Abstract
Scaffolding proteins of spherical prokaryotic and eukaryotic viruses have critical roles in capsid assembly. The primary scaffolding components of cytomegalovirus, called the assembly protein precursor (pAP, pUL80.5) and the maturational protease precursor (pPR, pUL80a), contain two nuclear localization sequences (NLS1 and NLS2), at least one of which is required in coexpression experiments to translocate the major capsid protein (MCP, pUL85) into the nucleus. In the work reported here, we have mutated NLS1 and NLS2, individually or together, in human cytomegalovirus (HCMV, strain AD169) bacmid-derived viruses to test their effects on virus replication. Consistent with results from earlier transfection/coexpression experiments, both single-mutant bacmids gave rise to infectious virus but the double mutant did not. In comparisons with the wild-type virus, both mutants showed slower cell-to-cell spread; decreased yields of infectious virus (3-fold lower for NLS1− and 140-fold lower for NLS2−); reduced efficiency of pAP, pPR, and MCP nuclear translocation (sixfold lower for NLS1− and eightfold lower for NLS2−); increased amounts of a 120-kDa MCP fragment; and reduced numbers of intranuclear capsids. All effects were more severe for the NLS2− mutant than the NLS1− mutant, and a distinguishing feature of cells infected with the NLS2− mutant was the accumulation of large, UL80 protein-containing structures within the nucleus. We conclude that these NLS assist in the nuclear translocation of MCP during HCMV replication and that NLS2, which is unique to the betaherpesvirus UL80 homologs, may have additional involvements during replication.
As with all herpesviruses, capsid assembly and DNA packaging for human cytomegalovirus (HCMV) takes place in the nucleus (6, 13, 29). The capsid is organized into a shell composed predominantly of the major capsid protein (MCP, pUL86; 150 kDa), which forms the capsomeres, and the minor capsid protein (mCP, pUL85; 35 kDa) and mCP-binding protein (mC-BP, pUL46; 33 kDa), which together form triplexes that interface with the capsomeres (4, 19, 28). The assembly protein precursor (pAP, pUL80.5; 38 kDa) and the related protease precursor (pPR, pUL80a; 74 kDa) (Fig. 1) coordinate the capsid assembly process as self-interacting elements of an internal scaffold and ultimately are eliminated to accommodate the viral DNA (7, 13, 20). Because all of these proteins must enter the nucleus on their own or in complex with an escort, the process of the nuclear transport of viral proteins is a potential control point in virus replication.
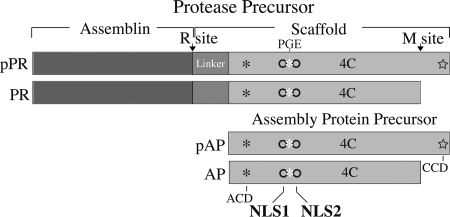
FIG. 1.
HCMV UL80a and UL80.5 proteins. Shown here are schematic representations of HCMV pPR and pAP and their M-site-cleaved products (PR and AP). The catalytic (Assemblin) and scaffolding (Scaffold) portions of pPR are shown, and the portion of the pAP amino acid sequence that is identical to that of the carboxyl 51% of pPR is indicated by light gray shading. A linker sequence (intermediate gray) is present between the assemblin and pAP sequences of pPR. NLS1 and NLS2 are represented as empty circles, with the conserved Pro-Gly-Glu sequence (25, 27) indicated as a white asterisk between them; the positions of the amino-conserved domain (ACD; asterisk), carboxyl-conserved domain (CCD; star), tetraCys motif (4C), and the release (R) and maturational (M) cleavage sites are also indicated.
Movement of the HCMV MCP from the cytoplasm into the nucleus, for example, requires its interaction with pAP or pPR (35). Work done with the simian CMV (SCMV) pAP homolog identified two simian virus 40 T-antigen-like nuclear localization signals (NLS) toward its carboxyl end. One of these (NLS1) has a counterpart in all herpesvirus pAP homologs, but the other (NLS2) appears to be present only in betaherpesvirus homologs (25). By using site-directed mutagenesis, we showed that the nuclear translocation of SCMV pAP (and the related pPR) requires either NLS1 or NLS2, and that when both were disabled its own nuclear translocation was blocked, as was its ability to mediate the nuclear translocation of MCP (25).
Based on these results and others from GAL4 two-hybrid assays, and supported by more recent studies with the purified proteins (21), a working model was proposed for the early functions of the CMV pAP (13, 35). Specific features of the model are that (i) pAP interacts with MCP in the cytoplasm, (ii) this interaction is mediated by the carboxyl-conserved domain of pAP, (iii) the pAP-MCP interaction is promoted or stabilized by an amino-conserved domain of pAP, and (iv) NLS in pAP are required to translocate pAP-MCP complexes into the nucleus (Fig. 1).
In the work reported here, we have mutated NLS1 and NLS2 (singly or together) in an HCMV bacmid and determined the impact of these mutations on the outgrowth of infectious virus from the bacmids, on virus assembly and infectivity, and on the nuclear translocation of pAP, pPR, and MCP (and five other virion proteins). Our results demonstrate the importance of these NLS in CMV replication; show that inactivating NLS2 is more detrimental to replication than inactivating NLS1, suggesting it has a different or additional function; and uncover an apparently site-specific cleavage of MCP correlating with its slower nuclear translocation in mutant-infected cells.
MATERIALS AND METHODS
Cells and transfection.
Human foreskin fibroblast (HFF) cells were maintained in Dulbecco's modified Eagle's medium containing high glucose (no. 11965; Gibco/BRL, Bethesda, MD), 10% fetal bovine serum (HyClone, Logan, UT) penicillin (100 U/ml), and streptomycin (100 U/ml) (complete DMEM). Transfections were done in 6-well plastic plates containing HFF cells seeded to a density of ∼50% confluence. The next day, plasmids were combined with FuGENE 6 transfection reagent (no. 11815091001; Roche, Indianapolis, IN) and added to the cells according to the manufacturer's suggested protocol.
Preparation of UL80 NLS mutant viruses.
WT and mutant viruses were derived from parental HCMV (strain AD169) bacmids, either encoding or not encoding an enhanced green fluorescent protein (GFP, regulated by the HCMV UL123 promoter) marker (AD169-US2-11-EGFP-loxP and AD169RVHB5/pHB5, respectively) (5). Both were obtained from U. Koszinowski, M. Messerle, and G. Hahn. The NLS1 and NLS2 sequences were mutated singly or together by homologous recombination using the GFP-encoding bacmid, appropriate mutagenic primers, and Gam-, Beta-, and Exo-expressing Escherichia coli DY380 (18). The NLS1 target sequence in UL80 was knocked out using a recombination cassette, composed of the streptomycin-sensitive RpsL gene linked to the neomycin gene, in plasmid pRpsL-neo (K002; Gene Bridges, Dresden, Germany) and the NLS1 flanking sequences. The RpsL-neomycin cassette was replaced in a second recombination by using a mutagenic double-strand oligonucleotide that changed only the NLS1 coding sequence (i.e., from AAACGCCGTAAG1539 to CAACAGCAGCAA1539), substituting glutamine (Gln) for lysine (Lys) and arginine (Arg) in the original NLS1 amino acid sequence KRRK513 (2, 8, 26, 36). The resulting mutant bacmid is called bHCMV-UL80NLS1−.
NLS2 was mutated in the same way, but with a mutagenic oligonucleotide that changed the NLS2 coding sequence from CGGGCACGAAAGCGTCTAAAA1629 to CAGGCACAACAGCAACTACAA1629, substituting Gln for Lys and Arg in the original NLS2 amino acid sequence RARKRLK543. The resulting mutant bacmid was called bHCMV-UL80NLS2−. A third bacmid, with both NLS1 and NLS2 mutated, was derived from bHCMV-UL80NLS2− by introducing the NLS1− mutation described above to give the double-mutant bacmid bHCMV-UL80NLS1−/2−.
Bacmids were then nucleofected into HFF cells by using the amaxa system (amaxa Biosystems, Cologne, Germany) with 100 μl of VPD-1001 solution (amaxa), 0.5 × 106 to 1.0 × 106 HFF cells, and 3 to 4 μg of DNA. Cytopathic effects (CPE) typical of HCMV were observed by 3 weeks in all cultures, except those nucleofected with the double mutant NLS1−/2− (see Results). Stocks of the viruses were prepared when extensive CPE was observed (usually within 4 to 5 weeks of transfection).
The mutant phenotypes were validated in three ways (data not shown). First, the UL80 open reading frames were PCR amplified from virus particles in sucrose gradient fractions following the rate-velocity sedimentation of material in the culture medium from cells infected with WT, NLS1−, or NLS2− virus. The DNA products were sequenced and found to have no nucleotide changes other than those intended for the mutations. Second, the NLS1− and NLS2− mutant bacmids were repaired to WT UL80 sequences, and virus reconstituted from each regained WT growth characteristics, including minimal production of the MCP fragment. Repair of the NLS1−/2− double mutant to the NLS1− single mutation restored its ability to replicate but left it with the characteristic overproduction of the MCP fragment. A silent nucleotide change was introduced into each revertant to distinguish it from the WT (i.e., NLS1 Arg codon 511, CGC→CGA; and NLS2 Lys codon 540, AAG→AAA). Third, the NLS1−, NLS2−, and NLS1−/2− mutants were all reconstructed using nonconservative Ala substitutions in place of the more conservative Gln substitutions. The resulting Ala-substituted NLS1− and NLS2− mutant viruses have the same growth phenotypes as those observed for the Gln-substituted mutants: WT > NLS1− >>> NLS2− (S. Fernandes, J. Wang, and W. Gibson, data not shown).
The NLS1− and NLS2− mutations described above also were introduced into a bacmid encoding UL80 proteins tagged with a tetracysteine (tetraCys) motif (CCPGCC) that enabled them to be stained with the biarsenical dye FIAsH (see below) in live infected cells (1, 32). This was done by repeating the cloning steps described above, but with a parental bacmid lacking the GFP coding sequence and encoding UL80 proteins that contain the tetraCys motif inserted between Ser474 and Gly476 (bHCMV-UL80-4Cys) (21). These mutant bacmids are called bHCMV-UL80-4Cys/NLS1− and bHCMV-UL80-4Cys/NLS2−.
All mutations and repairs were verified by the automated sequence analysis of PCR products prepared from the recombinant bacmid genes.
Plasmid construction.
Plasmids expressing the HCMV pAP fused to the carboxyl end of GFP were constructed for WT pAP and for the NLS1−, NLS2−, and NLS1−/2− pAP mutants (described above) by using the following two-step procedure. First, PCRs using SureTaq (no. CB-4095; Denville Scientific Inc., Metuchen, NJ) were performed to obtain full-length DNA sequences of the UL80.5 gene from the WT, NLS1−, NLS2−, and NLS1−/2− bacmids described above. The resulting 1.2-kb PCR products were gel purified and blunt-end ligated into Zero-blunt (no. 44-032; Invitrogen, Carlsbad, CA), a linearized vector with EcoRI and XbaI restriction sites flanking the insertion site. The WT and mutant genes were then subcloned into the EcoRI/XbaI sites of pEGFP-C2 (no. 6083-1; Clontech, Mountain View, CA) to make the four GFP-pAP fusion constructs, called GFP-pAP/WT, GFP-pAP/NLS1−, GFP-pAP/NLS2−, and GFP-pAP/NLS1−/2−. The pAP sequences of all four plasmids were verified for fidelity and correct in-frame fusion with the GFP coding region. The subcloning procedure resulted in an insertion of 14 amino acids (SGRTQISSSSFEFR) between the end of GFP and the starting Met of pAP for all constructs.
Infectivity assays.
Titers of GFP-marked virus in stock preparations cleared of large particulate material by microcentrifugation (16,000 × g, 1 min at room temperature) were determined by infecting HFF cells in 96-well plates with serial fivefold dilutions (50 μl of inoculum per well), as described before (21). Infected cells were monitored by fluorescence microscopy for the expression of the GFP marker from day 3 to 6 after infection, and the virus titer was calculated as the average number of foci expressing GFP (i.e., plaques) per well (in triplicate) multiplied by the dilution factor (i.e., PFU per ml).
The spread of infection by GFP-marked viruses was monitored by fluorescence microscopy after infecting HFF cells in 96-well culture plates at a low multiplicity of infection (MOI; ∼1), as described before (21).
A time course assay of virus production was done by infecting subconfluent cultures of HFF cells in 10-cm dishes with either WT or mutant virus at an MOI of ∼1 (i.e., 517 μl WT, 215 μl NLS1−, and 2,873 μl NLS2−). On days 1 to 8 after infection, 1 ml of growth medium was removed from each culture, clarified by microcentrifugation, serially diluted in fivefold increments, and stored at −80°C until all samples were collected. Fresh medium (1 ml) was added back to each culture after the sampling. Virus titers in samples from each time point were calculated, all as described before (21).
Pulse-chase radiolabeling and immunoprecipitation.
Cells in 24-well culture plates were infected with WT, NLS1−, or NLS2− virus or left uninfected. Seven days later, the culture medium was replaced with radiolabeling medium (60 μCi [35S]Met/Cys in DMEM containing 10% of the normal amounts of Met and Cys). One hour later, the labeling medium was removed and replaced with complete DMEM. Cells from one set of wells (i.e., uninfected, WT, NLS1−, and NLS2−) were dislodged into the medium and collected by microcentrifugation at 4°C. The resulting cell pellets were suspended in 50 μl of NP-40 buffer (0.5% NP-40, 40 mM phosphate buffer, 150 mM NaCl, pH 7.4), placed in ice for 5 min, and subjected to microcentrifugation at 4°C to pellet the NP-40 nuclear fraction (11). The NP-40 cytoplasmic fraction (supernatant) was removed and saved; the NP-40 nuclei were suspended in 50 μl of NP-40 buffer and dispersed by vortex mixing. The resulting, operationally defined nuclear and cytoplasmic fractions (24) were stored at −80°C.
To reduce pools of radioactive Met and Cys during the chase period, the growth medium in the remaining cultures was replaced again ∼10 and 30 min after radiolabeling and three more times before the 4-h time point. Four more sets of cells (i.e., uninfected, WT, NLS1−, and NLS2−) were similarly collected and separated into NP-40 cytoplasmic and nuclear fractions 4, 8, 24, and 48 h after removing the radiolabeling medium. All samples were stored at −80°C.
In preparation for immunoprecipitation, the NP-40 fractions were brought to final concentrations of 2% sodium dodecyl sulfate (SDS) and 50 mM dithiothreitol (DTT) and incubated at room temperature for ∼5 min to denature MCP and make it available to the antibody. After microcentrifugation at room temperature for 2 min to pellet insoluble material, SDS and DTT concentrations were reduced by subjecting 30-μl aliquots of each fraction to microcentrifugal desalting (spin column no. 89849; Pierce, Rockford, IL) per the manufacturer's instructions. The resulting material was combined with 20 μl of anti-MCP rabbit anti-peptide antibody (14) and incubated at room temperature for 90 min. Antibody complexes were recovered by adding 70 μl of a 1:1 slurry of protein A beads; incubating the mixture for 60 min at room temperature with rocking; and washing the beads four times with phosphate-buffered saline (PBS) lacking calcium and magnesium (CMF-PBS) but containing 0.5% deoxycholate and 1.0% NP-40, and then once with CMF-PBS (34). Proteins were released from the beads by adding 30 μl of protein sample buffer. The suspension was heated in a boiling water bath for 3 min to dissociate the immunoglobulin G molecules.
PAGE and Western immunoassay.
Proteins were separated by electrophoresis in 4% to 12% polyacrylamide gradient gels (NP0323BOX; Invitrogen) containing SDS and using 2-(N-morpholino)ethanesulfonic acid electrode buffer (MES; NP0002; Invitrogen) (SDS-PAGE). Following electrophoresis, the proteins were transferred to a polyvinyl difluoride membrane (no. LC2002; Invitrogen) for Western immunoassays. Western immunoassays were done as described before (21), using rabbit polyclonal antibodies to MCP (anti-MCP [12]), mCP (anti-mCP [12]), mC-BP (anti-mC-BP), pAP (anti-pAPhr [21]), BPP (anti-BPP [15]), and high-molecular-weight protein (HMWP, pUL48) (anti-UL48c [33]). Also used were mouse monoclonal antibodies to the lower matrix protein (LM, pp65, pUL83) (anti-LM) and to the smallest capsid protein (SCP, pUL48/49) (anti-SCP, no. 11-21-23) from Gary Pearson and Bill Britt, respectively. 125I-protein A (no. NEX146L; PerkinElmer Life and Analytical Sciences, Waltham, MA) was used as the secondary reagent (following rabbit anti-mouse immunoglobulin G for monoclonal antibodies) to visualize bound antibodies and was detected and quantified by phosphorimaging (BAS-2500 with ImageGauge and ImageQuant v4.22; Fuji Medical Systems, Stamford, CT). Membranes that were probed more than once were incubated in hot SDS-β-EtSH, rinsed, and blocked with bovine serum albumin between immunoassays (17).
Microscopy.
Fluorescence microscopy to document the spread of GFP-marked viruses was done using a Nikon Eclipse TE200 inverted microscope equipped with a Sony DKC5000 camera as described before (21).
The procedures used to visualize tetraCys-tagged proteins with FIAsH (Green Lumio; catalog no. 12589-057; Invitrogen), DNA with 4′,6′-diamidino-2-phenylindole (DAPI; 25 μg/ml in PBS), and virus particles by heavy-metal staining (i.e., osmium tetraoxide, uranyl acetate, and lead citrate) in 80- to 90-nm thin sections of Epon resin-embedded infected cells have been described before (21, 33). Confocal microscopy was done using an UltraViewTM LCI confocal system equipped with a Zeiss Axiovert 200 microscope. Transmission electron microscopy was done using a Hitachi 7600 microscope equipped with a DVC1412M-FW digital camera system and the AMTV542 software program.
RESULTS
NLS1 or NLS2 is required to translocate pAP-GFP fusion protein into the nucleus.
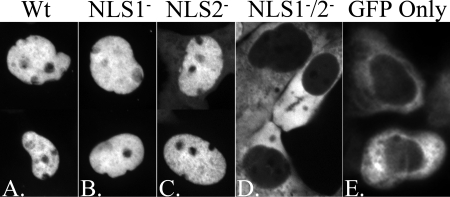
NLS1 and NLS2 were identified functionally in the SCMV homolog of pAP and were studied in transfection experiments using mutants with Ala substitutions for the NLS Lys and Arg residues (25). It was found that nuclear translocation was blocked only when both NLS were disabled. To verify the functionality of these NLS in the HCMV pAP, and to determine whether Gln could be used instead of Ala as a more conservative replacement for the NLS Arg and Lys residues (2, 8, 26, 36), WT and the NLS1− and NLS2− mutants of HCMV pAP were expressed as fusions with GFP and tested for nuclear translocation in transfection experiments. Fluorescence microscopy done on days 3 through 6 after transfection showed that WT GFP-pAP (∼69 kDa) was located predominantly within the nucleus, whereas GFP alone was predominantly cytoplasmic (Fig. 2A and E). Mutating either NLS1 or NLS2 alone was insufficient to block nuclear translocation (Fig. 2B and C), but when both were disabled the mutant GFP-pAP protein was excluded from the nucleus (Fig. 2D).
FIG. 2.
Disruption of NLS1 and NLS2 together (NLS1−/2−) is required to block the nuclear translocation of GFP-pAP fusion protein in transfected cells. HFF cells were transfected with plasmids encoding fusions of GFP with WT pAP (WT), NLS1−, NLS2−, or NLS1−/2− or encoding GFP only. Shown are fluorescence images of representative cells from each culture taken 3 to 6 days after transfection. (A to C) Composites of two different cells each, showing predominantly nuclear fluorescence. Although not easily seen in these images, cells expressing the NLS2− construct showed perceptibly more cytoplasmic fluorescence than those expressing the WT or NLS1− constructs. (D and E) Single fields with predominantly cytoplasmic fluorescence; nuclei (verified by phase-contrast microscopy) are dark ovals surrounded by GFP fluorescence.
Loss of NLS function affects virus growth.
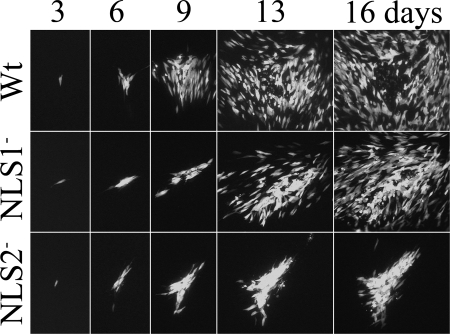
Since the Gln substitutions disabled the HCMV NLS and gave results (Fig. 2) comparable to those obtained for Ala-substituted SCMV pAP (25), we tested the effects of these mutations on HCMV replication. HCMV bacmids and viruses with the same NLS1−, NLS2−, and NLS1−/2− substitutions were tested for their ability to give rise to virus. A GFP marker to identify cells containing replicating viral DNA was expressed by all bacmids (and viruses) within 3 days of nucleofection. Infection failed to propagate from cells nucleofected with the NLSI−/2− double mutant, but by 6 days, the NLS1− and NLS2− mutants began spreading to adjacent cells, as indicated by the appearance of GFP fluorescence (Fig. 3). As the infection progressed, it became evident that the NLS1− mutant spread more slowly than the WT virus (e.g., ∼3-day lag) and that spread of the NLS2− mutant was severely impeded, appearing comparatively more dependent on direct cell-to-cell transmission (i.e., few foci arising away from the initially nucleofected cell), and showing slower progression to full CPE (e.g., cell rounding, detachment, and rupture) (Fig. 3). Bacmid-derived viruses having nonconservative Ala replacements for the NLS1 and NLS2 basic amino acids showed relative rates of spread comparable to those of the more conservative Gln replacements (i.e., WT > NLS1− >>> NLS2−; S. Fernandes et al., data not shown).
FIG. 3.
NLS2− mutant spreads more slowly than either WT virus or the NLS1− mutant. HFF cells were infected with low multiplicities of WT virus or the NLS1− or NLS2− mutant. All viral genomes contained an ectopic copy of the GFP gene as a marker for the presence and replication of viral DNA. Shown are fluorographic images of the cultures taken 3, 6, 9, 13, and 16 days after infection. The same plaques were photographed on each of the five days. Note comparatively larger WT focus on days 6 to 16, loss of fluorescent cells from the center of WT focus by day 16, and comparatively high density of fluorescent cells in the NLS2− focus on days 9 to 16.
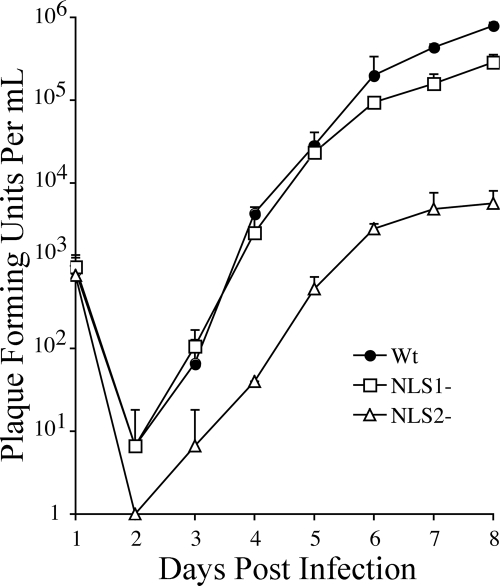
Stocks of WT virus and the NLS1− and NLS2− mutants were prepared, their titers were determined, and an experiment was done to compare the time course of virus production for each following low-multiplicity infections of ∼1 PFU/cell. The NLS2− mutant showed an ∼10-fold more pronounced drop in titer during eclipse than WT or the NLS1− mutant (Fig. 4). Eight days after infection, the titers of the NLS1− (2.9 × 105) and the NLS2− (5.7 × 103) mutants were ∼3-fold and ∼140-fold lower than that of WT (7.9 × 105). Thus, both NLS mutations reduced production of infectious virus, but the effect of mutating NLS2 was more severe (Fig. 4).
FIG. 4.
Production of infectious virus is reduced for both NLS mutants but more strongly for the NLS2− mutant. Samples collected on days 1 to 8 from HFF cultures infected at an MOI of ∼1 PFU/cell with virus encoding WT UL80 proteins (WT) or the NLS1− or NLS2− UL80 mutants (NLS1− or NLS2−) were processed and assayed for infectious virus as described in Results and Materials and Methods. The changing amount of virus present [(number of GFP-expressing foci)(dilution factor) = PFU/ml] in the culture medium each day following infection is shown. Data points and standard deviation bars represent the averages from three replicate samples.
In related pilot experiments for which data are not shown, several attempts were made to recover extracellular virus particles from cells infected with the NLS1− or NLS2− mutant. Following sedimentation in sucrose gradients (3), NLS1− preparations gave light-scattering bands of all three types of particles recovered from cells infected with WT virus (i.e., noninfectious enveloped particles [NIEPs], virions, and dense bodies; see reference 16). In contrast but consistent with the low level of infectious virus produced, cells infected with the NLS2− mutant yielded an indistinct virion band and a generally slow-sedimenting, heterogeneous population of light-scattering particles, suggestive of an assembly defect. When the gradients were collected by fractionation (3) and probed by Western immunoassay using anti-MCP to detect capsid-containing particles, the highest concentrations of MCP for the NLS1− mutant spanned approximately five fractions at the relative positions expected for NIEPs and virions, both of which contain capsids. The highest concentrations of MCP for the NLS2− mutant also were distributed over approximately five fractions but not as far into the gradient as those for the NLS1− mutant. Material taken from fractions of each gradient over the region considered most likely to include virions (i.e., based on the distribution of MCP) was subjected to PCR amplification of the UL80 gene. The full-length UL80 DNA product was obtained from both NLS1− and NLS2− particles, subjected to automated sequence analysis, and verified to contain the intended base changes and no others. We interpret these initial but reproducible findings to indicate that cells infected with either the NLS1− or NLS2− mutant release virus particles that contain viral DNA (e.g., particles and DNA cosediment), but that those released by the NLS2− mutant sediment aberrantly and therefore are likely to be structurally different than WT or NLS1− NIEPs and virions.
NLS mutations interfere with nuclear localization of pAP, pPR, and MCP.
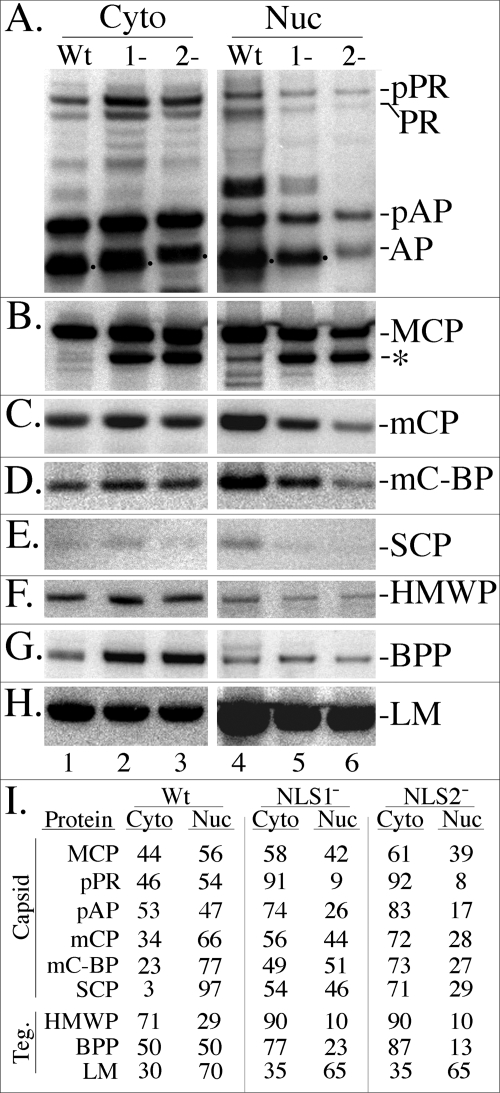
To determine how the NLS mutations affect the nuclear/cytoplasmic partitioning of the UL80 proteins, cells were infected at an MOI of ∼1 with WT virus or with the NLS mutants. Ten days later, NP-40 nuclear and cytoplasmic fractions were prepared from the cultures and subjected to SDS-PAGE, followed by Western immunoassay with anti-pAPhr to detect pAP and pPR. Phosphorimaging showed that pPR and pAP were about evenly distributed between the cytoplasmic and nuclear fractions in WT-infected cells, but a substantially greater percentage partitioned with the cytoplasmic fraction of cells infected with either mutant (∼90% for pPR and ∼75 to 85% for pAP), the difference being more pronounced for the NLS2− mutant (Fig. 5A, I).
FIG. 5.
Steady-state, intracellular distribution of pPR, pAP, MCP, and other capsid and tegument (Teg.) proteins in cells infected with WT virus or the UL80 NLS mutants. HFF cells in 6-well plates were infected with WT, NLS1−, or NLS2− virus, scraped into the growth medium, collected by microcentrifugation at 4°C, and separated into NP-40 cytoplasmic and nuclear fractions as described in Materials and Methods for cells from 24-well plates. The samples were subjected to SDS-PAGE and analyzed by Western immunoassay as described in Results and Materials and Methods. Shown are images of the resulting membranes probed with antibodies as follows. (A) The UL80 proteins, including pPR and pAP and their cleavage products (PR and AP), were detected with anti-pAPhr; (B) MCP and a 120-kDa fragment (asterisk) were detected with anti-MCP; (C) mCP was detected with anti-mCP; (D) mC-BP was detected with anti-mC-BP; (E) SCP was detected with anti-SCP; (F) HMWP was detected with anti-pUL48c; (G) BPP/pp150 was detected with anti-BPP; and (H) LM/pp65 was detected with anti-LM. (I) Shown are the percentages of each protein in the NP-40 cytoplasmic (Cyto) and NP-40 nuclear (Nuc) fractions of the infected cells, calculated from the phosphorimages shown in the panels A to H.
It was concluded from transfection assays done with the SCMV pAP that mutating either NLS1 or NLS2 reduced its ability to translocate MCP into the nucleus (25). A replica membrane containing the samples described above was prepared and probed in parallel with antibodies to MCP to determine whether MCP nuclear translocation was also affected in cells infected with the UL80 NLS mutant viruses. Phosphorimaging showed that ∼60% of MCP was in the nuclear fraction and ∼40% in the cytoplasmic fraction of cells infected with WT virus (Fig. 5B, I). The distribution was reversed for both NLS mutants (∼40% nuclear, ∼60% cytoplasmic). In addition, a ∼120-kDa protein, cross-reactive with anti-MCP and present in small amounts in the nuclear fraction of WT-infected cells (2% of total MCP plus fragment), was dramatically increased in both the nuclear and cytoplasmic fractions of cells infected with the NLS mutants (NLS1−, 40% of total MCP plus fragment; NLS2−, 50% of total MCP plus fragment) (Fig. 5B).
Nuclear translocation of MCP is slower for NLS mutants.
A pulse-chase radiolabeling experiment was done to determine whether the altered nuclear/cytoplasmic distribution of MCP is due to its slower translocation in mutant-infected cells and to investigate the origin of the cross-reactive, 120-kDa fragment. It was necessary to use immunoprecipitation with anti-MCP to resolve MCP and the fragment from cellular proteins for quantification and to first denature MCP (with SDS and DTT) to expose the peptide sequence recognized by anti-MCP. Results of the experiment show that most of the newly synthesized MCP (96% of the total) was in the cytoplasmic fraction of the samples taken immediately after the 60-min labeling period and then decreased (to 32% at 48 h) in correspondence with an increase in the nuclear fraction (Fig. 6A, B). By 6.5 h after the labeling period, 50% of MCP was in the nuclear fraction of cells infected with WT virus, and a steady-state distribution of 40% cytoplasmic to 60% nuclear was established ∼8 h after labeling (Fig. 6B and 5B and I). It took nearly six times longer for 50% of MCP to associate with the nuclear fraction in cells infected with the NLS1− mutant and approximately eight times longer in cells infected with the NLS2− mutant (Fig. 6B). Interestingly, but for undetermined reasons, the rate of MCP translocation during the initial 8 h of these biphasic plots was faster and more similar between viruses. In WT virus-infected cells, the amount of MCP in the nuclear fraction increased at approximately 8% per hour, and in cells infected with either mutant it increased at one-half to one-third that rate (∼3% per hour).
FIG. 6.
Movement of MCP from the cytoplasmic to the nuclear fraction slowed in cells infected with NLS mutants. HFF cells infected with WT, NLS1−, or NLS2− virus were pulse radiolabeled for 1 h with [35S]Met/Cys, collected, separated into NP-40 cytoplasmic (Cyto) and nuclear (Nuc) fractions, subjected to immunoprecipitation using anti-MCP, and analyzed by phosphorimaging following SDS-PAGE, all as described in Results and Materials and Methods. (A) Phosphorimages of the resulting cytoplasmic and nuclear fractions prepared immediately after the 1-h radiolabeling period (0) and after additional chase periods of 4, 8, 24, and 48 h. Positions of MCP and the immunologically cross-reactive 120-kDa fragment (frag.) are indicated. (B) The percentage of MCP plus fragment in the cytoplasmic (filled symbols) and nuclear (empty symbols) fractions at each time point. (C) The relative amount of fragment per amount of MCP in the cytoplasmic (filled symbols) and nuclear (empty symbols) fractions at each time point.
The 120-kDa fragment was absent from or present in only trace amounts in the sample prepared immediately following the labeling period but was detected 4 h later and increased slowly and continuously in cells infected with either the NLS1− or NLS2− mutant (Fig. 6A, C). There was a somewhat greater amount of fragment relative to MCP in the nuclear fraction than in the cytoplasmic fraction and for the NLS2− mutant versus the NLS1− mutant, but the fragment appeared at about the same time and increased in both cell fractions for both mutants (Fig. 6C). These data indicate that the nuclear translocation of MCP is less efficient for the NLS mutants than for the WT and that the 120-kDa fragment is a slow-appearing (1 to 4 h after pulse) cleavage product of MCP (i.e., reacts with anti-MCP).
NLS mutations of UL80 proteins affect intracellular distribution of other capsid and tegument proteins.
The effect of these NLS mutations on the nuclear/cytoplasmic distribution of other viral proteins was examined for comparison by reprobing the membranes shown in Fig. 5A and B as follows. The membranes were (i) stripped and reprobed with antibodies to either the triplex mCP (pUL85) or to the tegument BPP (pp150; pUL32); then they were (ii) stripped again and reprobed with antibodies to the tegument LM (pp65; pUL83) or to the other triplex component, mCBP (pUL46); and finally, they were (iii) stripped again and reprobed with antibodies to the tegument HMWP (pUL48) or to the SCP (pUL48/49). The resulting data are presented in Fig. 5C, G, H, D, F, and E, respectively, and are quantified in Fig. 5I. With the exception of LM, all other proteins tested showed increased abundance in the cytoplasmic fraction of mutant-infected cells relative to that in WT virus-infected cells (Fig. 5I). Although these proteins were affected to different extents between the WT and the mutants, generally their differences were stronger for NLS2− than NLS1−. Other noteworthy features of these data are that (i) the HMWP (pUL48) and BPP (pUL32) tegument proteins both accumulated to significantly higher amounts in mutant-infected cells, with the majority of each partitioning with the NP-40 cytoplasmic fraction (Fig. 5G, I), and (ii) the comparatively weak intensity of the SCP (pUL48/49) signal reduced confidence in the absolute values obtained but not in the relative abundance of cytoplasmic versus nuclear accumulation.
We conclude from these comparisons that mutating the UL80 NLS affects the distribution of other capsid proteins and several capsid-associated tegument proteins such that they partition more strongly with the NP-40 cytoplasmic fraction. We suggest, without further discussion, that this is due to (i) downstream effects on capsid assembly, tegumentation, and subsequent envelopment resulting from there being fewer and apparently aberrant capsids formed in cells infected with these mutants (Fig. 7A to I), or to (ii) a generalized effect on nuclear translocation resulting from congestion caused by the slowed transport of pAP-MCP/pPA-MCP complexes in mutant-infected cells.
FIG. 7.
Cells infected with NLS2− mutant virus have few DNA-containing capsids and show an altered distribution of tetraCys-tagged UL80 proteins. HFF cells infected with WT, NLS1−, or NLS2− virus were processed for electron microscopy (EM) (A to I), for confocal microscopy (FIAsH) (J to L), or standard fluorescence microscopy (DAPI) (M to O), as described in Results and Materials and Methods. (A, D, and G) Representative areas of nuclei (capture magnification, ×6,000); capsids appear as small, uniformly dark black specks (see white arrows). (B, C, E, F, H, and I) Representative intranuclear capsid forms (capture magnification, ×50,000). (J to L) Nuclear inclusions in live cells infected with WT or mutant virus expressing tetraCys-tagged UL80 proteins and stained with FIAsH (capture magnification, ×100). (M to O) Shown are the cells in panels J to L after fixation and DAPI staining for DNA. Representative spheres fluorescing brightly with FIAsH (numbered 1 to 3 in panel L) correspond to areas of relatively weak DAPI fluorescence (replicate numbering pattern, 1 to 3 in panel O).
NLS mutations of UL80 proteins affect virus assembly.
We compared WT virus-infected cells to NLS mutant-infected cells by electron microscopy to determine whether the differences in titers and in the intracellular distribution and cleavage of MCP (see above) reflect or relate to the effects on virus formation. Overall, we found fewer virus particles (i.e., intranuclear capsids and tegumented or enveloped cytoplasmic capsids) in cells infected with the NLS1− mutant than in those infected with WT virus and still fewer in cells infected with the NLS2− mutant. One consistent difference in the appearance of virus particles was that the double-ringed nuclear capsids typical of infections with WT virus were relatively rare or absent from mutant-infected cells. Examples of intranuclear capsids considered representative for each infection are shown in Fig. 7. Because these infections progress at different rates, as gauged by relatively subjective CPE, meaningful normalization of particle numbers was considered infeasible, and quantification was not done.
A distinctive feature of nuclei in cells infected with the NLS2− mutant was the presence throughout of electron-dense spherical structures, ranging broadly in diameter from ∼500 to >1,500 nm. The spongiform intranuclear inclusions typical of cells infected with WT virus (Fig. 7A), and also seen with the NLS1− mutant (Fig. 7D), were not evident in cells infected with the NLS2− mutant (Fig. 7G). To determine whether the intranuclear spheres in cells infected with the NLS2− mutant reflect an aberrant distribution of the mutant UL80 proteins, we constructed mutant viruses expressing the UL80/NLS1− or NLS2− proteins tagged with a tetraCys motif for visualization by microscopy in living cells (see Materials and Methods). The same tetraCys insertion was used previously to reveal the presence of UL80 proteins in intranuclear tubules formed in cells infected with a different pAP mutant (21).
The tetraCys motif itself had no evident effect on the replication of either WT (21) or NLS mutant viruses (data not shown from the virus outgrowth following bacmid nucleofection). Cells infected with WT virus and stained live with the biarsenical dye FIAsH gave the same spongiform pattern reported before (21) (Fig. 7J). A similar pattern was seen in the nuclei of cells infected with the NLS1− mutant (Fig. 7K), but the predominant staining pattern in cells infected with the NLS2− mutant was one of large spherical structures ranging in diameter and distributed throughout the nucleus (Fig. 7L), reminiscent of the electron-dense structures observed by electron microscopy (Fig. 7G). Images of the same cells fixed and stained with DAPI to detect DNA showed that with WT and the NLS1− mutant, the regions most intensely stained for DNA correspond with those stained most intensely for UL80 proteins (Fig. 7M, N). In contrast, the spherical structures stained for UL80 proteins in cells infected with the NLS2− mutant appeared to be in regions with relatively little DNA staining (Fig. 7L, O). Thus, the UL80 proteins distribute differently in nuclei of cells infected with the NLS2− mutant than in nuclei of cells infected with WT virus or the NLS1− mutant.
DISCUSSION
We have tested the biological importance of two NLS (NLS1 and NLS2) in the CMV UL80 proteins that are proposed to function during the early stages of virus assembly. These NLS were first recognized in the SCMV pAP and pPR and studied using alanine substitution mutants in transfection/immunofluorescence assays (25). The results of that work led to the hypothesis that the pUL80 NLS are required to translocate the viral MCP, in complexes with pAP and pPR, into the nucleus for incorporation into capsids. We now have disabled these NLS in mutant viruses of HCMV and determined the effects on replication. Our findings (i) support the proposed involvement of both UL80 NLS in translocating MCP into the nucleus; (ii) reveal an MCP cleavage that is enhanced by delayed translocation; and (iii) indicate that NLS1 and NLS2 are functionally nonequivalent, with the consequences of mutating NLS2 being more extreme.
Inactivating UL80 NLS disrupts virus replication.
The use of glutamine as a conservative but functionally inactivating substitution in NLS1 and NLS2 was validated by transfection experiments, which showed that the HCMV pAP mutants had intracellular distributions similar to those of the alanine-substituted SCMV pAP and pPR mutants tested before (25). The NLS1− and NLS2− mutant proteins were predominantly nuclear, like the WT, and the double NLS1−/2− mutant was predominantly, if not exclusively, cytoplasmic (Fig. 2).
The proposed key role of these NLS during early steps of the virus assembly pathway was supported by results showing that inactivating either NLS1 or NLS2 alone reduced the spread of infection in cell culture and the production of infectious virus (Fig. 3, 4), and that inactivating both was lethal. Our conclusion that these NLS are required to translocate MCP-pAP and MCP-pPR complexes into the nucleus was also supported. Pulse-chase assays and steady-state measurements of MCP intracellular distribution showed a six- to eightfold slower nuclear translocation of MCP in cells infected with mutant versus WT virus (Fig. 6) and a higher percentage of total intracellular MCP in the NP-40 cytoplasmic fraction of mutant-infected cells (Fig. 5). Our finding that MCP translocation is less efficient in mutant virus-infected cells is consistent with expectations that decreasing the number of NLS per protein would slow its transport (30). Further, our calculations indicate a close relationship between the twofold-decreased number of NLS in the mutant proteins and the approximately threefold-decreased nuclear translocation rate of their MCP cargo during the first 8 h of the chase.
MCP cleavage increased by NLS mutations.
The 120-kDa fragment of MCP that accumulates in cells infected with the NLS1− or NLS2− mutant virus is not strictly a consequence of these mutations. Its relative amount also increased, though less dramatically, in cells infected with a virus having a point mutation in the UL80 protein amino-conserved domain (L382A; black asterisk in Fig. 1) that is thought to influence nuclear translocation (21), and it is detected in small amounts even in WT virus-infected cells (Fig. 5, 6). The size of the fragment, its reactivity with antibodies to the carboxyl end of MCP, and its delayed appearance in pulse-chase radiolabeling experiments (Fig. 6) indicate that it is derived from MCP by a proteolytic cleavage that removes ∼20% of its amino end.
Our unpublished observations that MCP is relatively stable against proteolytic attack when expressed alone in plasmid-transfected mammalian cells or in recombinant baculovirus-infected insect cells where it remains cytoplasmic, suggest that this cleavage requires changes in MCP conformation or intracellular localization that depend on other viral proteins. Such changes could be triggered by interactions with pAP or higher-order MCP-pAP complexes or by trafficking from the cytoplasmic site of complex formation into the nucleus. In WT virus-infected cells, in which the MCP-pAP interaction and nuclear translocation are optimal, the percentage of MCP cleaved is minimal. The percentage of MCP cleaved is increased in cells infected with the L382A mutant virus where self interaction of the UL80 proteins is abolished and MCP-pAP interaction is reduced (e.g., by ∼75% in GAL4 two-hybrid assays) (21, 35), possibly compromising both the pAP-induced conformational change and the nuclear translocation of MCP (21). As shown here, the percentage of MCP cleaved was dramatically increased when either NLS1 or NLS2 was mutated, resulting in nuclear translocation of MCP being slowed and its time of exposure to attack by putative cytoplasmic or translocation-associated proteases consequently increased. These observations imply a relationship between MCP cleavage and nuclear translocation.
If MCP cleavage is due to a cellular protease, a potential link between cleavage and translocation raises the consideration that the activity is directly associated with the nuclear translocation process, conceivably as a mechanism to protect the pore against blockage by complexes engaged but unable to pass through. Alternatively, if MCP cleavage is caused by a viral enzyme, the UL80a maturational protease pPR is an obvious candidate. Although MCP contains no self-evident pPR cleavage sequence in the region of its amino end that would trim it by ∼20 kDa, that end of MCP is predicted to interact with the carboxyl terminus of the UL80 proteins, including pPR (9). If so, the maturational cleavage site at the carboxyl end of pAP and pPR (M site) (Fig. 1), the primary substrate of pPR, may be brought into proximity with a susceptible site on MCP and attract the catalytic portion of pPR to attack that MCP site under enabling conditions. Thus, MCP cleavage may result from encountering a random or specific cellular protease during trafficking to the nucleus or from a circumstantial association of the viral pPR with a region of MCP that is normally less accessible.
Mutating NLS2 interferes more severely with virus replication than mutating NLS1.
Inactivating NLS2 had more pronounced effects than inactivating NLS1 in all comparisons of the mutant proteins and viruses, including efficiency of MCP nuclear translocation, production of infectious virus, and organization of intranuclear inclusions. This difference could result from the more disruptive intramolecular effects of the NLS2− mutation on the overall protein fold rather than on intermolecular interactions of the NLS2 amino acid sequence per se. It also could result from the stronger effects of the NLS2− mutation on MCP nuclear translocation and cleavage, acting in combination to interfere with capsid formation (e.g., slowing the accumulation of MCP for assembly; yielding more MCP fragment to complete normal interactions) and giving a comparatively more pronounced phenotype than that of NLS1−.
Although neither of these possibilities can be conclusively eliminated by available data, the 45-fold stronger interference of the NLS2− mutation with virus production (compared with NLS1− mutation) and its dramatic impact on the distribution of UL80 proteins in the nucleus raises interest in a third interpretation. Namely, that the NLS2 sequence has a functional requirement beyond serving as an NLS, and that disrupting this additional function underlies the more extreme effects of the NLS2− mutation on replication. In this connection, we note a recently identified interaction of the herpes simplex virus (HSV) homolog of pAP (i.e., pUL26.5, preVP22a, scaffolding protein) with the capsid portal protein (HSV pUL6) (23). The region of HSV pUL26.5 involved in the interaction (27) includes a conserved Pro-Gly-Glu sequence just upstream of the CMV pAP NLS2 (Fig. 1; also see Fig. 11 in reference 25). Mutating NLS2 near or overlapping that region could account for the severity of the NLS2− mutation if it disrupts a corresponding critical interaction between the CMV pAP and portal protein (HCMV pUL104), which forms a structure enabling viral DNA to enter and leave the capsid (10, 22, 31). Perhaps, in the absence of such an interaction, the UL80 proteins are improperly targeted within the nucleus, giving rise to the aberrant spherical structures seen following infection with the NLS2− mutant.
In summary, our results support the proposed role of the CMV pUL80 NLS1 and NLS2 sequences in nuclear translocation of MCP and capsid assembly. They also uncover a cleavage of MCP that appears to be exaggerated by slowing its nuclear translocation and establish that NLS2 is more than a dose-amplifying functional duplication of NLS1. The comparatively more disruptive effects of mutating NLS2 suggests that it has a major function other than or in addition to its role as an NLS.
Acknowledgments
Electron and confocal microscopies were done in the School of Medicine Microscope Facility with advice from Mike Delannoy and assistance from Jian Zhang. We thank Ron Schnaar for the use of his inverted fluorescence microscope, Bill Britt for monoclonal antibody to HCMV pUL48/49 (no. 11-2-23), Gary Pearson for monoclonal antibody to HCMV pUL83, and Neal Copeland for E. coli DY380.
N.N. was supported by NCI training grant CA09243. This work was aided by USPHS research grants AI13718 and AI32957 to W.G.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Adams, S. R., R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis, and R. Y. Tsien. 2002. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 1246063-6076. [DOI] [PubMed] [Google Scholar]
- 2.Anstrom, D. M., L. Colip, B. Moshofsky, E. Hatcher, and J. Remington. 2005. Systematic replacement of lysine with determine and alanine in Escherichia coli malate synthase G: effect on crystallization. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 611069-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 756865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter, M. K., and W. Gibson. 1997. The putative human cytomegalovirus triplex proteins, minor capsid protein (mCP) and MCP-binding protein (mC-BP), form a heterotrimeric complex that localizes to the cell nucleus in the absence of other viral proteins, abstr. 168. Presented at the 22nd International Herpesvirus Workshop, La Jolla, CA.
- 5.Borst, E., and M. Messerle. 2000. Development of a cytomegalovirus vector for somatic gene therapy. Bone Marrow Transplant. 25(Suppl. 2)S80-S82. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. C., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids, p. 111-153. In A. H. E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, New York, NY.
- 7.Chan, C. K., E. J. Brignole, and W. Gibson. 2002. Cytomegalovirus assemblin (pUL80a): cleavage at internal site not essential for virus growth; proteinase absent from virions. J. Virol. 768667-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cinti, C., P. P. Claudio, C. M. Howard, L. M. Neri, Y. Fu, L. Leoncini, G. M. Tosi, N. M. Maraldi, and A. Giordano. 2000. Genetic alterations disrupting the nuclear localization of the retinoblastoma-related gene RB2/p130 in human tumor cell lines and primary tumors. Cancer Res. 60383-389. [PubMed] [Google Scholar]
- 9.Desai, P., and S. Person. 1999. Second site mutations in the N-terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology 261357-366. [DOI] [PubMed] [Google Scholar]
- 10.Dittmer, A., and E. Bogner. 2005. Analysis of the quaternary structure of the putative HCMV portal protein PUL104. Biochemistry 44759-765. [DOI] [PubMed] [Google Scholar]
- 11.Gibson, W. 1981. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology 111516-537. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39389-400. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, W. 2008. Structure and function of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol. 235187-204. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, W., K. S. Clopper, W. J. Britt, and M. K. Baxter. 1996. Human cytomegalovirus smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J. Virol. 705680-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greis, K. D., W. Gibson, and G. W. Hart. 1994. Site-specific glycosylation of the human cytomegalovirus tegument basic phosphoprotein (UL32) at serine 921 and serine 952. J. Virol. 688339-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130118-133. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann, S. H., C. M. Ewing, and J. H. Shaper. 1987. The erasable western blot. Anal. Biochem. 16189-95. [DOI] [PubMed] [Google Scholar]
- 18.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 19.Liu, F., and Z. H. Zhou. 2007. Comparative virion structures of human herpesviruses, p. 27-43. In A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 20.Loveland, A. N., C. K. Chan, E. J. Brignole, and W. Gibson. 2005. Cleavage of human cytomegalovirus protease pUL80a at internal and cryptic sites is not essential but enhances infectivity. J. Virol. 7912961-12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loveland, A. N., N. L. Nguyen, E. J. Brignole, and W. Gibson. 2007. The amino-conserved domain of human cytomegalovirus UL80a proteins is required for key interactions during early stages of capsid formation and virus production. J. Virol. 81620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 7510923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb, W. W., D. R. Thomsen, F. L. Homa, and J. C. Brown. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 779862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penman, S. 1966. RNA metabolism in the HeLa cell nucleus. J. Mol. Biol. 17117-130. [DOI] [PubMed] [Google Scholar]
- 25.Plafker, S. M., and W. Gibson. 1998. Cytomegalovirus assembly protein precursor and proteinase precursor contain two nuclear localization signals that mediate their own nuclear translocation and that of the major capsid protein. J. Virol. 727722-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in the nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64615-623. [DOI] [PubMed] [Google Scholar]
- 27.Singer, G. P., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 2005. Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal. J. Virol. 79132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spencer, J. V., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 1998. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J. Virol. 723944-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 30.Timney, B. L., J. Tetenbaum-Novatt, D. S. Agate, A. J. Williams, J. Zhang, B. T. Chait, and M. P. Rout. 2006. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 175579-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 7812668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67509-544. [DOI] [PubMed] [Google Scholar]
- 33.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 806003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welch, A. R., L. M. McNally, M. R. Hall, and W. Gibson. 1993. Herpesvirus proteinase: site-directed mutagenesis used to study maturational, release, and inactivation cleavage sites of precursor and to identify a possible catalytic site serine and histidine. J. Virol. 677360-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zacksenhaus, E., R. Bremner, R. A. Phillips, and B. L. Gallie. 1993. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol. Cell. Biol. 134588-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]