Abstract
Since the number of human cases of infection with avian H5N1 influenza viruses is ever increasing, a pandemic outbreak caused by these viruses is feared. Therefore, in addition to virus-specific antibodies, there is considerable interest in immune correlates of protection against these viruses, which could be a target for the development of more universal vaccines. After infection with seasonal influenza A viruses of the H3N2 and H1N1 subtypes, individuals develop virus-specific cytotoxic T-lymphocyte responses, which are mainly directed against the relatively conserved internal proteins of the virus, like the nucleoprotein (NP). Virus-specific cytotoxic T lymphocytes (CTL) are known to contribute to protective immunity against infection, but knowledge about the extent of cross-reactivity with avian H5N1 influenza viruses is sparse. In the present study, we evaluated the cross-reactivity with H5N1 influenza viruses of polyclonal CTL obtained from a group of well-defined HLA-typed study subjects. To this end, the recognition of synthetic peptides representing H5N1 analogues of known CTL epitopes was studied. In addition, the ability of CTL specific for seasonal H3N2 influenza virus to recognize the NP of H5N1 influenza virus or H5N1 virus-infected cells was tested. It was concluded that, apart from some individual epitopes that displayed amino acid variation between H3N2 and H5N1 influenza viruses, considerable cross-reactivity exists with H5N1 viruses. This preexisting cross-reactive T-cell immunity in the human population may dampen the impact of a next pandemic.
Since the first documentation of bird-to-human transmissions of highly pathogenic avian H5N1 influenza viruses, these viruses have spread from Southeast Asia to other regions of the world (2, 3, 22, 27). Since 2003 the number of human cases has continued to increase; as of 28 February 2008, 369 human cases have been reported, of which 234 were fatal (28). It is feared that an H5N1 virus may cause the next influenza pandemic when it is able to replicate in mammalian species by adaptation through genetic reassortment or accumulation of point mutations in relevant gene segments (11). Although neuraminidase subtype 1 cross-reactive antibodies have been demonstrated in human subjects, antibodies to H5 molecules are hardly existent in the human population as a result of limited exposure to H5N1 viruses, which contributes to a scenario for these viruses to become pandemic (20). In general, the exposure history and the immune status of the human population will influence the size and the severity of pandemics (4, 8, 9, 14). The presence of T-cell immunity induced by infection with human influenza virus strains may provide some degree of cross-protective immunity against the H5N1 viruses. Cytotoxic T-lymphocyte (CTL) responses are predominantly directed to internal viral proteins, the nucleoprotein (NP) in particular (23, 29), which is much more conserved than the surface hemagglutinin and neuraminidase glycoproteins (5, 13, 15, 29). It has been suggested that cross-reactive CD8+ T cells may temper the impact a pandemic potentially could have on the human population (9, 14, 19). In humans, the presence of cross-reactive CTL responses inversely correlated with the amount of shedding of a heterosubtypic strain that was used for experimental infection of study subjects (14). Although preexisting CTL immunity against influenza virus may be of importance in the face of the current H5N1 pandemic threat, our knowledge of the cross-reactive nature of the human CTL response is limited (9).
In the present study we tested the cross-reactivity of polyclonal virus-specific CD8+ T-cell populations obtained from well-defined HLA-typed study subjects with H5N1 virus. The recognition of target cells pulsed with peptide variants, transfected with the NP gene from a human or an avian influenza virus, or infected with viruses of the H3N2 or H5N1 subtype was tested.
It was concluded that the human CTL response displays a high degree of cross-reactivity with avian H5N1 influenza viruses and could reduce morbidity and mortality during a pandemic caused by these H5N1 strains.
MATERIALS AND METHODS
Cells.
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood obtained from fifteen HLA-typed healthy blood donors (Sanquin Bloodbank, Rotterdam, The Netherlands) by density gradient centrifugation using lymphoprep (Axis-Shield PoC AS, Oslo, Norway) and then cryo-preserved at −135°C. Genetic subtyping was performed in the laboratory for Histocompatibility and Immunogenetics at the Sanquin Bloodbank using a commercial typing system (Genovision, Vienna, Austria). Three groups of study subjects were selected on the basis of their major histocompatibility complex class I (MHC-I) alleles, for which influenza CTL epitopes were identified. Within groups the subjects shared identical HLA-A and -B alleles; between groups there were differences of one or two alleles. The groups were as follows: group I, HLA A*0101, A*0201, B*0801, and B*3501; group II, HLA A*0101, A*0201, B*0801, and B*2705 (2702); and group III, HLA A*0101, A*0301, B*0801, and B*3501 (3503) (1). Subject 15 was not tested since PBMC of this donor were no longer available.
Peptides.
Amino acid sequences of all known human influenza A virus CTL epitopes were compared with their counterparts in H5N1 influenza viruses isolated since 2003, which were obtained from the influenza sequence database (12). All possible variants that could be identified in the H5N1 sequences are listed in Table 1. A set of immunograde peptides representing immunodominant CTL epitopes and the most prevalent analogues in H5N1 strains were synthesized and analyzed by mass spectrometry and were found to be >70% pure (Eurogentec, Seraing, Belgium). Variant peptide analogues from the NP, which is the main target for CTL responses, were synthesized when they had a prevalence in H5N1 strains of >0.25%, with the exception of the NP383-391 epitope since the G384K mutation observed in H5N1 viruses was known to abrogate recognition by specific CTLs completely (26). For the remaining viral proteins, all variants with a prevalence of >2.25% were synthesized and tested. Only peptides that matched the HLA alleles of the study subjects were considered.
TABLE 1.
Variant sequences of known CTL epitopes in H5N1 viruses
| Sequence name | Variant sequence of the CTL epitope at the indicated HLA allelea
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-A1 PB1 (591-599) | HLA-A3 M1 (13-21) | HLA-A*0201 M1 (58-66) | HLA-B*3501 M1 (128-135) | HLA-A*0201 NS1 (122-130) | HLA-A*0201 NS1 (123-132) | HLA-B*44 NS1 (158-166) | HLA-A1 NP (44-52) | HLA-A*6801 NP (91-99) | HLA-B*1402 NP (146-154) | HLA-B*2705 NP (174-184) | HLA-A3 NP (188-198) | HLA-A3 NP (265-273) | HLA-B*3701 NP (339-347) | HLA-B*08 NP (380-388) | HLA-B*2702 NP (381-388) | HLA-B*2705 NP (383-391) | HLA-B*3501 NP (418-426) | |
| Epitope | VSDGGPNLY | SIIPSGPLK | GILGFVFTL | ASCMGLIY | AIMDKNIIL | IMDKNIILKA | GEISPLPSL | CTELKLSDY | KTGGPIYKR | TTYQRTRAL | RRSGAAGAAVK | TMVMELVRMIK | ILRGSVAH | EDLRVLSFI | ELRSRYWAI | LRSRYWAI | SRYWAIRTR | LPFEKSTVM |
| Variant | ||||||||||||||||||
| 1 | I-------- | -V------- | -M------- | ------S- | -----T--- | ----T----- | ------H-- | --------H | -------R- | S-------- | ---------I- | ------I---- | -----I--- | -----S--- | ----K---- | ---K---- | -K------- | ----RA-I- |
| 2 | -------P- | --V------ | --W------ | -----V--- | ----V----- | -------F- | --------Q | -----V--- | A-------- | -I------L-- | --A-------- | -----M--- | ----IS--- | D-LGK---K | -LGK---K | ------K-- | ----R--I- | |
| 3 | ------IP- | F-------- | -------T- | ------T--- | ---L----- | ------N-- | -----F-RG | AA------- | --F-------- | -----QI---- | V-------- | -----H--- | ----H--- | -------P- | ----RAAI- | |||
| 4 | G-------- | -T------- | -----TV-- | ----TV---- | --------I | ------T-- | --------G | A--H----- | Q----VI---- | --------K | ----RV-I- | |||||||
| 5 | -A------- | -L------- | -----A--- | ----A----- | -----S--- | ---F----- | AP------- | --------P | ----RA--- | |||||||||
| 6 | ATS------ | -T-----T- | T-----T--- | -------Y- | A-------V | GK---KWMI | ||||||||||||
| 7 | --VR----Q | -----D--- | ----D----- | A-------- | A-SQ----- | --H------ | ||||||||||||
| 8 | --VL----- | --V--T--- | -V--T----- | -----I--- | --------G | |||||||||||||
| 9 | S----T--- | ----T-S--- | ||||||||||||||||
| 10 | -----T-S- | --N------- | ||||||||||||||||
| 11 | ---N----- | -V--T-T--- | ||||||||||||||||
| 12 | --V--T-T- | -V----T--- | ||||||||||||||||
| 13 | --V----T- | ----T-L--- | ||||||||||||||||
| 14 | -----T-L- | ----I----- | ||||||||||||||||
| 15 | -----I--- | -V--TV---- | ||||||||||||||||
| 16 | --V--TV-- | ----T--W-- | ||||||||||||||||
| 17 | -----T--W | ----T----G | ||||||||||||||||
Variant sequences of known CTL epitopes were ranked according to their relative prevalence. Anchor residues of the epitopes are underlined. Amino acid residue positions are given in parentheses.
Target cells.
B-lymphoblastoid cell lines (BLCL) were established as described previously (18) and used as target or stimulator cells. A total of 30,000 cells were incubated in the absence or presence of 10 μM peptide for 1 h at 37°C, washed once, and resuspended in RPMI 1640 medium (Cambrex, East Rutherford, NJ) containing antibiotics, l-glutamine, and 10% fetal calf serum (R10F medium). Cells of the BLCL were also infected at a multiplicity of infection of five 50% tissue culture infective doses/cell (17) with influenza viruses A/Netherlands/18/94 (A/NL/18/94) (H3N2) or A/Vietnam/1194/04 (A/VN/1194/04) (H5N1), which were propagated and titrated in MDCK cells using standard procedures. After an incubation period of 1 h at 37°C, the cells were washed and resuspended in R10F medium and incubated for 16 to 18 h at 37°C prior to their use for the stimulation of CD8+ T cells. Infection rates were determined by an immunofluorescence assay and were similar for both viruses (data not shown). The human influenza virus A/NL/18/94 (H3N2) was used as a representative of seasonal influenza viruses, while influenza virus A/VN/1194/04 was used as an example for H5N1 influenza virus.
T-cell clones.
CD8+ T-cell clones directed against the HLA-A1-restricted NP44-52 (CTELKLSDY) epitope, HLA-A3-restricted NP265-273 (ILRGSVAHK) epitope, HLA-B27-restricted NP174-184 (RRSGAAGAAVK) epitope, and the HLA-B*3501-restricted NP418-426 (LPFEKSTVM) epitope were generated as described previously (26).
In vitro expansion of influenza A virus-specific T-cell populations.
PBMC were stimulated with influenza virus A/NL/18/94-infected cells as previously described (1). Eight days after stimulation, cells were harvested and used as effector cells in an enzyme-linked immunospot (ELISPOT) or fluorescent antigen-transfected target cell (FATT)-CTL assay. For the ELISPOT assays, CD8+ T cells were purified from the in vitro expanded PBMC by magnetic bead cell sorting (Miltenyi Biotec, Bergisch Gladbach, Germany). Typically, a purity of >96% was obtained.
FATT-CTL assay.
The NP genes of influenza viruses A/NL/18/94 and A/VN/1194/04 without their stop codons were cloned into the plasmid pEGFP-N1 (Becton Dickinson, Alphen a/d Rijn, The Netherlands) in frame with the open reading frame of the green fluorescent protein (GFP) as previously described (25). Plasmid DNA was purified using a Genopure plasmid midi kit (Roche, Woerden, The Netherlands). Nucleotide sequences of the recombinant plasmids were confirmed using a Big Dye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Primer and plasmid sequences are available on request.
The plasmids were used in the FATT-CTL assay for the detection of lytic activity of virus-specific CTLs as described previously (25). In brief, BLCL were nucleofected using cell line nucleofector kit V (Amaxa Biosystems, Cologne, Germany) with program T16 and subsequently incubated in R10F medium for 4 h at 37°C. Then, they were cocultured for another 4 h in triplicate with PBMC cultures and in vitro expanded after stimulation with influenza virus A/NL/18/94 at various effector-to target (E:T) cell ratios. The number of viable GFP-positive cells was measured using a FACSCalibur (Becton Dickinson). The percent nucleoprotein-specific lysis was then calculated by the following formula: 100 × [(number of viable GFP-positive cells in the sample without effector − number of viable GFP-positive cells in the sample with effector)/number of viable GFP-positive cells in the sample without effector].
Gamma interferon (IFN-γ) assay.
ELISPOT assays were performed with in vitro expanded CD8+ T cells as effector cells and peptide-pulsed or virus-infected HLA-matched BLCL as stimulator cells as described previously (1). The number of spots was determined using an ELISPOT reader and image analysis software (Aelvis, Sanquin Reagents, Amsterdam, The Netherlands), and the average number was calculated of triplicate wells.
RESULTS
Comparison of amino acid sequences of known influenza A virus CTL epitopes.
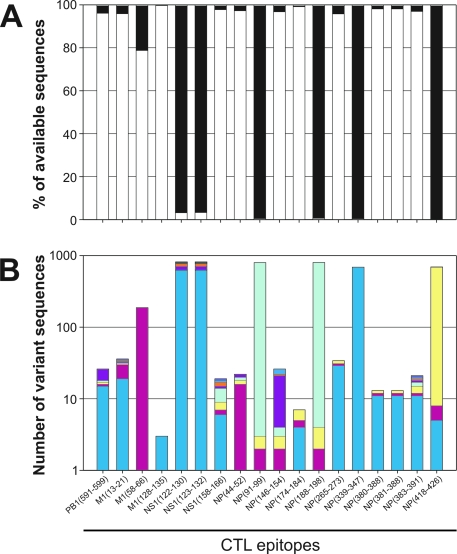
The amino acid sequences of known human influenza A virus CTL epitopes were compared with the corresponding sequences in approximately 900 H5N1 viruses obtained from the influenza sequence database (12). As shown in Fig. 1A, the epitope sequences were identical in >95% of the H5N1 viruses for the majority of the known epitopes analyzed including PB1591-599, M113-21, M1128-135, NS1158-166, NP44-52, NP146-154, NP174-184, NP265-273, NP380-388, NP381-388, and NP383-391. For some of the other epitopes the percentage of H5N1 viruses with identical sequences was variable and ranged from 79% for epitope M158-66 to 4% for epitope NS1122-130. For the epitopes NP91-99, NP188-198, NP339-347, and NP418-426, no identical sequences were found in the H5N1 viruses. In order to identify the most prevalent variant sequences in H5N1 viruses, the number of individual variants was analyzed (Fig. 1B). In some cases a single variant was identified that accounted for almost all variant sequences observed in H5N1 viruses (Table 1 and Fig. 1B). For other epitopes multiple variants were identified, although for some of these the number was low, and the number of major variants was limited (12).
FIG. 1.
The presence of known CTL epitopes in H5N1 strains. The percentage of H5N1 viruses with an epitope sequence identical to human influenza viruses (white bars) is shown. The black bars indicate the percentage of H5N1 viruses with one or more amino acid substitutions in the epitope sequence. The absolute numbers of each variant of an epitope are shown in panel B, where each color represents a single variant (sequences can be found in Table 1). For this analysis almost 900 H5N1 viruses for which sequence information was available in the influenza sequence database (12) were analyzed.
The recognition of known CTL epitopes and their avian analogues.
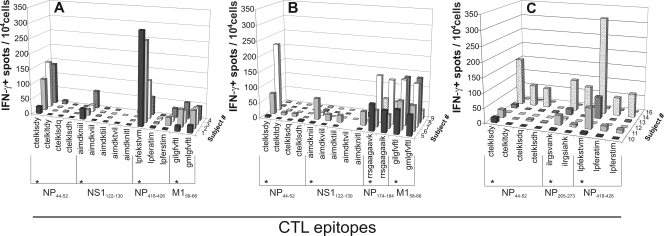
All subjects in group I (HLA A*0101, A*0201, B*0801, and B*3501) displayed T-cell reactivity with the epitopes NP44-52, NS1122-130, NP418-426, and M158-66 as they are present in human influenza A viruses although the frequency of specific CTLs varied between study subjects and the peptides tested (Fig. 2A). In none of the subjects of this group was reactivity observed with the peptide variants of epitopes NP44-52 and NS1122-130, obtained from H5N1 influenza viruses. Three out of four subjects responded to the NP418-426 variant LPFERSTIM, and all subjects responded to the M158-66 variant GMLGFVFTL. Of group II (HLA A*0101, A*0201, B*0801, and B*2705 [2702]), most subjects responded to the peptides representing epitopes from human influenza A viruses (Fig. 2B) although the magnitudes of the responses varied considerably. Only one subject in this group had an appreciable response to the H5N1 analogue sequence of the NS1122-130 epitope. Four out of five subjects responded to the H5N1 variants of the HLA B*2705-restricted NP174-184 epitope whereas all five responded to the M158-66 variant. The subjects of group III (HLA A*0101, A*0301, B*0801, and B*3501 [3503]) responded to the original epitopes to various extents. Some subjects were poor responders and hardly displayed CTL reactivity with some of these epitopes (Fig. 2C). However, the in vitro expanded PBMC of subject 14 responded strongly to the NP418-426 and also reacted with both epitope variants from H5N1 viruses, indicating that at least a fraction of the CTL population was capable of cross-recognizing these analogues. The same holds true for the NP44-52- and NP265-273-specific CTL responses in this study subject. Clones were used as positive and negative controls, and the clonal responses supported the results obtained with the polyclonal populations (data not shown).
FIG. 2.
Epitope-specific IFN-γ production by CTLs after stimulation with peptide-pulsed BLCL. The number of IFN-γ-producing cells per 10,000 CD8+ T cells (5,000 cells for subject 3) from subjects from HLA groups I (A), II (B), and III (C) were measured by ELISPOT assay. CD8+ T cells were isolated from PBMC populations expanded in vitro with influenza virus A/NL/18/94 and subsequently stimulated with peptide variants as indicated. (*, peptide sequence of the known human influenza virus CTL epitopes).
Cross-recognition of the NP derived from influenza virus A/VN/1194/04.
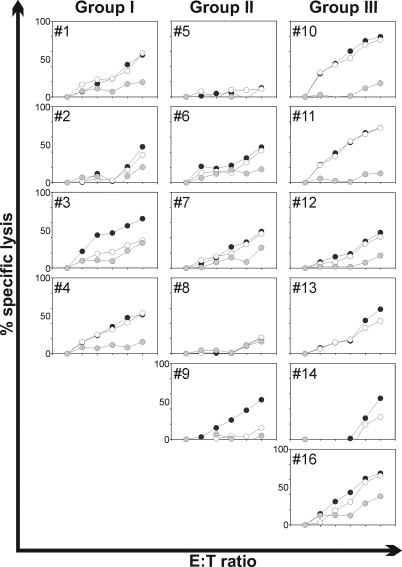
The capacity of polyclonal T-cell populations directed to the human influenza virus A/NL/18/94 to cross-react with the NP of influenza virus A/VN/1194/04 was assessed in the FATT-CTL assay. PBMC from all study subjects were stimulated with influenza virus A/NL/18/94 and allowed to proliferate. As shown in Fig. 3, 2 out of the 15 subjects tested (subjects 5 and 8) were low responders or nonresponders (Fig. 3) since no NP-specific lytic activity could be demonstrated. In the remaining 13 subjects, lytic activity was observed against the homologous NP. In most cases the PBMC cross-reacted with the NP of influenza virus A/VN/1194/04 to a considerable extend (Fig. 3). Only for subject 3 (group I) and subject 9 (group III) did the influenza virus A/NL/18/94 NP-specific CTLs fail to recognize the NP of influenza virus A/VN/1194/04 (Fig. 3).
FIG. 3.
Recognition of NP derived from H3N2 and H5N1 influenza virus by in vitro expanded PBMC specific for influenza virus A/NL/18/94 (H3N2). The lytic activity of in vitro expanded PBMC was tested with MHC-I matched BLCL nucleofected with NP-GFP coding plasmid (NP of either influenza virus A/NL/18/94 [black circles] or A/VN/1194/04 [open circles]) or empty control GFP plasmid (gray circles). The NP-specific lytic activity was tested with PBMC of the subjects from group I (subjects 1 to 4), II (subjects 5 to 9), and III (subjects 10 to 14 and 16). E:T ratios were 0, 3.125, 6.25, 12.5, 25, and 50 for all subjects except numbers 13 and 14, for whom E:T ratios were 0, 0.3, 1, 3, 10 and 30. The lytic activities against control plasmid-transfected target cells are not visible for subjects 13 and 14 as a result of negative values for the percentages of specific lysis, which were caused by slight increases in the number of GFP-positive viable cells. Standard deviation of the means was <10%.
Cross-recognition of BLCL infected with influenza virus A/VN/1194/04.
Next, we wished to assess the cross-reactive nature of the whole repertoire of CD8+ T lymphocytes specific for the human influenza virus A/NL/18/94. To this end, PBMC were stimulated with this virus, and after 8 days the CD8+ cells were isolated to obtain virus-specific polyclonal CTL populations. These cells were used as effector cells in an IFN-γ ELISPOT assay using MHC-I-matched BLCL infected with influenza virus A/NL/18/94 or A/VN/1194/04 as stimulator cells.
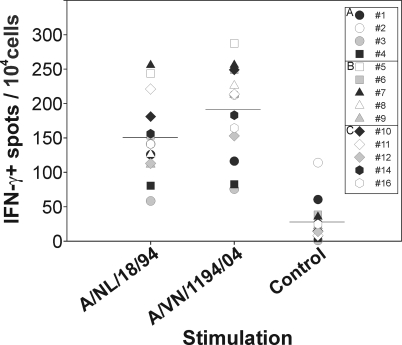
As shown in Fig. 4, the in vitro expanded PBMC population that recognized cells infected with influenza virus A/NL/18/94 also recognized cells infected with influenza virus A/VN/1194/04. The average number of IFN-γ-positive spots per 104 cells observed after stimulation with A/NL/18/94-infected cells was 151 (standard deviation, 58), and the number observed after stimulation with A/VN/1194/04 was even slightly higher at 192 (standard deviation, 65) although this difference was not statistically significant.
FIG. 4.
Recognition of influenza virus-infected BLCL by CTLs. The number of IFN-γ-producing cells per 10,000 CD8+ T cells was measured by ELISPOT assay after stimulation with BLCL infected with influenza virus A/NL/18/94 or A/VN/1194/04. Each symbol represents an individual subject from group I (A), II (B), or III (C). Uninfected BLCL were used as negative controls. The horizontal bars represent the average responses of all study subjects in groups I, II, and III.
DISCUSSION
In the present paper the cross-reactive nature of the human influenza virus-specific CTL response was investigated. It was concluded that a considerable portion of CTL populations specific for the H3N2 influenza virus A/NL/18/94 cross-reacted with the H5N1 strain A/VN/1194/04.
For most CTL epitopes, it was found that a vast majority of the H5N1 strains contained epitope sequences identical to those present in human influenza A viruses. This conservation of epitopes is responsible for the cross-reactive nature of CTL responses in humans against seasonal influenza A viruses of the H3N2 and H1N1 subtypes. However, some variation in these epitopes was observed also, and for a number of CTL epitopes the H5N1 strains did not contain identical sequences. Apart from the NP174-184 and M158-66 epitope restricted by HLA-B*2705 and HLA-A*0201, respectively, very little cross-reaction was observed of polyclonal CTL populations with variant peptides derived from H5N1 viruses.
However, as indicated above, most epitopes are relatively conserved, including those located in the NP, which contributed to the cross-reactive nature of the NP-specific CTL response. Most of the study subjects that responded to NP derived from seasonal H3N2 influenza viruses also responded to the NP derived from influenza virus A/VN/1194/04 (H5N1). The polyclonal virus-specific T-cell populations of two of these subjects failed, however, to cross-react with the NP of influenza virus A/VN/1194/04 for reasons that are unclear. Possibly, the most immunodominant responses in these subjects were directed to CTL epitopes in the NP that were not conserved.
To account for the full repertoire of virus-specific CD8+ T lymphocytes, the reactivity with MHC-I-matched cells infected with influenza virus A/NL/18/94 or A/VN/1194/04 was also analyzed. In all cases A/VN/1194/04-infected target cells were recognized to a similar extent as A/NL/18/94-infected cells, indicating that the level of cross-reactivity of human CTL responses to seasonal H3N2 influenza viruses with H5N1 strains is substantial.
Thus, apart from some individual epitopes that display amino acid sequence variation between H3N2 and H5N1 influenza A viruses, the level of cross-reactivity is considerable and does not seem to be influenced by the HLA phenotype of the study subjects. The NP383-391 epitope is present in H5N1 viruses but has disappeared from human H3N2 viruses during decades of virus evolution (7, 26). Since a virus was used for stimulation of the PBMC that does not contain this epitope, this does not give a false sense of cross-reactivity in HLA-B*2705-positive subjects. It is unknown whether the presence of the NP383-391 epitope interferes with the presentation of other HLA-B*2705-restricted epitopes. Interference has been observed only with the overlapping HLA-B*0801-restricted NP380-388 epitope (1, 24).
Although it is unknown to what extent preexisting T-cell immunity can dampen the impact of a next influenza pandemic, it is speculated that the protective effect of cross-reactive CTL responses has a beneficial effect on the outcome of infection with new pandemic influenza virus strains. This speculation is supported by a number of different observations. First, in animal models it has been shown that virus-specific CTLs contribute to heterosubtypic immunity (6, 10, 16); secondly, it was found in 1957 that individuals that had experienced documented infections with H1N1 influenza A viruses were less likely to develop severe disease or succumb to infection with the pandemic strain of the H2N2 subtype (4). In this respect it is of interest that during the current outbreak of H5N1 infections in humans, younger individuals are especially at risk for severe disease and a fatal outcome of infection (21). It can be hypothesized that younger individuals are less likely to have been exposed to seasonal influenza A viruses of the H3N2 and H1N1 subtypes and thus have not mounted a (cross-reactive) CTL response to an alternative subtype. However, it cannot be excluded that confounding factors play a role in the observed disproportionate age distribution of severe H5N1 human cases. Last but not least, the human CTL response against epidemic strains is largely cross-reactive with H5N1 influenza virus strains, as was demonstrated in the present study. Although these cross-reactive CTL populations may not prevent infection with pandemic strains, they may contribute to a certain degree of heterosubtypic immunity and facilitate a more rapid clearance of the infection than in immunologically naïve individuals who lack cross-reactive T-cell populations. This may determine the difference between life and death during a pandemic outbreak. In addition, the induction of cross-reactive CTL responses may be an attractive target for the development of universal vaccines that could confer broadly protective immunity against influenza viruses of various subtypes.
Acknowledgments
This study was conducted under the auspices of The Netherlands Influenza Vaccine Research Center and financially supported in part by The Netherlands Organization for Health Research and Development (ZonMW; grant 91402008).
We thank T. Bestebroer and C. Baas for outstanding technical assistance.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Boon, A. C., G. de Mutsert, Y. M. Graus, R. A. Fouchier, K. Sintnicolaas, A. D. Osterhaus, and G. F. Rimmelzwaan. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 76582-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claas, E. C., A. D. Osterhaus, R. van Beek, J. C. De Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351472-477. [DOI] [PubMed] [Google Scholar]
- 3.de Jong, J. C., E. C. Claas, A. D. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein, S. L. 2006. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J. Infect. Dis. 19349-53. [DOI] [PubMed] [Google Scholar]
- 5.Fleischer, B., H. Becht, and R. Rott. 1985. Recognition of viral antigens by human influenza A virus-specific T lymphocyte clones. J. Immunol. 1352800-2804. [PubMed] [Google Scholar]
- 6.Flynn, K. J., G. T. Belz, J. D. Altman, R. Ahmed, D. L. Woodland, and P. C. Doherty. 1998. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity 8683-691. [DOI] [PubMed] [Google Scholar]
- 7.Gog, J. R., G. F. Rimmelzwaan, A. D. Osterhaus, and B. T. Grenfell. 2003. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc. Natl. Acad. Sci. USA 10011143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jameson, J., J. Cruz, and F. A. Ennis. 1998. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 728682-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jameson, J., J. Cruz, M. Terajima, and F. A. Ennis. 1999. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J. Immunol. 1627578-7583. [PubMed] [Google Scholar]
- 10.Kreijtz, J. H., R. Bodewes, G. van Amerongen, T. Kuiken, R. A. Fouchier, A. D. Osterhaus, and G. F. Rimmelzwaan. 2007. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25612-620. [DOI] [PubMed] [Google Scholar]
- 11.Kuiken, T., E. C. Holmes, J. McCauley, G. F. Rimmelzwaan, C. S. Williams, and B. T. Grenfell. 2006. Host species barriers to influenza virus infections. Science 312394-397. [DOI] [PubMed] [Google Scholar]
- 12.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection. Elsevier Science, Amsterdam, The Netherlands.
- 13.McMichael, A. 1994. Cytotoxic T lymphocytes specific for influenza virus. Curr. Top. Microbiol. Immunol. 18975-91. [DOI] [PubMed] [Google Scholar]
- 14.McMichael, A. J., F. M. Gotch, G. R. Noble, and P. A. Beare. 1983. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 30913-17. [DOI] [PubMed] [Google Scholar]
- 15.McMichael, A. J., C. A. Michie, F. M. Gotch, G. L. Smith, and B. Moss. 1986. Recognition of influenza A virus nucleoprotein by human cytotoxic T lymphocytes. J. Gen. Virol. 67719-726. [DOI] [PubMed] [Google Scholar]
- 16.O'Neill, E., S. L. Krauss, J. M. Riberdy, R. G. Webster, and D. L. Woodland. 2000. Heterologous protection against lethal A/HongKong/156/97 (H5N1) influenza virus infection in C57BL/6 mice. J. Gen. Virol. 812689-2696. [DOI] [PubMed] [Google Scholar]
- 17.Rimmelzwaan, G. F., M. Baars, E. C. Claas, and A. D. Osterhaus. 1998. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J. Virol. Methods 7457-66. [DOI] [PubMed] [Google Scholar]
- 18.Rimmelzwaan, G. F., N. Nieuwkoop, A. Brandenburg, G. Sutter, W. E. Beyer, D. Maher, J. Bates, and A. D. Osterhaus. 2000. A randomized, double blind study in young healthy adults comparing cell mediated and humoral immune responses induced by influenza ISCOM vaccines and conventional vaccines. Vaccine 191180-1187. [DOI] [PubMed] [Google Scholar]
- 19.Rimmelzwaan, G. F., and A. D. Osterhaus. 1995. Cytotoxic T lymphocyte memory: role in cross-protective immunity against influenza? Vaccine 13703-705. [DOI] [PubMed] [Google Scholar]
- 20.Sandbulte, M. R., G. S. Jimenez, A. C. Boon, L. R. Smith, J. J. Treanor, and R. J. Webby. 2007. Cross-reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med. 4e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smallman-Raynor, M., and A. D. Cliff. 2007. Avian influenza A (H5N1) age distribution in humans. Emerg. Infect. Dis. 13510-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279393-396. [DOI] [PubMed] [Google Scholar]
- 23.Townsend, A. R., and J. J. Skehel. 1984. The influenza A virus nucleoprotein gene controls the induction of both subtype specific and cross-reactive cytotoxic T cells. J. Exp. Med. 160552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tussey, L. G., S. Rowland-Jones, T. S. Zheng, M. J. Androlewicz, P. Cresswell, J. A. Frelinger, and A. J. McMichael. 1995. Different MHC class I alleles compete for presentation of overlapping viral epitopes. Immunity 365-77. [DOI] [PubMed] [Google Scholar]
- 25.van Baalen, C. A., D. Kwa, E. J. Verschuren, M. L. Reedijk, A. C. Boon, G. de Mutsert, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Gruters. 2005. Fluorescent antigen-transfected target cell cytotoxic T lymphocyte assay for ex vivo detection of antigen-specific cell-mediated cytotoxicity. J. Infect. Dis. 1921183-1190. [DOI] [PubMed] [Google Scholar]
- 26.Voeten, J. T., T. M. Bestebroer, N. J. Nieuwkoop, R. A. Fouchier, A. D. Osterhaus, and G. F. Rimmelzwaan. 2000. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J. Virol. 746800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. 2007. Affected areas with confirmed cases of H5N1 avian influenza since 2003, status as of 17.10.2007. http://gamapserver.who.int/mapLibrary/Files/Maps/Global_H5N1inHumanCUMULATIVE_FIMS_20071017.png. Accessed 26 October 2007.
- 28.World Health Organization. 2007. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_02_28/en/index.html. Accessed 3 March 2008.
- 29.Yewdell, J. W., J. R. Bennink, G. L. Smith, and B. Moss. 1985. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 821785-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]