1989. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J. Virol. 63101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julien, M. D., Z. Polonskaya, and J. Hearing. 2004. Protein and sequence requirements for the recruitment of the human origin recognition complex to the latent cycle origin of DNA replication of Epstein-Barr virus oriP. Virology 326317-328. [DOI] [PubMed] [Google Scholar]
- 14.Knutson, J. C., and D. Yee. 1987. Electroporation: parameters affecting transfer of DNA into mammalian cells. Anal. Biochem. 16444-52. [DOI] [PubMed] [Google Scholar]
- 15.Leight, E. R., and B. Sugden. 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell. Biol. 214149-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leight, E. R., and B. Sugden. 2001. The cis-acting family of repeats can inhibit as well as stimulate establishment of an oriP replicon. J. Virol. 7510709-10720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menezes, J., W. Leibold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B-cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine. 22276-284. [PubMed] [Google Scholar]
- 18.Nanbo, A., A. Sugden, and B. Sugden. 2007. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 264252-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niller, H. H., G. Glaser, R. Knuchel, and H. Wolf. 1995. Nucleoprotein complexes and DNA 5′ ends at oriP of Epstein-Barr virus. J. Biol. Chem. 27012864-12868. [DOI] [PubMed] [Google Scholar]
- 20.Noguchi, K., A. Vassilev, S. Ghosh, J. L. Yates, and M. L. Depamphilis. 2006. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 255372-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polonskaya, Z., C. J. Benham, and J. Hearing. 2004. Role for a region of helically unstable DNA within the Epstein-Barr virus latent cycle origin of DNA replication oriP in origin function. Virology 328282-291. [DOI] [PubMed] [Google Scholar]
- 22.Pulvertaft, J. V. 1965. A study of malignant tumours in Nigeria by short-term tissue culture. J. Clin. Pathol. 18261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42859-868. [DOI] [PubMed] [Google Scholar]
- 24.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 1163971-3984. [DOI] [PubMed] [Google Scholar]
- 25.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 204588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 7811487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomae, A. W., D. Pich, J. Brocher, M. P. Spindler, C. Berens, R. Hock, W. Hammerschmidt, and A. Schepers. 2008. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc. Natl. Acad. Sci. USA 1051692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, J., S. E. Lindner, E. R. Leight, and B. Sugden. 2006. Essential elements of a licensed, mammalian plasmid origin of DNA synthesis. Mol. Cell. Biol. 261124-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 744512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.