Abstract
We report here the construction of a triply fluorescent-tagged herpes simplex virus 1 (HSV-1) expressing capsid protein VP26, tegument protein VP22, and envelope protein gB as fusion proteins with monomeric yellow, red, and cyan fluorescent proteins, respectively. The recombinant virus enabled us to monitor the dynamics of these capsid, tegument, and envelope proteins simultaneously in the same live HSV-1-infected cells and to visualize single extracellular virions with three different fluorescent emissions. In Vero cells infected by the triply fluorescent virus, multiple cytoplasmic compartments were found to be induced close to the basal surfaces of the infected cells (the adhesion surfaces of the infected cells on the solid growth substrate). Major capsid, tegument, and envelope proteins accumulated and colocalized in the compartments, as did marker proteins for the trans-Golgi network (TGN) which has been implicated to be the site of HSV-1 secondary envelopment. Moreover, formation of the compartments was correlated with the dynamic redistribution of the TGN proteins induced by HSV-1 infection. These results suggest that HSV-1 infection causes redistribution of TGN membranes to form multiple cytoplasmic compartments, possibly for optimal secondary envelopment. This is the first real evidence for the assembly of all three types of herpesvirus proteins—capsid, tegument, and envelope membrane proteins—in TGN.
Herpes simplex virus 1 (HSV-1) consists of three morphologically distinct structures: nucleocapsid, tegument, and envelope (58). The nucleocapsid contains the linear double-stranded DNA viral genome in an icosahedral capsid (58). The nucleocapsid is surrounded by a proteinaceous layer designated the tegument, which is enclosed by an envelope that is a host cell-derived lipid bilayer containing virus-encoded glycoproteins (58). After virus entry into the host cell, the deenveloped nucleocapsids reach the nucleopores, and the viral genome enters the nucleus (58). Subsequently, viral DNA replication and packaging of nascent progeny virus genomes into assembling capsids take place within nuclear compartments, called replication compartments (12, 56, 58). The progeny nucleocapsids then acquire primary envelopes by budding through the inner nuclear membrane into the space between the inner and the outer nuclear envelopes, called the perinuclear space (39, 58). While assembly of progeny nucleocapsids within the nucleus and primary envelopment of nucleocapsids at the inner nuclear membrane have been well documented, the route of the nascent virions from the perinuclear space to the extracellular space is controversial (7, 34, 41, 42, 58, 66). It is now generally accepted that perinuclear virions lose their envelopes by fusion with the outer nuclear membrane, thereby releasing naked nucleocapsids in the cytoplasm (40, 41, 60). These nucleocapsids must acquire tegument proteins in the cytoplasm and/or upon budding through the nuclear membrane and must be enveloped again at cytoplasmic membranes (secondary envelopment), probably at the trans-Golgi network (TGN) or endosomal membranes (22, 38, 64). HSV infection induces redistribution and reorganization of membrane-enclosed organelles and cytoskeletons in the cytoplasm. In HSV-infected cells, the Golgi apparatus, TGN, and microtubules are redistributed and/or reorganized (2, 6, 64, 68). Although HSV-1 maturation has been the subject of various studies in recent years, there are still many aspects of the virus assembly pathway, especially in the cytoplasm, as well as the biological significance of changes in cellular organelles and the cytoskeleton mediated by HSV infection, that remain to be elucidated.
Virus replication is often associated with specific intracellular compartments (48, 67). These cellular compartments are thought to provide a physical scaffold for concentrating progeny virus structural proteins, thereby increasing the efficiency of virion assembly and/or genome replication. For example, African swine fever virus, a large cytoplasmic DNA virus, induces a compartment in infected cells at a juxtanuclear cytoplasmic region that closely resembles aggresomes (23). Several RNA viruses, including mouse hepatitis virus and poliovirus, induce compartments in autophagosomes (25, 55). Formation of the compartments has been suggested to be important for virus replication, based on observations that virus replication is impaired when the compartments are not made or when viruses are not able to access the structures (23, 25, 55). As described above, HSV viral DNA replication and packaging into nascent capsids within nuclear compartments (replication compartments) have been well documented, but distinct compartments in the cytoplasm involved in virus envelope and/or tegument assembly have not yet been identified. The only HSV-induced compartment in the cytoplasm that has been reported is one formed at the juxtanuclear cytoplasmic domain (18, 47, 50, 64). Concentration of envelope, tegument, and capsid proteins in the juxtanuclear cytoplasmic domain and accumulation of virus particles in the compartment have been reported. Although the role of the juxtanuclear cytoplasmic compartment in HSV virion assembly remains largely unknown, a similar cytoplasmic compartment has also been reported to be induced by human cytomegalovirus infection (59).
During tegument and envelope assembly, the nucleocapsid must encounter tegument and envelope proteins at the site(s) of virus assembly. Therefore, information on subcellular localization of viral structural proteins, especially simultaneous observations of multiple viral structural proteins in the same infected cells, would help address a range of issues about HSV-1 maturation. Most studies on subcellular localization of HSV proteins in infected cells have been carried out by immunofluorescent analysis of fixed cells. Considering the process of HSV maturation as described above, viral structural proteins often exhibit dynamic localization as infection progresses, which cannot be analyzed using fixed-cell preparations. However, dynamic localization of viral proteins can be studied by live-imaging procedures using recombinant viruses expressing fluorescently tagged structural proteins, thereby enabling the visualization and localization of these proteins in live cells and the monitoring of changes in their localization in the same cells as infection progresses. Live-cell imaging of cells infected with recombinant fluorescent viruses expressing a fusion protein consisting of a viral protein and a fluorescent protein (FP) or two viral protein-FP fusion proteins has been reported for HSV-1 and another alphaherpesvirus pseudorabies virus (1, 13-15, 18, 27, 38, 53, 63-65). Such recombinant viruses should enable analysis of intracellular trafficking of viral proteins and dynamic aspects of the interactions between viral proteins. In particular, recombinant doubly fluorescent viruses expressing a capsid and a tegument protein or a capsid and an envelope protein, with both proteins fused to different FPs, should be useful to investigate specific events in HSV maturation, such as tegument and envelope assembly. However, use of such doubly fluorescent viruses for analyzing HSV maturation pathways is limited because HSV maturation involves the concurrent assembly of three structural domains: capsid, tegument, and envelope. Therefore, recombinant triply fluorescent HSV expressing capsid, tegument, and envelope proteins, each fused to a different FP, is needed for studies of the continuous events in virus assembly (e.g., capsid assembly, nuclear egress, tegument assembly, cytoplasmic envelopment, and virion release) in live cells as the infection progresses.
We report here the construction of a recombinant HSV-1 strain expressing three fluorescent fusion proteins: HSV-1 capsid protein VP26 fused to VenusA206K (a monomeric variant of an enhanced yellow FP [EYFP] carrying the mutation A206K), HSV-1 tegument protein VP22 fused to mRFP1 (a monomeric variant of a red FP [RFP], DSRed), and HSV-1 envelope protein gB fused to ECFPA206K (a monomeric enhanced cyan FP [ECFP]). This triply fluorescent recombinant HSV-1, together with several other singly and doubly fluorescent HSV-1 recombinants constructed for these studies, has enabled simultaneous observations of viral capsid, tegument, and envelope proteins in the same live cells during infection. In addition, using the triply fluorescent virus, we have identified multiple cytoplasmic compartments, where capsid, tegument, and envelope proteins and TGN proteins concentrate.
MATERIALS AND METHODS
Cells and viruses.
Vero, rabbit skin, HeLa, and SK-N-SH cells were described previously (28, 62). HEp-2 and human embryonic lung (HEL) fibroblast cells were kindly provided by Bernard Roizman and Shinya Watanabe, respectively, and were maintained in Dulbecco's modified Eagle's medium containing 5% and 10% fetal calf serum, respectively. HSV-1 wild-type strain HSV-1(F) was described previously (16, 30). Recombinant virus YK304 was reconstituted from pYEbac102 containing a complete HSV-1(F) sequence with the bacterial artificial chromosome (BAC) sequence inserted into the HSV intergenic region between UL3 and UL4 (62). YK304 has been shown to have an identical phenotype to wild-type HSV-1(F) in cell cultures and in mouse models (62).
Plasmids.
(i) The transfer plasmid pBS-VenusA206K-UL35, for generating a recombinant HSV-1 expressing a fusion protein of the VenusA206K FP and HSV-1 VP26 capsid protein, was constructed as follows. Plasmid pBS-Venus was constructed by amplifying the Venus open reading frame (ORF) (44) without its stop codon from Venus/pCS2 (a generous gift from A. Miyawaki) by PCR using the primer pair 1 (Table 1) and cloning the fragment into the EcoRI and SpeI sites of pBluescript KS(+) (Stratagene). Using pBS-Venus, the pBS-VenusA206K construct, in which alanine at Venus residue 206 was replaced with lysine, was generated using a QuickChange site-directed mutagenesis kit according to manufacturer's instructions (Stratagene). Since green fluorescent protein (GFP) and its variants form dimers at high concentrations, the A206K mutation was used to prevent dimerization without significant alternation in FP spectral properties (69). The 1-kb sequences flanking the HSV-1 UL35 start codon were amplified by PCR, using the primer pairs 2 and 3 (Table 1), from pBC1014 (a generous gift from B. Roizman) (32) and sequentially cloned into the pBS-VenusA206K EcoRI and HindIII sites and the NotI and SpeI sites to produce pBS-VenusA206K-UL35. UL35 encodes the VP26 capsid protein. The resultant plasmid consisted of VenusA206K-UL35 bounded by UL35 flanking sequences. (ii) The transfer plasmid pBS-mRFP1-UL49, for generating a recombinant HSV-1 expressing a fusion protein of the mRFP1 FP and HSV-1 VP22 tegument protein, was constructed as follows. Plasmid pBS-mRFP1 was constructed by amplifying the mRFP1 (8) ORF without its stop codon from pRSETB-mRFP1 (a generous gift from R. Y. Tsien) using the primer pair 4 (Table 1) and cloning it into the EcoRI and SpeI sites of pBluescript KS(+). The 1-kb sequences flanking the HSV-1 UL49 start codon were amplified using the primer pairs 5 and 6 (Table 1) from pBC1007 (a generous gift from B. Roizman) (32) and sequentially cloned into the EcoRI and HindIII sites and the NotI and SpeI sites of pBS-mRFP1 to produce pBS-mRFP1-UL49. UL49 encodes the VP22 tegument protein. The resultant plasmid consisted of mRFP1-UL49 bounded by UL49 flanking sequences. (iii) The transfer plasmid pBS-mRFP1-UL47, used to generate a recombinant HSV-1 expressing a fusion protein of the mRFP1 FP and HSV-1 VP13/14 tegument protein, was constructed by sequentially cloning the 1-kb sequences flanking the UL47 start codon, amplified using the primer pairs 7 and 8 (Table 1), from pBC1007 into the EcoRI and HindIII sites and the NotI and SpeI sites of pBS-mRFP1. UL47 encodes the VP13/14 tegument protein. This plasmid consisted of part of the UL47 ORF fused to mRFP1 and the 1-kb sequence upstream of the UL47 start codon. (iv) The transfer plasmid pBS-VenusA206K-UL47, used to generate a recombinant HSV-1 expressing a fusion protein of the VenusA206K FP and HSV-1 VP13/14 tegument protein, was constructed as described for the construction of pBS-mRFP1-UL47 except that pBS-VenusA206K was used instead of pBS-mRFP1. (v) The transfer plasmid pBS-ECFPA206K-UL27, used to generate a recombinant HSV-1 expressing a fusion protein of the ECFPA206K FP and HSV-1 gB envelope protein, was constructed as follows. pBluescript II KS(+) was digested with NotI, treated with T4 DNA polymerase, and religated to disrupt its NotI site. The resultant plasmid was designated pBSΔNotI. To construct pBS-UL27, a 2-kb sequence containing part the UL27 ORF and its upstream region was amplified from pBC1014 by PCR using primer pair 10 (Table 1) and cloned into the XbaI and HindIII sites of pBSΔNotI. UL27 encodes the HSV-1 gB envelope glycoprotein. To construct pBS-ECFP, the cyan ECFP ORF without its stop codon was amplified from pECFP-C1 (Clontech) by PCR and cloned into the EcoRI and SpeI sites of pBluescript KS(+). pBS-ECFPA206K, in which alanine at ECFP residue 206 was replaced with lysine, was generated using a QuickChange site-directed mutagenesis kit according to manufacturer's instructions. pBS-ECFPA206K-UL27 was then constructed by an in-frame insertion of the ECFPA206K coding sequence without a stop codon, which had been amplified from pBS-ECFPA206K by PCR using primer pair 9 (Table 1), into the NotI site of the UL27 gene in pBS-UL27. The resultant plasmid consisted of part of the UL27 ORF fused to ECFPA206K and the 1-kb region upstream of the UL27 start codon. In the plasmid, ECFPA206K was inserted at UL27 amino acid 43. (vi) Plasmids pEYFP-Golgi, pEYFP-ER (where ER is endoplasmic reticulum), and pECFP-Mito were purchased from Clontech. Plasmid pECFP-Golgi was constructed from pECFP-Mito by replacing its NheI-BamHI fragment, which contains a mitochondrial target sequence, with the NheI-BamHI fragment of pEYFP-Golgi, which contains a TGN target sequence. (vii) To construct pGEX-UL35 encoding VP26 fused to glutathione S-transferase (GST), the entire UL35 coding sequence was amplified from pBC1007 by PCR and cloned into pGEX4T-1 (GE Healthcare) in frame with GST.
TABLE 1.
Primer sequences used for the construction of the transfer plasmids
| Name | Amplified region | Sequence |
|---|---|---|
| Pair 1 | Venus | 5′-GCACTAGTCGCCACCATGGTGAGCAAGGGC-3′ |
| 5′-GCGAATTCCTTGTACAGCTCGTCCATGC-3′ | ||
| Pair 2 | UL35-upstream | 5′-GCACTGCGGCCGCTTAACCGGTTCCTGGACCTG-3′ |
| 5′-GCACTAGTCGGGACCGGAGGTCGGGAAG-3′ | ||
| Pair 3 | UL35-downstream | 5′-GCGAATTCGCCGTCCCGCAATTTCACCG-3′ |
| 5′-GCGAAGCTTTTCCGCGTCTTCCACAAATC-3′ | ||
| Pair 4 | mRFP | 5′-GCACTAGTCGCCACCATGGTGAGCAAGGGC-3′ |
| 5′-GCGAATTCCTTGTACAGCTCGTCCATGC-3′ | ||
| Pair 5 | UL49-upstream | 5′-GCACTGCGGCCGCTCGCCAGGATGTCCAGGAAC-3′ |
| 5′-GCACTAGTGGTTCCACGAACACGCTAGG-3′ | ||
| Pair 6 | UL49-downstream | 5′-GCGAATTCACCTCTCGCCGCTCCGTGAA-3′ |
| 5′-GCGAAGCTTTCCCCGTCGGATTGGGAAAC-3′ | ||
| Pair 7 | UL47-upstream | 5′GCACTGCGGCCGCAGCACCTTAACCTCCCGCTG-3′ |
| 5′-GCACTAGTGGTGGCGATAGACGCGGGTT-3′ | ||
| Pair 8 | UL47-downstream | 5′-GCGAATTCTCGGCTCGCGAACCCGCGGG-3′ |
| 5′-GCGAAGCTTATTGAGGCGGGCGAGCAGGG-3′ | ||
| Pair 9 | ECFP | 5′-GCACTGCGGCCGCAGTGAGCAAGGGCGAGGAGCT-3′ |
| 5′-GCACTGCGGCCGCCTTGTACAGCTCGTCCATGC-3′ | ||
| Pair 10 | UL27 | 5′-GCTCTAGACCTGACGAAGCGGTCGTTGG-3′ |
| 5′-GCGAAGCTTGAGAATCGGAAGGAGCCGCC-3′ |
Construction of recombinant viruses.
To construct recombinant viruses encoding fusion proteins consisting of an FP and an HSV-1 structural protein, rabbit skin cells were cotransfected with each transfer plasmid and intact YK304 viral DNA by using a calcium phosphate precipitation technique as described previously (32). Viral DNAs were extracted from infected cells and purified on 5 to 20% potassium acetate gradients as described previously (32). At 3 days posttransfection, the transfected cells were harvested and subjected to freeze-thawing and sonication. The cell lysates were diluted and inoculated on Vero cells, and plaques were screened for fluorescence with an inverted fluorescent microscope (Olympus IX71). Recombinants were plaque purified three times on Vero cells. In this way, transfer plasmids pBS-VenusA206K-UL35, pBS-mRFP1-UL49, pBS-mRFP1-UL47, pBS-ECFPA206K-UL27, and pBS-VenusA206K-UL47 were used to construct recombinant viruses YK601, YK602, YK603, YK604, and YK610, respectively. Recombinant viruses encoding two different FP-tagged structural proteins were constructed by coinfection of Vero cells with two singly FP-tagged fluorescent viruses, which were then screened for progeny virus producing the two FP-tagged proteins (i.e., doubly fluorescent viruses). In this way, YK605, YK606, and YK607 were constructed by coinfection with YK602, YK603 or YK604, and YK601, respectively. YK612 was constructed by coinfection with YK602 and YK604. The genotypes of the singly and doubly fluorescent viruses were confirmed by Southern blotting (data not shown). Expression of the predicted fusion proteins was confirmed by infecting Vero cells with each singly or doubly fluorescent virus and assaying the infected cells and resulting extracellular progeny virions for the appropriate fusion protein by immunoblotting (data not shown). Recombinant viruses encoding three FP-tagged structural proteins were constructed by coinfection with a double-tagged fluorescent virus, YK605 or YK606, and the single-tagged fluorescent virus YK604 and screening for progeny virus producing the three FP-tagged proteins (i.e., triply fluorescent viruses). Descriptions of the recombinant viruses constructed for this study are in Table 2.
TABLE 2.
Virus strains used in this study
| HSV strain | HSV-1 protein and fused FPa
|
BAC insertion | |||
|---|---|---|---|---|---|
| VP26 | VP22 | VP13/14 | gB | ||
| HSV-1(F) | − | ||||
| YK304 | + | ||||
| YK601 | VenusA206K | + | |||
| YK602 | mRFP1 | + | |||
| YK603 | mRFP1 | + | |||
| YK604 | ECFPA206K | + | |||
| YK605 | VenusA206K | mRFP1 | + | ||
| YK606 | VenusA206K | mRFP1 | + | ||
| YK607 | VenusA206K | ECFPA206K | + | ||
| YK608 | VenusA206K | mRFP1 | ECFPA206K | + | |
| YK609 | VenusA206K | mRFP1 | ECFPA206K | + | |
| YK610 | VenusA206K | + | |||
| YK612 | mRFP1 | ECFPA206K | + | ||
Each FP was fused to the amino terminus of the indicated HSV-1 protein, with the exception of ECFPA206K, which was inserted in frame at position 43 of gB.
Southern blotting and immunoblotting.
Southern blotting and immunoblotting were performed as described previously (31, 62).
Growth analysis of recombinant viruses.
Nearly confluent Vero cells grown in 35-mm-diameter plastic dishes (approximately 106 cells per dish) were infected with each of the recombinant viruses at a multiplicity of infection (MOI) of 5, and residual virus was removed after virus adsorption. At the indicated times (see Fig. 3A), total virus from the cell culture supernatants and the infected cells was harvested. In the other experiments, intracellular virus from the infected cells (Fig. 3B) and extracellular virus from the cell culture supernatants (Fig. 3C) were separately harvested at the indicated times. The titers of each sample were determined by standard plaque assays on Vero cells.
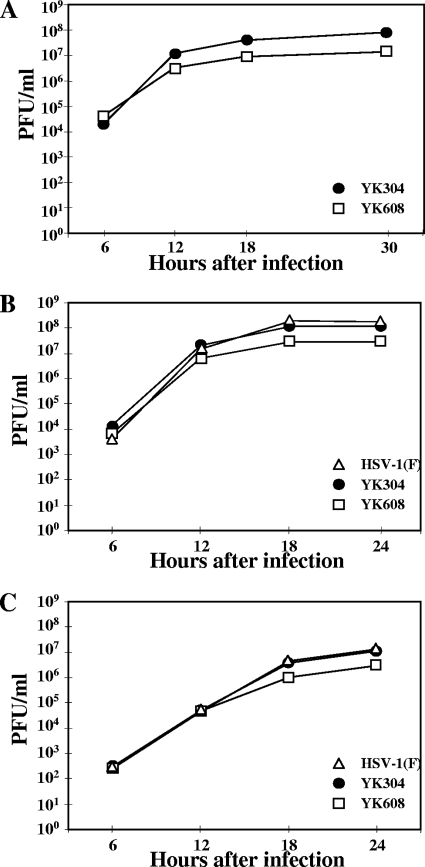
FIG. 3.
Growth curves of YK608. Vero cells were infected at an MOI of 5 with wild-type YK304, triply fluorescent virus YK608, and wild-type HSV-1(F). Total virus from the cell culture supernatants and the infected cells (A), cell-associated virus from the infected cells (B), and extracellular virus from the cell culture supernatants (C) were harvested at the indicated times and assayed on Vero cells.
Antibodies.
To generate rabbit polyclonal antibody to VP26, two rabbits were immunized with purified GST-VP26 fusion protein by the standard protocol at MBL (Nagoya, Japan). For these studies, GST-VP26 was expressed in Escherichia coli that had been transformed with pGEX-UL35 and purified as described previously (29). Rabbit polyclonal antibody to VP22 has been described previously (49). Mouse monoclonal antibodies to gB, a Golgi protein with a molecular weight of 58,000 (58K), GM130, and EEA1 were purchased from the Goodwin Institute, Sigma, and BD Transduction. Rabbit polyclonal antibodies to GFP and DsRed were purchased from MBL and Clontech, respectively. Sheep polyclonal antibody to TGN46 was purchased from AbD Serotec. Rabbit polyclonal antibody to β-catenin and mouse monoclonal antibody to vinculin were purchased from Sigma.
Purification of virions.
Virions were purified as described previously (1, 33, 63). Briefly, Vero cells were infected with YK608 at an MOI of 0.01 for 48 h. Cell culture supernatants were then harvested by low-speed centrifugation. The HSV-containing supernatant was centrifuged for 2 h at 25,000 rpm in an SRP28S rotor (Hitachi). The pellet was resuspended in 0.5 ml of TBSal (200 mM NaCl, 2.6 mM KCl, 10 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 1.8 mM CaCl2), layered onto a 9-ml discontinuous sucrose gradient (30%, 40%, and 50%) in TBSal, and centrifuged for 2 h at 18,000 rpm in a P40ST rotor. Aliquots of peak virion-containing fractions were pelleted by centrifugation for 2 h at 29,800 rpm in a P40ST rotor.
Indirect immunofluorescence assay.
Indirect immunofluorescence assays were performed as described previously (27).
Transfection.
Vero cells were transfected with appropriate plasmids using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen).
Live-cell imaging.
Cells for live analysis (approximately 5 × 104 cells per dish) were cultured on 35-mm-diameter glass-bottomed dishes (MatTek or Matsunami). Prior to imaging analyses, the culture medium was replaced with medium 199 that does not contain the pH indicator phenol red (Invitrogen) and supplemented with 1% fetal calf serum. The dish was then placed in a humidified chamber, mounted on an LSM5 inverted confocal microscope (Carl Zeiss), supplied with 5% CO2, and heated to 37°C (CZI-3 controller; Carl Zeiss). The LSM5 microscope was used with a Plan-Apochromat 63× (1.4 numerical aperture) objective, an argon laser (458, 488, and 514 nm), and HeNe lasers (543 and 633 nm) (Carl Zeiss). ECFPA206K fluorescence was imaged using 458-nm excitation and a BP467.5-497.5 emission filter. EYFP and VenusA206K fluorescence was imaged using 488-nm excitation and a BP515-545 emission filter. mRFP1 fluorescence was imaged using 543-nm excitation and an LP620 or BP615-675 emission filter. The secondary antibody AlexaFluor 680 (Invitrogen) for indirect immunofluorescence assay was imaged using 633-nm excitation and an LP690 emission filter.
Electron microscopic analysis.
For negative staining, a drop of the purified virus described above was applied onto Formvar-coated 400-mesh copper grids (EM Sciences) for 5 min, excess liquid was blotted away, and a drop of 2% phosphotungstic acid was added for 2 min. Samples were then examined by transmission electron microscopy (Hitachi H-7500; Tokyo, Japan).
RESULTS
Construction of triply fluorescent viruses.
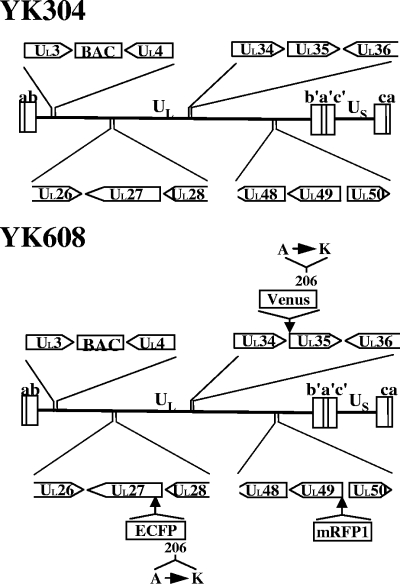
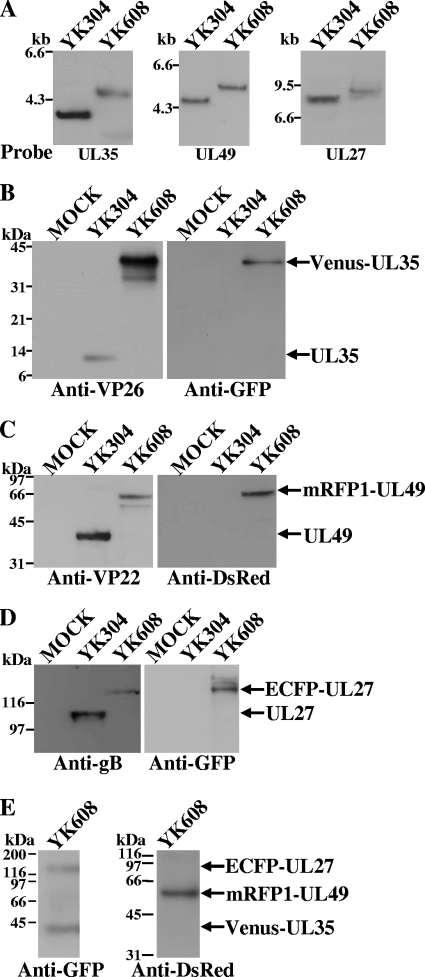
To study HSV-1 maturation in live cells, triply fluorescent recombinant viruses were constructed that expressed capsid protein VP26 tagged with VenusA206K (VenusA206K-VP26), tegument protein VP22 or VP13/14 tagged with mRFP1 (mRFP1-VP22 or mRFP1-VP13/14), and envelope protein gB tagged with ECFPA206K (ECFPA206K-gB) (Fig. 1). The parental virus YK304 used to generate these recombinant viruses carried a BAC in its genome (Fig. 1), and, therefore, the fluorescent viruses generated in this study can be easily manipulated further for future studies using the BAC system (62). To construct triply fluorescent viruses, we first constructed singly fluorescent viruses with an FP fused to an HSV-1 structural protein: VenusA206K fused to the amino terminus of VP26 (YK601), mRFP1 fused to the amino terminus of VP22 (YK602) or VP13/14 (YK603), and ECFPA206K fused to gB at amino acid 43 (YK604) (Table 2). We then constructed doubly fluorescent viruses expressing VenusA206K-VP26 and mRFP1-VP22 (YK605), VenusA206K-VP26 and mRFP1-VP13/14 (YK606), VenusA206K-VP26 and ECFPA206K-gB (YK607), and mRFP1-VP22 and ECFPA206K-gB (YK612) (Table 2). The growth curves of YK602, YK604, and YK612 were almost identical to that of the parental YK304 at an MOI of 5 on Vero cells (data not shown). YK601, YK603, YK605, YK606, and YK603 showed similar growth curves to the YK304 growth curve, although these recombinant viruses produced progeny virus titers 5- to 10-fold lower than YK304. Finally, we constructed triply fluorescent viruses that expressed VenusA206K-VP26, mRFP1-VP22, and ECFPA206K-gB (YK608) or VenusA206K-VP26, mRFP1-VP13/14, and ECFPA206K-gB (YK609) (Table 2). The genotypes of YK608 and YK609 were confirmed by Southern blotting (Fig. 2A and data not shown). Expression of the predicted fusion proteins in Vero cells infected with YK608 and YK609 was confirmed by immunoblotting (Fig. 2B to D and data not shown). Incorporation of each fusion protein into progeny virions was confirmed by immunoblotting (Fig. 2E and data not shown).
FIG. 1.
Schematic diagram of the genome structures of YK304 and YK608 and locations of viral genes relevant for these studies. The positions of insertion of the BAC and each fluorescent protein are indicated.
FIG. 2.
(A) Southern blots of electrophoretically separated SacI (left), EcoRV (middle), and BamHI (right) digests of YK304 and YK608 DNAs, hybridized to a radiolabeled probe specific for the UL35 gene, UL49 gene, or UL27 gene, respectively. (B to D) Immunoblots of electrophoretically separated lysates from Vero cells mock infected (lane 1) or infected with YK304 (lane 2) or YK608 (lane 3) at an MOI of 5. Infected cells were harvested 24 h postinfection and analyzed by immunoblotting with antibody to VP26 or GFP, VP22 or DsRed, or gB or GFP. (E) Immunoblots of electrophoretically separated YK608 virions purified by sucrose gradient centrifugation and reacted with antibody to GFP or DsRed. Molecular sizes are shown on the left of each figure.
Growth properties of the triply fluorescent viruses.
The growth curves of the triply fluorescent recombinant viruses were compared with the parental YK304 by infecting Vero cells with each virus at an MOI of 5, harvesting total virus from the cell culture supernatants and the infected cells at the indicated times (Fig. 3A) postinfection, and assaying each sample by standard plaque assay on Vero cells. We have previously reported that the growth curve of total virus obtained with YK304 was identical to that with HSV-1(F) (62). YK608 showed similar growth kinetics to YK304, but produced a progeny virus yield fivefold lower than YK304 (Fig. 3A). In contrast, the YK609 virus yield was approximately 100-fold less than that of YK304 (data not shown). To investigate the growth properties of YK608 in more detail, Vero cells were infected with HSV-1(F), YK304, or YK608 at an MOI of 5, and extracellular secreted virus and cell-associated virus were separately harvested. The titers of each sample were determined by standard plaque assay. As shown in Fig. 3B and C, the growth curves of cell-associated and extracellular viruses obtained with HSV-1(F) were almost identical to the YK304 growth curve. YK608 showed similar growth kinetics to YK304 but produced fourfold lower progeny yield than YK304 (Fig. 3B and C). These results indicate that the quadruple insertion of three FPs and BAC in YK608 has relatively little effect on viral replication in Vero cells at a high MOI.
Simultaneous observation of capsid, tegument, and envelope proteins in live infected cells and in single virion particles.
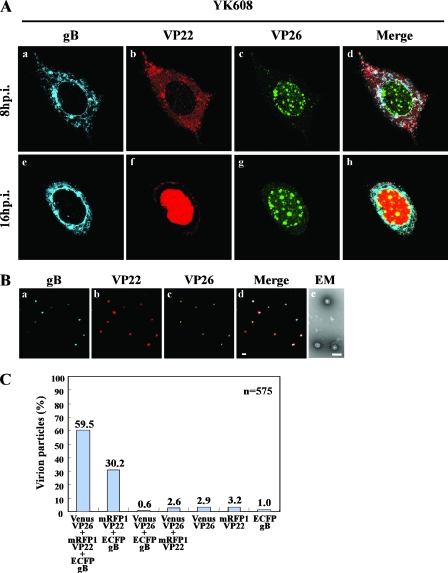
To determine whether we could examine the subcellular localization of capsid, tegument, and envelope proteins in the same live cells simultaneously, Vero cells on a glass-bottomed dish were infected with YK608 at an MOI of 10 and examined by confocal microscopy at 8 and 16 h postinfection. Figure 4A shows typical images of live YK608-infected Vero cells. VenusA206K-VP26 (capsid protein) accumulated in both the nucleus and cytoplasm throughout the infection. In the nucleus VenusA206K-VP26 was detected as granular structures. ECFPA206K-gB (envelope protein) accumulated mainly in the cytoplasm and the nuclear rim throughout the infection. Similar results for VenusA206K-VP26 and ECFPA206K-gB localization were obtained with Vero cells infected with YK609 (data not shown). In Vero cells infected with YK609, mRFP1-VP13/14 (tegument protein) localized mainly in the nucleus, and, at later times postinfection, an increase in cytoplasmic fluorescence was observed (data not shown). These results for recombinant viruses with FP-tagged proteins in infected cells were consistent with those published previously (14, 15, 54). However, in contrast to a previous report (17, 18) that GFP-tagged VP22 (tegument protein) translocates from the cytoplasm to the nucleus only during cell division, in the studies reported here most VP22 localized in the cytoplasm at 8 h postinfection and in the nucleus at 16 h postinfection (Fig. 4A, frames b and f). The translocation of mRFP1-VP22 from the cytoplasm to the nucleus observed in this study is consistent with the immunofluorescence studies of fixed cells infected with wild-type HSV-1 previously reported (52).
FIG. 4.
(A) Time-lapse microscopy of live Vero cells infected with YK608. Vero cells seeded on a glass-bottomed dish were infected with YK608 at an MOI of 10 and transferred to a heated chamber at 3 h postinfection. A single field was chosen for analysis, and images were collected by confocal microscopy at 8 and 16 h postinfection. Single-color images were captured separately and are shown in frames a, b, c, e, f, and g. Frames d and h show simultaneous acquisitions of the three colors. (B) Visualization of singly fluorescent virion particles. YK608 virion particles purified by sucrose gradient centrifugation were placed on a coverslip. Single-color images were then captured separately by confocal microscopy and are shown in frames a, b, and c. Frame d shows simultaneous acquisition of the three colors. Frame e shows an image of purified YK608 virions negatively stained with phosphotungstic acid and examined by transmission electron microscopy. Scale bars: 1 μm (d) and 200 nm (e). (C) The relative percentages of purified YK608 virion particles emitting VenusA206K-VP26, mRFP1-VP22 and ECFPA206K-gB, mRFP1-VP22 and ECFPA206K-gB, VenusA206K-VP26 and ECFPA206K-gB, VenusA206K-VP26 and mRFP1-VP22, VenusA206K-VP26 alone, mRFP1-VP22 alone, or ECFPA206K-gB alone. Virion particles were purified and visualized as described in panel B.
Extracellular YK608 virions purified on a sucrose gradient were mainly intact when analyzed by negative-stain electron microscopy (Fig. 4B), enabling single-particle analysis. With confocal microscopy, purified YK608 virions appeared as discrete fluorescent puncta with VenusA206K-VP26, mRFP1-VP22, and ECFPA206K-gB fluorescent emissions (approximately 60% of the total) (Fig. 4B and C). These were likely HSV infectious “heavy particles,” which are complete virions. Occasionally, fluorescent puncta with only mRFP1-VP22 and ECFPA206K-gB emissions were also observed (approximately 30% of the total) (Fig. 4B and C). These were likely HSV “light particles,” which are tegument and envelope but without a capsid (36, 61). The other fluorescent puncta (approximately 10% of the total) (Fig. 4C) are probably due to variable emission intensities from particle to particle and virus assembly intermediates, as reported previously (13, 35).
Visualization of multiple cytoplasmic compartments formed in Vero cells infected with fluorescent viruses where envelope, tegument, and capsid proteins concentrate.
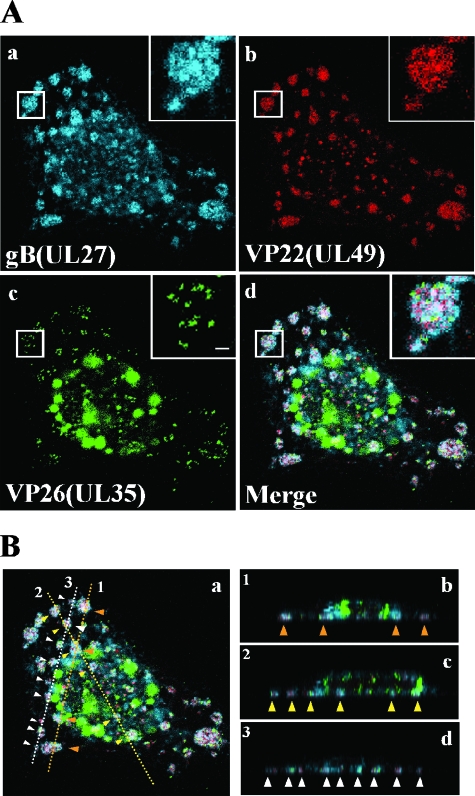
During imaging studies of live YK608-infected Vero cells, we noted that VenusA206K-VP26, mRFP1-VP22, and ECFPA206K-gB accumulate in cytoplasmic compartments at late infection times (at 8 h postinfection and thereafter). The compartments were observed close to the basal surfaces of the infected cells (the adhesion surfaces of the infected cells on the solid growth substrate) (Fig. 5B). In the compartments at 12 h postinfection, mRFP1-VP22 and ECFPA206K-gB were diffusely distributed, while VenusA206K-VP26 particles were detected as discrete puncta (Fig. 5A). The sizes of VenusA206K-VP26 puncta were similar to those observed with purified extracellular virions, suggesting that mature capsid structures accumulate in these compartments. Similar cytoplasmic compartments, with accumulations of capsid, tegument, and envelope fusion proteins, were also observed in other cell lines infected with YK608, including HEL (Fig. 6A), rabbit skin, HEp-2, HeLa, and SK-N-SH cells (data not shown). We refer to the cytoplasmic compartments as assembly sites here.
FIG. 5.
Visualization of assembly sites induced close to the basal surfaces of YK608-infected cells. Vero cells were infected with YK608 at an MOI of 10 and live cells were examined 12 h postinfection by confocal microscopy. Confocal Z-sections were acquired through the entire thickness of the cells. (A) Single-color images were captured separately and are shown in frames a, b, and c. Frame d shows simultaneous acquisition of the three colors. Insets show magnified images of individual assembly sites. Scale bar, 1 μm. (B) Frames b, c, and d show Z-stack sections at lines 1, 2, and 3 indicated in frame a, respectively. Individual assembly sites are indicated by arrowheads.
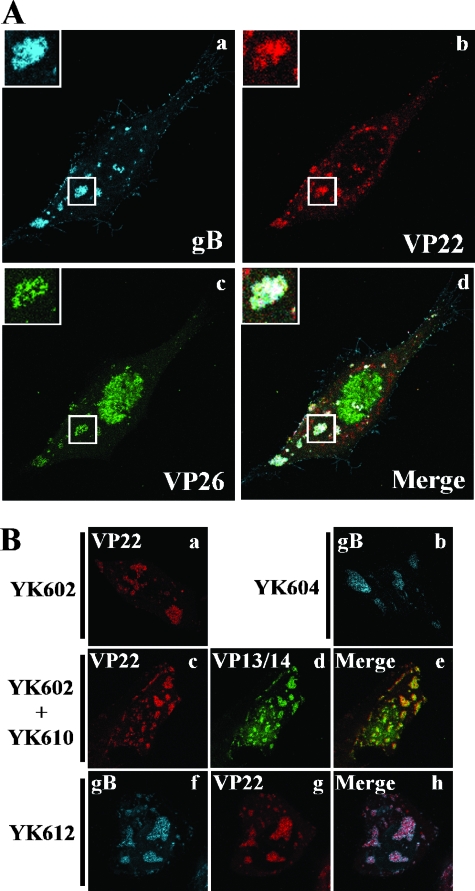
FIG. 6.
(A) Visualization of the assembly sites induced in HEL cells infected with YK608 at an MOI of 10. At 12 h postinfection, live infected cells were examined by confocal microscopy. Single-color images were captured separately and are shown in frames a, b, and c. Frame d shows simultaneous acquisitions of the three colors. Insets show magnified images of individual assembly sites. (B) Visualization of the assembly sites induced in Vero cells infected with singly and doubly fluorescent viruses. Vero cells were infected with YK602 (a), YK604 (b), and YK612 (f to h) or coinfected with YK602 and YK610 (c to e) at an MOI of 10. At 12 h postinfection, live infected cells were examined by confocal microscopy. Single-color images were captured separately and are shown in frames a, b, c, d, f, and g. Frames e and h show simultaneous acquisitions of both colors.
Since the triply fluorescent virus YK608 did not grow as well as wild-type virus, these studies were repeated with singly (YK602, YK604, and YK610) and doubly (YK612) fluorescent viruses, whose growth was almost identical to wild-type virus (data not shown). In Vero cells infected with singly fluorescent viruses YK602 and YK604 or doubly fluorescent virus YK612, the fusion proteins accumulated in compartments close to the basal surfaces of the infected cells (Fig. 6B, frames a, b, f, g, and h). Therefore, formation of the compartments was not related to the decrease in virus growth of triply fluorescent viruses. Furthermore, mRFP1-VP22 and VenusA206K-VP13/14 colocalized in the compartments in Vero cells coinfected with singly fluorescent viruses YK602 and YK610 (Fig. 6B, frames c, d, and e), showing that multiple tegument proteins accumulate in the compartments.
Time-lapse analysis of formation of the assembly sites at the cell periphery in live infected Vero cells.
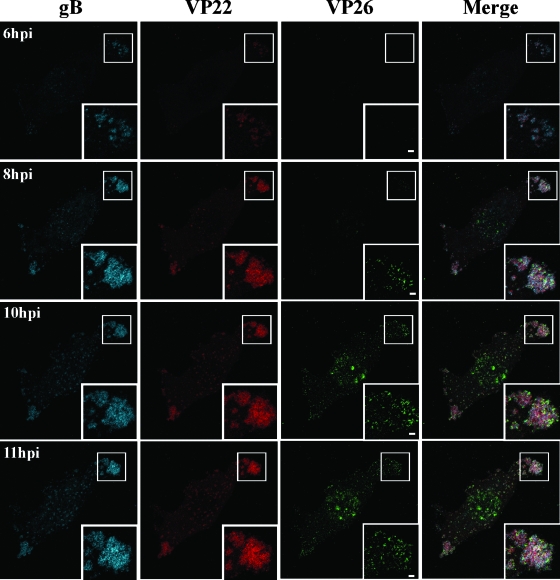
To monitor formation of the assembly sites in individual living cells, Vero cells growing on a glass-bottomed dish were infected with YK608 at an MOI of 10 and examined by confocal microscopy at various times postinfection. As shown in Fig. 7, ECFPA206K-gB and mRFP1-VP22 began to accumulate in compartments formed close to the basal surfaces of the infected cells at 6 h postinfection. ECFPA206K-gB and mRFP1-VP22 were colocalized and appeared as punctate structures at this time point, but VenusA206K-VP26 was not detectable. As infection progressed, both of these punctate structures expanded and coalesced into globular structures. The globular structures were irregular in size and shape, and both ECFPA206K-gB and mRFP1-VP22 were diffusely distributed in them. Interestingly, gB and VP22 colocalized in the compartments throughout infection. VenusA206K-VP26 particles became detectable as discrete puncta at 7 h postinfection (data not shown). As the infection progressed, the foci of VenusA206K-VP26 puncta had similar sizes, and the number of these punctate foci increased. The observations that the timing of detection and patterns of localization of VenusA206K-VP26 in the compartments were different from those of ECFPA206K-gB and mRFP1-VP22 suggest that capsid structures may be transported separately from the envelope and tegument proteins into the compartments. However, we could not eliminate the possibility that the later detection of VenusA206K-VP26 fluorescence in the assembly sites simply reflects its lower amounts in the compartments rather than later delivery.
FIG. 7.
Time-lapse analysis of formation of the assembly sites in live HSV-1-infected cells. Vero cells were infected with YK608 at an MOI of 10, transferred to a heated growth chamber at 5 h postinfection, and examined by confocal microscopy hourly to 11 h postinfection. Images at 6, 8, 10, and 11 h postinfection (hpi) are shown. The same confocal microscope settings were used at each time point. Insets show magnified images of individual assembly sites. Scale bar, 2 μm.
Accumulation of TGN markers in the assembly sites.
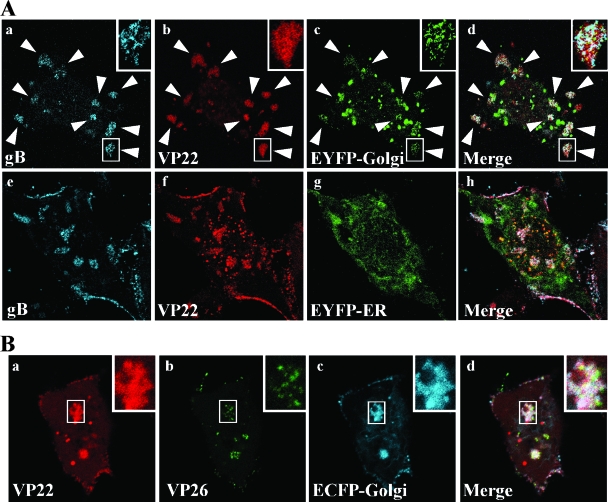
To characterize the assembly sites formed close to the basal surfaces of the infected cells in more detail, we attempted to identify cellular organelle(s) related to the compartments. Therefore, Vero cells were transfected with pEYFP-Golgi, which encodes EYFP fused to a sequence from human GlcNAc β-1,4-galactosyltransferase (GT), and subsequently infected with YK612 at an MOI of 10. The living infected cells were then examined by confocal microscopy at 12 h postinfection. GlcNAc β-1,4-GT is known to localize to trans-Golgi and TGN (51). EYFP-GlcNAc β-1,4-GT accumulated and colocalized with ECFPA206K-gB and mRFP1-VP22 in most of the compartments formed close to the basal surface of the infected cells (Fig. 8A). Similarly, ECFP-GlcNAc β-1,4-GT accumulated and colocalized with mRFP1-VP22 and VenusA206K-VP26 in the compartments of Vero cells transiently transfected with pECFP-Golgi and subsequently infected with YK605 (Fig. 8B). The accumulation of EYFP-GlcNAc β-1,4-GT and ECFP-GlcNAc β-1,4-GT in the compartments is specific, since EYFP fused to the calreticulin ER-targeting sequence, encoded by pEYFP-ER, did not accumulate in the compartments of YK612-infected Vero cells (Fig. 8A, frame g).
FIG. 8.
Visualization of the accumulation of markers for Golgi and TGN in assembly sites formed close to the basal surfaces of live Vero cells infected with HSV-1. (A) Vero cells were transfected with pEYFP-Golgi (a to d) or pEYFP-ER (e to h) that encodes EYFP fused to a Golgi- or ER-targeting sequence, respectively, and subsequently infected with YK612 at an MOI of 10 at 24 h posttransfection. At 12 h postinfection, the live infected cells were examined by confocal microscopy. Single-color images were then captured separately and are shown in panels a, b, c, e, f, and g. Frames d and g show simultaneous acquisitions of the three colors. Individual assembly sites are indicated by arrowheads. Insets show magnified images of individual assembly sites. (B) Vero cells were transfected with pECFP-Golgi and subsequently infected with YK605 at an MOI of 10. At 12 h postinfection, the cells were processed as described in panel A. Single-color images were captured separately and are shown in frames a, b, and c. Frame d shows simultaneous acquisition of the three colors. Insets show magnified images of individual assembly sites.
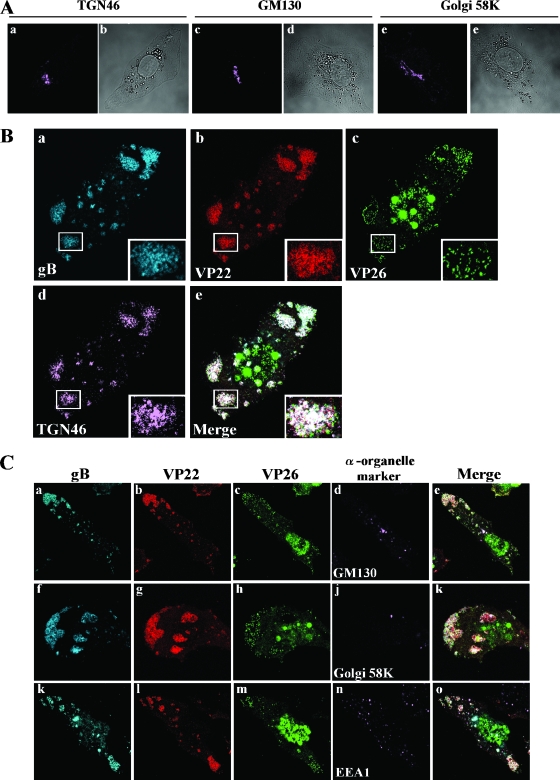
To investigate whether the assembly sites are associated with the Golgi or TGN or both and the relationship between the dynamics of the cellular organelles and formation of the compartments in infected cells, Vero cells were mock infected or infected with YK608 at an MOI of 10 for 12 h; fixed; treated with anti-TGN46, anti-GM130, anti-Golgi 58K, and anti-EEA1 antibodies; and examined by confocal microscopy. TGN46, a type 1 membrane protein found predominantly in the TGN, rapidly cycles back to the TGN from endosomes and the cell surface (3, 20, 53, 57). GM130 is a specific marker for cis-Golgi (45), whereas Golgi 58K is a maker for cis-, medial-, and trans-Golgi (5, 19). EEA1 is an endosomal marker (43). In mock-infected Vero cells, Golgi proteins detected by the anti-GM130 or the anti-Golgi 58K antibodies and TGN detected by the anti-TGN46 antibodies were morphologically intact, were mainly detected at the juxtanuclear domain (Fig. 9A), and were barely detected close to the basal surface of the cells (data not shown). As described previously (6, 64, 68), the integrity of Golgi proteins and TGN was altered in YK608-infected Vero cells at 12 h postinfection, and TGN46, GM130, and Golgi 58K proteins were dispersed (data not shown). Interestingly, the anti-TGN46 antibodies specifically accumulated and colocalized with VenusA206K-VP26, ECFPA206K-gB, and mRFP1-VP22 in the compartments formed close to the basal surfaces of the infected cells, while the anti-GM130, anti-Golgi 58K, and anti-EEA1 antibodies did not (Fig. 9B and C). Thus, the assembly sites are induced where TGN46 proteins are redistributed in HSV-infected cells. These results suggest that the assembly sites formed close to the basal surfaces of infected cells are TGN-associated compartments.
FIG. 9.
Visualization of recruitment of TGN markers into assembly sites formed close to the basal surfaces of HSV-1-infected Vero cells. Vero cells mock infected (A) or infected with YK608 at an MOI of 10 (B and C) were fixed at 12 h postinfection, permeabilized, and then stained with antibody to TGN46, GM130, Golgi 58K, or EEA1, which were detected with AlexaFluor 680-conjugated donkey anti-sheep immunoglobulin G or goat anti-mouse immunoglobulin G and examined by confocal microscopy. Confocal Z-sections were acquired through the entire thickness of the cells. Images of sections at the middle and differential interference contrast of the mock-infected cells are shown in panel A, and those at the bottom of the cells infected with YK608 are shown in panels B and C. (A) Mock-infected Vero cells. Localization of TGN46 (a), GM130 (c), and Golgi 58K (e) and differential interference contrast (b, d, and f) of each cell are shown. (B and C) YK608-infected Vero cells. Single-color images were captured separately, and localization of gB, VP22, VP26, or each organelle marker is shown. Frames e, k, and o show simultaneous acquisitions of the four colors. Insets show magnified images of individual assembly sites.
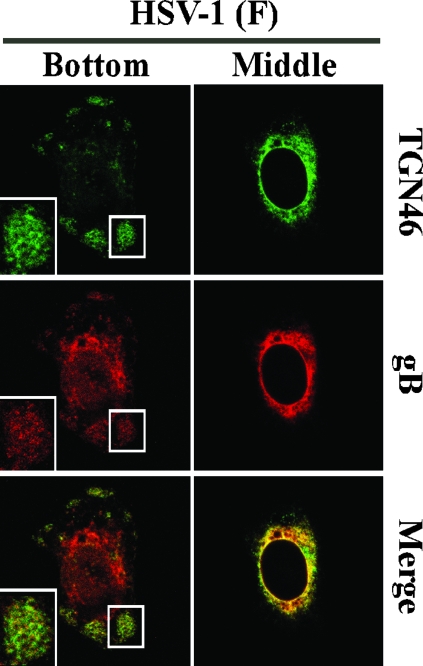
It has been reported that TGN46 proteins are redistributed to adhesion junctions in HSV-1-infected cells (37). The assembly sites observed in these studies were not associated with adhesion junctions, based on the observation that antibodies to β-catenin and vinculin, both of which are markers for adherence junctions, did not react with the compartments (data not shown). We also note that in agreement with the data on Vero cells infected with YK608 (Fig. 8 and 9), wild-type HSV-1(F) infection induced dispersal of TGN46 proteins throughout the cytoplasm of the infected cells (Fig. 10, upper panels), and TGN46-associated compartments (Fig. 10, lower panels), where gB proteins also accumulated, formed close to the basal surface of the infected Vero cells. These results suggest that wild-type HSV-1, as well as FP-tagged recombinant viruses, is able to induce the assembly sites close to the basal surface of infected Vero cells.
FIG. 10.
Localization of TGN46 in Vero cells infected with wild-type HSV-1. Vero cells were infected with HSV-1(F) at an MOI of 10 for 12 h, fixed, permeabilized, and then stained with antibody to TGN46 and gB. Anti-TGN46 antibodies were detected with AlexaFluor 488-conjugated donkey anti-sheep immunoglobulin G, and anti-gB antibodies were detected with AlexaFluor 546-conjugated goat anti-mouse immunoglobulin G. Samples were then examined by confocal microscopy. Confocal Z-sections were acquired through the entire thickness of the cells, and images of sections at the bottom and middle of the cell are shown. Single-color images were captured separately and are also shown (rows TGN46 or gB). Merged frames show simultaneous acquisition of the two colors. Insets show magnified images of assembly sites.
DISCUSSION
The only cytoplasmic compartment in which HSV structural proteins concentrate and virions accumulate that has been reported to date is a compartment formed at the juxtanuclear cytoplasmic domain (18, 47, 50, 64). It has been reported that a single relatively packed compartment is induced at the juxtanuclear domain of some cell lines infected with HSV (18, 47, 50, 64). In the present studies, we demonstrated that multiple assembly sites are induced close to the basal surfaces of the infected cells by using three-dimensional analyses of live cells infected with the triply fluorescent recombinant HSV-1 (YK608). The salient features that suggest that these compartments play a role in virion maturation are as follows.
(i) The major capsid, tegument, and envelope proteins (24, 46) analyzed simultaneously in these studies accumulated in the multiple assembly sites formed close to the basal surface of infected cells. Virion assembly sites are thought to be scaffolds for virion assembly, and, therefore, viral structural proteins concentrate in the compartments (48, 67). Since the HSV-1 virion consists of a nucleocapsid, tegument, and envelope, components of these structures are expected to accumulate in the same cellular compartments for HSV-1 virion assembly. In the studies presented here, we have shown that capsid protein VP26, tegument protein VP22, and envelope protein gB, all of which are major virion components (24, 58), colocalized in multiple assembly sites formed close to the basal surfaces of Vero cells infected with YK608 (Fig. 5). Other viral structural proteins probably also accumulate in the compartments, based on the result that VP13/14 colocalized with VP22 in the compartments in Vero cells coinfected with a recombinant virus expressing mRFP1-VP22 (YK602) and one expressing VenusA206K-VP13/14 (YK610) (Fig. 6).
(ii) Time-lapse analysis of the formation of assembly sites showed that the patterns of localization of capsid proteins in the multiple assembly sites were different from those of tegument and envelope proteins. Consistent with the present study, capsid-independent accumulation of glycoproteins in TGN-associated cellular compartments has been demonstrated previously using a temperature-sensitive mutant of HSV-1 (64, 68). Interestingly, the localization patterns of mRFP1-VP22 and ECFPA206K-gB were identical at each time studied postinfection, suggesting that VP22 and gB are transported together into the multiple compartments. VP22 and gB may form scaffold structures in the multiple compartments, and capsid proteins (VP26) separately reach the assembly sites.
(iii) TGN markers accumulated and colocalized with viral structural proteins in multiple assembly sites formed close to the basal surfaces of infected cells. It has been reported that HSV-1 acquires its final envelope from the cytoplasmic membrane, probably the TGN or endosomal membranes (22, 38, 64, 65). In the studies presented here, marker proteins for TGN specifically localized in multiple assembly sites formed close to the basal surfaces of Vero cells infected with YK608 (Fig. 8 and 9). The TGN markers seemed to be recruited into the assembly sites by HSV-1 infection, based on the observations that the TGN markers were barely detectable close to the basal surfaces of mock-infected Vero cells. It has been reported that HSV-1 infection induces dispersal of TGN, with the degree of TGN dispersal dependent on the cell line (64, 68). However, the role(s) of HSV-induced dispersal of TGN in virion assembly still remains largely unknown. In the present study, we demonstrated that redistribution of a TGN marker, TGN46, induced by HSV infection was correlated with the formation of multiple assembly sites, and these compartments were detected where dispersed TGN46 proteins were recruited (Fig. 8 and 9). From these observations, it is reasonable to speculate that HSV-1 infection causes the redistribution of TGN membranes into the multiple assembly sites formed close to the basal surfaces of infected cells, possibly for secondary envelopment. In addition, the observations support the hypothesis suggested in previous reports (22, 38, 64, 65) that TGN is one of the sites of secondary envelopment.
In the studies presented here, we have constructed a recombinant HSV-1 (YK608) expressing a capsid, tegument, and envelope protein, each fused to a different monomeric fluorescent protein. To our knowledge, this is the first report of the construction of a triply fluorescent herpesvirus. This recombinant virus in combination with the time-lapse imaging system described here provides a unique experimental system for investigating virion maturation pathways because it can detect three distinct virion components simultaneously and monitor their localization within the same living cells as the infection progresses. YK608 should be more useful for studies of the viral life cycle than singly or doubly fluorescent viruses because the triply fluorescent virus enables continuous analysis of events in virion maturation (e.g., capsid assembly, nuclear egress, tegumentation, cytoplasmic envelopment, and virion egress) as well as the events involved in virus entry (e.g., fusion of the viral envelope and cell membrane, release of tegument proteins from the capsid, and capsid transport to the nucleus). Since the YK608 genome contains a BAC, it is easy to construct recombinant virus combining YK608 with mutations in other genes. Such recombinants could be used to study viral protein functions in the steps in virus entry and maturation. Furthermore, YK608 could be used for development of new anti-HSV agents with different mechanisms of action than acyclovir, the current standard anti-HSV drug. Acyclovir is phosphorylated by HSV thymidine kinase, resulting in formation of an active inhibitor of viral DNA replication (11). Interestingly, monensin and brefeldin A, which have been reported to inhibit HSV replication (9, 10, 21, 26), dramatically changed the localization patterns and expression levels of viral proteins in cells infected with YK608 (data not shown). The imaging system described here, modified for rapid data acquisition and high throughput, combined with HSV-1 YK608 could be the basis for development of new screening systems for anti-HSV agents that target virion maturation pathways.
It has been reported that the nucleotide sequence similarity of GFP variants can result in recombination and exchange of fluorescence properties in cotransfected epithelial cells (4). This phenomenon has been termed GFP walking. GFP walking should not have been a problem in the studies reported here, even though VenusA206K and ECFPA206K are GFP variants, for several reasons. In Vero cells infected with YK608, the localization patterns of VenusA206K-VP26 were quite different from those of ECFPA206K-gB (Fig. 4A, 5, 6A, 7, and 9), suggesting that GFP walking either did not occur or was at a very low frequency. Furthermore, we confirmed the results obtained with YK608 (Fig. 4A, 5, 6A, 7, and 9) by experiments with two doubly fluorescent viruses, YK605 expressing VenusA206K-VP26 and mRFP1-VP22 and YK612 expressing ECFPA206K-gB and mRFP1-VP22 (data not shown), showing that the results with the triply fluorescent virus (YK608) were not due to GFP walking. Recruitment of TGN markers fused to GFP variants into the assembly sites formed close to the basal surfaces of cells infected with recombinant HSV-1 expressing other GFP variants cannot be a result of GFP walking because an ER marker fused to EYFP was not recruited into the compartments and because recruitment of another TGN marker (TGN46) into the compartments was demonstrated by immunofluorescence assays in fixed cells (Fig. 8 and 9). Thus, the possibility that the observations obtained with the triply fluorescent viruses resulted from GFP walking was eliminated by experiments with dually fluorescent viruses and immunofluorescence assays as described above. Although it is necessary to eliminate the possibility of GFP walking in studies using triply fluorescent viruses, such viruses are still important tools for investigating dynamic events in the HSV-1 life cycle.
In conclusion, live-cell image analyses using the triply fluorescent virus constructed in this study enabled us to determine that multiple assembly sites are induced close to the basal surfaces of HSV-1-infected cells. This finding also suggests the role of TGN reorganization induced by HSV infection in the formation of multiple assembly sites. Thus, the triply fluorescent virus is a useful tool for investigating the virion maturation pathways of HSV-1 and for further understanding the life cycle of HSV-1.
Acknowledgments
We thank B. Roizman for pBC1007, pBC1014, and HEp-2 cells; R. Tsien for pRSETB-mRFP1; A. Miyawaki for Venus/pCS2; and S. Watanabe for human embryonic lung fibroblasts. We thank S. Koyama and K. Kiriyama for excellent technical assistance. We thank K. Maeda for advice on construction of recombinant viruses.
This study was supported in part by Grants for Scientific Research and Grants for Scientific Research in Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Asai, R., A. Kato, K. Kato, M. Kanamori-Koyama, K. Sugimoto, T. Sairenji, Y. Nishiyama, and Y. Kawaguchi. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 805125-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., S. Di Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 697472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banting, G., and S. Ponnambalam. 1997. TGN38 and its orthologues: roles in post-TGN vesicle formation and maintenance of TGN morphology. Biochim. Biophys. Acta 1355209-217. [DOI] [PubMed] [Google Scholar]
- 4.Bestvater, F., T. A. Knoch, J. Langowski, and E. Spiess. 2002. Construct conversions caused by simultaneous co-transfection: “GFP-walking.” BioTechniques 32844, 846, 848-850 passim. [PubMed] [Google Scholar]
- 5.Bloom, G. S., and T. A. Brashear. 1989. A novel 58-kDa protein associates with the Golgi apparatus and microtubules. J. Biol. Chem. 26416083-16092. [PubMed] [Google Scholar]
- 6.Campadelli, G., R. Brandimarti, C. Di Lazzaro, P. L. Ward, B. Roizman, and M. R. Torrisi. 1993. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 902798-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume, G., and B. Roizman. 2006. The egress of herpesviruses from cells: the unanswered questions. J. Virol. 806716-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 997877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee, S., and S. Sarkar. 1992. Studies on endoplasmic reticulum-Golgi complex cycling pathway in herpes simplex virus-infected and brefeldin A-treated human fibroblast cells. Virology 191327-337. [DOI] [PubMed] [Google Scholar]
- 10.Cheung, P., B. W. Banfield, and F. Tufaro. 1991. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J. Virol. 651893-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coen, D. M., and D. D. Richman. 2007. Antiviral Agents, p. 447-485. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 12.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55857-868. [DOI] [PubMed] [Google Scholar]
- 13.del Rio, T., T. H. Ch'ng, E. A. Flood, S. P. Gross, and L. W. Enquist. 2005. Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J. Virol. 793903-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 727563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnelly, M., and G. Elliott. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 752575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 17.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 742131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 734110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, Y. S., C. Alvarez, D. S. Nelson, and E. Sztul. 1998. Molecular cloning, characterization, and dynamics of rat formiminotransferase cyclodeaminase, a Golgi-associated 58-kDa protein. J. Biol. Chem. 27333825-33834. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, R. N., W. G. Mallet, T. T. Soe, T. E. McGraw, and F. R. Maxfield. 1998. An endocytosed TGN38 chimeric protein is delivered to the TGN after trafficking through the endocytic recycling compartment in CHO cells. J. Cell Biol. 142923-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh-Choudhury, N., A. Graham, and H. P. Ghosh. 1987. Herpes simplex virus type 2 glycoprotein biogenesis: effect of monensin on glycoprotein maturation, intracellular transport and virus infectivity. J. Gen. Virol. 681939-1949. [DOI] [PubMed] [Google Scholar]
- 22.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 751236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 431102-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanamori, M., S. Watanabe, R. Honma, M. Kuroda, S. Imai, K. Takada, N. Yamamoto, Y. Nishiyama, and Y. Kawaguchi. 2004. Epstein-Barr virus nuclear antigen leader protein induces expression of thymus- and activation-regulated chemokine in B cells. J. Virol. 783984-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 801476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 711019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 772359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawaguchi, Y., K. Nakajima, M. Igarashi, T. Morita, M. Tanaka, M. Suzuki, A. Yokoyama, G. Matsuda, K. Kato, M. Kanamori, and K. Hirai. 2000. Interaction of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) with HS1-associated protein X-1: implication of cytoplasmic function of EBNA-LP. J. Virol. 7410104-10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 717328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 768820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leuzinger, H., U. Ziegler, E. M. Schraner, C. Fraefel, D. L. Glauser, I. Heid, M. Ackermann, M. Mueller, and P. Wild. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 7913047-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 1025832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73269-276. [DOI] [PubMed] [Google Scholar]
- 37.McMillan, T. N., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 751928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106167-180. [DOI] [PubMed] [Google Scholar]
- 39.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113163-169. [DOI] [PubMed] [Google Scholar]
- 41.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C., and T. Minson. 2006. Egress of alphaherpesviruses. J. Virol. 801610-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mu, F. T., J. M. Callaghan, O. Steele-Mortimer, H. Stenmark, R. G. Parton, P. L. Campbell, J. McCluskey, J. P. Yeo, E. P. Tock, and B. H. Toh. 1995. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 27013503-13511. [DOI] [PubMed] [Google Scholar]
- 44.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2087-90. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1311715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232499-511. [DOI] [PubMed] [Google Scholar]
- 47.Norrild, B., I. Virtanen, B. Pedersen, and L. Pereira. 1983. Requirements for transport of HSV-1 glycoproteins to the cell surface membrane of human fibroblasts and Vero cells. Arch. Virol. 77155-166. [DOI] [PubMed] [Google Scholar]
- 48.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nozawa, N., Y. Kawaguchi, M. Tanaka, A. Kato, A. Kato, H. Kimura, and Y. Nishiyama. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J. Virol. 796947-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nozawa, N., Y. Yamauchi, K. Ohtsuka, Y. Kawaguchi, and Y. Nishiyama. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp. Cell Res. 299486-497. [DOI] [PubMed] [Google Scholar]
- 51.Paulson, J. C., and K. J. Colley. 1989. Glycosyltransferases. Structure, localization, and control of cell type-specific glycosylation. J. Biol. Chem. 26417615-17618. [PubMed] [Google Scholar]
- 52.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 736769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ponnambalam, S., M. Girotti, M. L. Yaspo, C. E. Owen, A. C. Perry, T. Suganuma, T. Nilsson, M. Fried, G. Banting, and G. Warren. 1996. Primate homologues of rat TGN38: primary structure, expression and functional implications. J. Cell Sci. 109675-685. [DOI] [PubMed] [Google Scholar]
- 54.Potel, C., K. Kaelin, I. Gautier, P. Lebon, J. Coppey, and F. Rozenberg. 2002. Incorporation of green fluorescent protein into the essential envelope glycoprotein B of herpes simplex virus type 1. J. Virol. Methods 10513-23. [DOI] [PubMed] [Google Scholar]
- 55.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 27910136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36857-868. [DOI] [PubMed] [Google Scholar]
- 57.Rajasekaran, A. K., J. S. Humphrey, M. Wagner, G. Miesenbock, A. Le Bivic, J. S. Bonifacino, and E. Rodriguez-Boulan. 1994. TGN38 recycles basolaterally in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 51093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2501-2602. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 59.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 755697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szilagyi, J. F., and C. Cunningham. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72661-668. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 771382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanaka, M., Y. Nishiyama, T. Sata, and Y. Kawaguchi. 2005. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology 341301-312. [DOI] [PubMed] [Google Scholar]
- 64.Turcotte, S., J. Letellier, and R. Lippe. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 798847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Genderen, I. L., R. Brandimarti, M. R. Torrisi, G. Campadelli, and G. van Meer. 1994. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology 200831-836. [DOI] [PubMed] [Google Scholar]
- 66.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Mueller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 791071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312875-878. [DOI] [PubMed] [Google Scholar]
- 68.Wisner, T. W., and D. C. Johnson. 2004. Redistribution of cellular and herpes simplex virus proteins from the trans-Golgi network to cell junctions without enveloped capsids. J. Virol. 7811519-11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296913-916. [DOI] [PubMed] [Google Scholar]