Abstract
A novel variant of hepatitis B virus was identified in Vietnam. This strain (HBV-VH24) had a novel intergenotypic recombination between genotypes A, C, and G. VH24 showed high similarity (98.3 to 98.9%) to the “aberrant strains” among Vietnamese isolates reported by Hannoun et al. (C. Hannoun et al., J. Gen. Virol. 81:2267-2272, 2000) and also had similar breakpoints of recombination. Phylogenetic analysis of the complete genome of these strains formed a separate clade. Furthermore, their pre-S/S gene-encoded seven unique conserved amino acid residues were not present in other genotypes. These findings support the designation of the new genotype I.
Hepatitis B virus (HBV) infection is a global health problem. Approximately 350 million people worldwide are chronic carriers of the virus (12). Eight HBV genotypes from A to H are defined by a divergence in the entire nucleotide sequence of more than 8% (1, 14, 18, 19, 23). The prevalence and distribution of HBV genotypes vary geographically (6, 8, 9). Some genotypes have been split into subgroups. In particular, Asian strains of HBV (genotypes B and C) vary genetically and can be classified into various subgroups (21). In addition to these classifications, intergenotypic recombinant of HBV has been observed in geographical regions where there are cocirculating genotypes (2, 3, 5, 11, 20, 25, 26). We report here the complete genomic sequence and phylogenetic analyses of an HBV strain isolated in Vietnam that has shown a high divergence from these genotypes and also a novel complex recombinant.
A serum sample was obtained from a 31-year-old male Vietnamese patient who lived in Hanoi, Vietnam. He was diagnosed with acute hepatitis B at Bach Mai Hospital, Hanoi, Vietnam, in 2003. This patient was seropositive for HBsAg and immunoglobulin M anti-HBc antibody with a high level of alanine transaminase (1,990 UI/liter). PCR using HBV genotype-specific primers (16) was used to amplify the virus genotype. The full-length 3.2 kb was amplified by using single-round amplification of the full HBV genome, using the primer pair HBV4 5′-CCGGAAAGCTTATGCTCTTCTTTTTCACCTCTGCCTAATCATC-3′ (sense; underlining of the primer sequence indicates the HindIII site) and HBV4R 5′-CCGGAGAGCTCATGCTCTTCAAAAAGTTGCATGGTGCTGGTG-3′ (antisense; underlining of the primer sequence indicates the SacI site) (4). PCR and sequencing analyses were done as described previously (17). Briefly, viral DNA was extracted from 100 μl of serum by using a DNA/RNA extraction kit (SepaGene RV-R; Sanko Junyaku Co., Ltd., Tokyo, Japan). The resulting pellet was resuspended in 50 μl of RNase-free water and kept at −20°C until use. The PCR conditions consisted of preincubation at 94°C and a 2-min activation of Blend Taq-Plus DNA polymerase (Toyobo, Ltd., Tokyo, Japan), followed by 40 cycles of PCR (94°C for 15 s, 55°C for 45 s, and 72°C for 3 min 20 s), with a final extension for 7 min at 72°C. The PCR products were separated by 1% agarose gel electrophoresis and purified by using a QIAquick gel extraction kit (Qiagen, Inc., Chatsworth, CA). Purified DNA was subjected to direct sequencing using an ABI Prism BigDye terminator cycle sequencing ready reaction kit and automated an ABI 3130 DNA sequencer (Applied Biosystems, Foster City, CA). Evolutionary and phylogenetic tree analysis were performed by using MEGA software version 3.0 (10), using the neighbor-joining method, with 1,000 bootstrapped data sets. Genetic distance calculation and pairwise distance comparisons used the Kimura two-parameter model integrated into the MEGA software. Intergenotype recombination of HBV strains was investigated by using the SIMPLOT program version 3.5 (http://sray.med.som.jhmi.edu/SCRoftware/simplot/ [13]), which identified phylogenetically informative sites supporting alternative tree topologies. The recombination detection was performed by considering four sequences at a time: one putative recombinant sequence, two or three reference sequences of the original genotypes, and one sequence of a known outgroup. Each informative site supported one of three possible phylogenetic relationships among the four taxa; contiguous sites suggesting a single phylogeny were inferred to represent regions between recombination breakpoints.
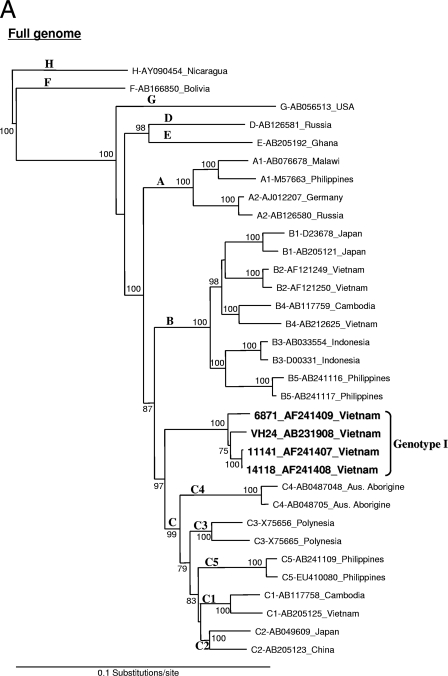
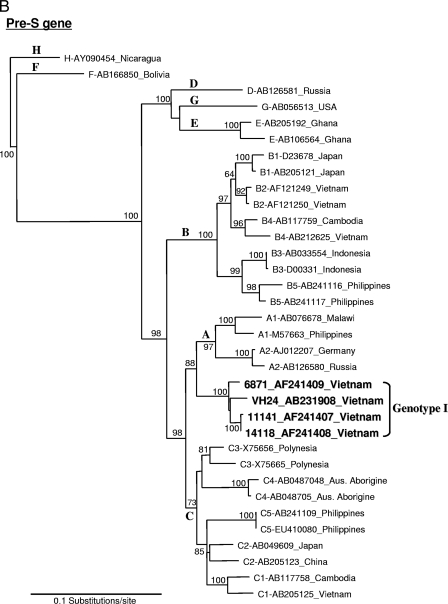
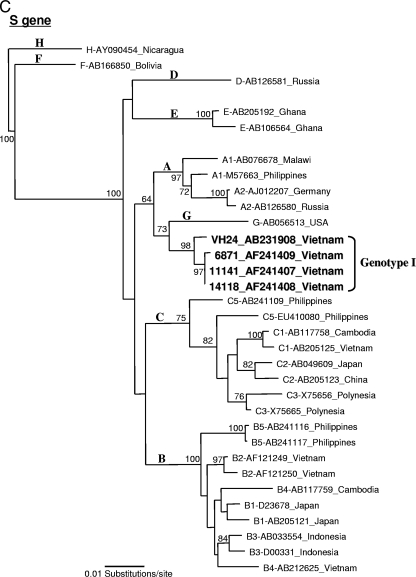
Initially, the genotyping result obtained by PCR indicated genotype A. Based on this finding, we attempted to obtain a full-length sequence of the Vietnamese HBV genotype A strain, which is rarely observed in this country. The full-length HBV genome was amplified by the long PCR method covering the entire genome by one-round PCR to avoid artificial recombination from amplifications of overlapping fragments. The complete genomic sequence of 3,215 nucleotides (nt) was recovered and denoted as HBV-VH24 (accession no. AB231908). However, phylogenetic analysis of this VH24 isolate with a full-length genome revealed a new clade suggesting a novel variant. To compare the similarity to known HBV sequences, the VH24 sequence was subjected to a homology search with BLAST (http://www.ddbj.nig.ac.jp/search/top-e.html) revealing that it had very high similarity (>98%) to the “aberrant HBV Vietnamese strains” (AF241407, AF241408, and AF241409) submitted by Hannoun et al. in 2000 (5). Further phylogenetic analysis of full-length sequences from VH24 and these three aberrant strains revealed that they formed a clade separating first from the major trunk of genotype C with a 100% bootstrap value (Fig. 1A). In addition, phylogenetic analysis using each region of pre-S and S showed that in the case of pre-S, this novel group was more closely related to genotype A, with a 100% bootstrap value, which accorded with the PCR genotyping result (Fig. 1B). On the other hand, the S gene was more closely related to the genotype G strain with a 98% bootstrap value (Fig. 1C).
FIG. 1.
Phylogenetic neighbor-joining tree constructed using full-length genome (A), pre-S gene (B), and S gene (C) of HBV, including VH24 (AB231908 in the present study) and three aberrant strains from the database (AF241407, AF241408, and AF241409). Bootstrap values were indicated at each tree roots (percentage of 1,000 bootstrap replicates that support the branch). The sequence AY090454 of genotype H was used as an outgroup. Only bootstrap values of >60% are indicated. The horizontal bar provides a genetic distance.
Compared to these three aberrant strains from the database, VH24 had very high similarity (98.3 to 98.9%) over the complete genome at the nucleotide level. Furthermore, pairwise comparison between these aberrant clade and known genotypes showed that it was most similar to genotype C (7.0% ± 0.4%, difference) but differed at least by 9% (9.5 to 14.4%) from other genotypes (Table 1). It was also noted that this new clade was the farthest cluster within genotype C. The HBV subgenotype C4 strain from Australian aborigines was considered to be the most divergent strain in genotype C (24), which has the serological subtype of ayw3. All strains in the new clade had Ile110, Thr126, and Lys160 within the S gene, which are characteristic of the serological subtype adw and thus differed from most genotype C strains, which were subtype adr. However, the S-gene nucleotide sequence of this novel clade was found to be most similar to genotype G (3.0% ± 0.6% difference) by pairwise comparison analysis and was not close to genotype C (5.7% ± 0.8%). The pre-S/S gene and the P gene of these aberrant strains showed high similarity to genotype A and clustered on the branch of this genotype (data not shown). These results suggested a recombination between genotypes A, C, and G and support the concept that this aberrant clade can be considered as a separate new genotype rather than a subgenotype. For this reason, we propose to designate this new clade genotype I.
TABLE 1.
Nucleotide distances between genotype 1 (VH24 and aberrant Vietnamese strains) and other reference genotype strains from the databasea
| Genotype | % Nucleotide distance (mean ± SD)
|
|
|---|---|---|
| Complete genome | S gene | |
| A | 9.5 ± 0.5 | 3.5 ± 0.7 |
| B | 9.7 ± 0.5 | 4.6 ± 0.8 |
| C | 7.0 ± 0.4 | 5.7 ± 0.8 |
| D | 10.4 ± 0.5 | 3.7 ± 0.7 |
| E | 10.4 ± 0.5 | 4.1 ± 0.7 |
| F | 13.8 ± 0.7 | 7.0 ± 1.0 |
| G | 10.9 ± 0.5 | 3.0 ± 0.6 |
| H | 14.4 ± 0.6 | 7.2 ± 1.0 |
| I (this study) | 1.1 ± 0.2 | 0.4 ± 0.2 |
The pairwise comparisons were performed over the complete genome and S gene.
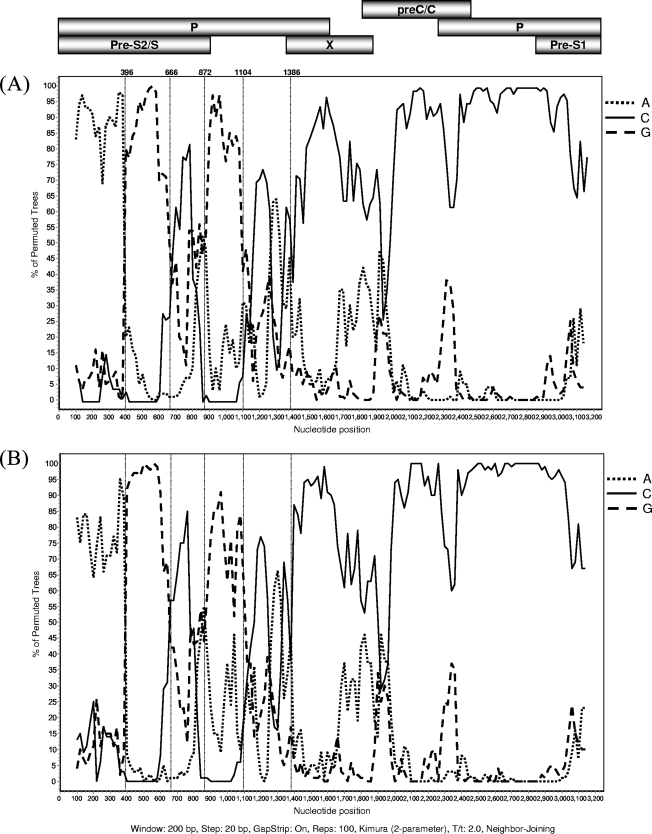
Further analysis using the Simplot program confirmed this complex recombination. The recombinant segments and breakpoints were the same in the VH24 isolate and the representative aberrant strain (AF241407) (Fig. 2). The bootscanning result showed that part of the genome (estimated to be at nt 1386 to 3215) was more similar to genotype C than to the other genotypes. The remaining part (nt 1 to 1385) differed substantially from all genotypes, although it was most similar to genotypes A and G. In segments from nt 396 to 666 and nt 872 to 1104, these strains were more similar to genotype G, and in segments from nt 1 to 396, these strains were more similar to genotype A. These recombination events were supported by high bootstrap values (P < 0.05).
FIG. 2.
Bootscan analysis demonstrating the complex recombination between genotypes A, C, and G in isolate VH24 (A) and reported aberrant Vietnamese isolate AF241407 (B). These two strains were subjected to bootscan analysis over the complete genome using the Simplot program with a 200-bp window size, 20-bp step size, and 100 bootstrap replicates, using gap-stripped alignments and neighbor-joining analysis. Strains were compared to three representative HBV genotypes: A (accession no. AB126580), C (accession no. AB049609), and G (accession no. AB056513). The dotted vertical lines showed the common estimated breakpoints of recombination in both analyzed strains.
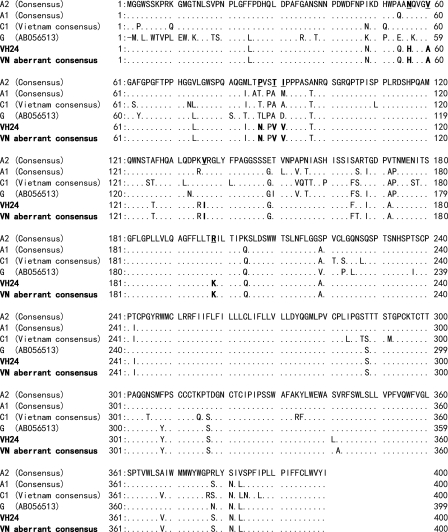
Furthermore, their pre-S or S gene, where sequence-classifying genotypes are present, encoded distinctive conserved amino acids such as His56, Ala60, Asn87, Val90, Val91, Ile136, and Lys198, which did not belong to genotypes A, C, or G (Fig. 3). This finding also supported the notion that this aberrant strain should belong to a new genotype.
FIG. 3.
Multi-alignment of deduced amino acid sequence the large S gene (amino acids 1 to 400) of genotypes A (subgenotypes A1 and A2), C (subgenotype C1 in Vietnam), and G and new variant strains. The VH24 and other aberrant strains showed identical deduced amino acids in the large S gene. Boldface characters show specific amino acid changes found in this new variant group compared to genotypes A, C, and G.
In this study, we examined an HBV isolate with a complex recombination between genotypes A, C, and G. A novel variant group comprising our isolate and three aberrant strains derived from the database were most related to the genotype C strain but had high divergence from 7 to 14.4%. Since pairwise comparisons of complete HBV genomes of intragenotypic and intergenotypic divergences ranged from 0% ± 1% to 7% ± 4% and from 6% ± 8% to 17% ± 1%, respectively (1), the four aberrant strains appear to represent a new genotype. Furthermore, in the pre-S/S gene where important region classifying genotypes are present there were seven unique conserved amino acid residues not present in other genotypes. The presence of unusual amino acids in the variant is interesting and suggests that it has undergone independent evolution, possibly since the inferred recombination events. These findings strongly support the idea of a new genotype rather than a subtype of genotype C, which is the most closely related genotype, and we proposed naming it genotype I.
Interestingly, all of the novel variant HBV strains originated from Hanoi, which is in the northern part of Vietnam, although we also investigated the southern part of Vietnam. There was a 1.1% nucleotide distance within the group with the same recombined segments and breakpoints, suggesting that the recombination may be recent or locally distributed. Thus far, the reason for the recombination of segments from different genotypes of HBV has not been discussed. Simmonds and Midgley reported the evidence of frequent intragenotypic recombination between genotypes A, D, and F/H and gibbon variants but not in B, C, or the Asian B/C recombinant group (22). Furthermore, in many cases described in that report, favored positions for both inter- and intragenotype recombination matched the positions of phylogenetic reorganization between human and ape genotypes, such as the end of the surface gene and the core gene, where sequence relationships between genotypes changed in the TreeOrder scan. To date, intergenotype recombination with genotype B/C (15, 25), C/D (3), A/D (20), A/G (7), and A/E (11) occurred in geographical regions where a number of genotypes cocirculate and resulted from mixed infections between two different genotypes. It is a notable finding that our results presented here showed a more complicated recombinant with genotype A/C/G. This finding was surprising, because the circulating HBV genotypes in Vietnam are B, C, and rarely A (27, 28). More interestingly, genotype G is quite rare in Asia and undetectable in Vietnam (6, 28). With the limited data available, the mechanism of this complex intergenotype is still unclear, although there is a report of HBV (DQ078791) with the C/G recombinant genotype in Thailand (26). By phylogenetic tree analysis of the complete genome, it was found that the DQ078791 isolate belongs to genotype G and not to the new genotype I (data not shown). In addition, the recombination breakpoints are quite different between them: an overlapping region (S gene) in Vietnamese strains and a nonoverlapping region (precore/core gene) in a Thai strain. Indeed, the sites and patterns of recombination seem to be different depending on the corresponding genotypes and the geographical area (3, 11, 25). Therefore, additional data on this complex intergenotypic recombination are still needed to ascertain the order of recombination events.
In conclusion, we reported here a novel variant of HBV (HBV-VH24) isolated in Vietnam. The VH24 had a novel intergenotypic recombinant with genotype A/C/G. The VH24 and the “aberrant strains” in Vietnam reported in 2000 had similar breakpoints of recombination among them. The findings of the phylogenetic analysis, pairwise comparison, and unique conserved amino acids in the pre-S/S gene suggested the designation of a new genotype, for which the nomenclature of genotype I is proposed.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 832059-2073. [DOI] [PubMed] [Google Scholar]
- 2.Bowyer, S. M., L. van Staden, M. C. Kew, and J. G. Sim. 1997. A unique segment of the hepatitis B virus group A genotype identified in isolates from South Africa. J. Gen. Virol. 781719-1729. [DOI] [PubMed] [Google Scholar]
- 3.Cui, C., J. Shi, L. Hui, H. Xi, et al. 2002. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J. Gen. Virol. 832773-2777. [DOI] [PubMed] [Google Scholar]
- 4.Gunther, S., B. C. Li, S. Miska, D. H. Kruger, H. Meisel, and H. Will. 1995. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 695437-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannoun, C., H. Norder, and M. Lindh. 2000. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J. Gen. Virol. 812267-2272. [DOI] [PubMed] [Google Scholar]
- 6.Huy, T. T., and K. Abe. 2004. Molecular epidemiology of hepatitis B and C virus infections in Asia. Pediatr. Int. 46223-230. [DOI] [PubMed] [Google Scholar]
- 7.Kato, H., E. Orito, R. G. Gish, N. Bzowej, M. Newsom, F. Sugauchi, S. Suzuki, R. Ueda, Y. Miyakawa, and M. Mizokami. 2002. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology 35922-929. [DOI] [PubMed] [Google Scholar]
- 8.Kidd-Ljunggren, K., Y. Miyakawa, and A. H. Kidd. 2002. Genetic variability in hepatitis B viruses. J. Gen. Virol. 831267-1280. [DOI] [PubMed] [Google Scholar]
- 9.Kramvis, A., M. Kew, and G. Francois. 2005. Hepatitis B virus genotypes. Vaccine 232409-2423. [DOI] [PubMed] [Google Scholar]
- 10.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 11.Kurbanov, F., Y. Tanaka, K. Fujiwara, F. Sugauchi, D. Mbanya, L. Zekeng, N. Ndembi, C. Ngansop, L. Kaptue, T. Miura, E. Ido, M. Hayami, H. Ichimura, and M. Mizokami. 2005. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J. Gen. Virol. 862047-2056. [DOI] [PubMed] [Google Scholar]
- 12.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 3371733-1745. [DOI] [PubMed] [Google Scholar]
- 13.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnius, L. O., and H. Norder. 1995. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology 3824-34. [DOI] [PubMed] [Google Scholar]
- 15.Morozov, V., M. Pisareva, and M. Groudinin. 2000. Homologous recombination between different genotypes of hepatitis B virus. Gene 26055-65. [DOI] [PubMed] [Google Scholar]
- 16.Naito, H., S. Hayashi, and K. Abe. 2001. Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J. Clin. Microbiol. 39362-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima, A., M. Usui, T. T. Huy, N. Masaki, T. Sata, and K. Abe. 2005. Full-length sequence of hepatitis B virus belonging to genotype H identified in a Japanese patient with chronic hepatitis. Jpn. J. Infect. Dis. 58244-246. [PubMed] [Google Scholar]
- 18.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198489-503. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto, H., F. Tsuda, H. Sakugawa, R. I. Sastrosoewignjo, M. Imai, Y. Miyakawa, and M. Mayumi. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 692575-2583. [DOI] [PubMed] [Google Scholar]
- 20.Owiredu, W. K., A. Kramvis, and M. C. Kew. 2001. Hepatitis B virus DNA in serum of healthy black African adults positive for hepatitis B surface antibody alone: possible association with recombination between genotypes A and D. J. Med. Virol. 64441-454. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer, S. 2007. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J. Gastroenterol. 1314-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmonds, P., and S. Midgley. 2005. Recombination in the genesis and evolution of hepatitis B virus genotypes. J. Virol. 7915467-15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 8167-74. [DOI] [PubMed] [Google Scholar]
- 24.Sugauchi, F., M. Mizokami, E. Orito, T. Ohno, H. Kato, S. Suzuki, Y. Kimura, R. Ueda, L. A. Butterworth, and W. G. Cooksley. 2001. A novel variant genotype C of hepatitis B virus identified in isolates from Australian aborigines: complete genome sequence and phylogenetic relatedness. J. Gen. Virol. 82883-892. [DOI] [PubMed] [Google Scholar]
- 25.Sugauchi, F., E. Orito, T. Ichida, H. Kato, H. Sakugawa, S. Kakumu, T. Ishida, A. Chutaputti, C. L. Lai, R. Ueda, Y. Miyakawa, and M. Mizokami. 2002. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J. Virol. 765985-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suwannakarn, K., P. Tangkijvanich, A. Theamboonlers, K. Abe, and Y. Poovorawan. 2005. A novel recombinant of hepatitis B virus genotypes G and C isolated from a Thai patient with hepatocellular carcinoma. J. Gen. Virol. 863027-3030. [DOI] [PubMed] [Google Scholar]
- 27.Thuy, L. T. T., H. Ryo, L. Van Phung, K. Furitsu, and T. Nomura. 2005. Distribution of genotype/subtype and mutational spectra of the surface gene of hepatitis B virus circulating in Hanoi, Vietnam. J. Med. Virol. 76161-169. [DOI] [PubMed] [Google Scholar]
- 28.Tran, H. T., H. Ushijima, V. X. Quang, N. Phuong, T. C. Li, S. Hayashi, X. Lien, T. Sata, and K. Abe. 2003. Prevalence of hepatitis virus types B through E and genotypic distribution of HBV and HCV in Ho Chi Minh City, Vietnam. Hepatol. Res. 26275-280. [DOI] [PubMed] [Google Scholar]