Abstract
The membrane-spanning domain (MSD) of the human immunodeficiency virus type 1 (HIV-1) gp41 glycoprotein is critical for its biological activity. Previous C-terminal truncation studies have predicted an almost invariant core structure of 12 amino acid residues flanked by basic amino acids in the HIV-1 MSD that function to anchor the glycoprotein in the lipid bilayer. To further understand the role of specific amino acids within the MSD core, we initially replaced the core region with 12 leucine residues and then constructed recovery-of-function mutants in which specific amino acid residues (including a GGXXG motif) were reintroduced. We show here that conservation of the MSD core sequence is not required for normal expression, processing, intracellular transport, and incorporation into virions of the envelope glycoprotein (Env). However, the amino acid composition of the MSD core does influence the ability of Env to mediate cell-cell fusion and plays a critical role in the infectivity of HIV-1. Replacement of conserved amino acid residues with leucine blocked virus-to-cell fusion and subsequent viral entry into target cells. This restriction could not be released by C-terminal truncation of the gp41 glycoprotein. These studies imply that the highly conserved core residues of the HIV Env MSD, in addition to serving as a membrane anchor, play an important role in mediating membrane fusion during viral entry.
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) is a trimeric complex composed of three noncovalently linked dimers of gp120, the receptor-binding surface (SU) component, and gp41, the membrane-spanning, transmembrane (TM) component (8, 21, 43, 44). The gp120 and gp41 glycoproteins are synthesized as a precursor gp160 glycoprotein, which is encoded by the env gene. The gp160 precursor is cotranslationally glycosylated and, following transport to the trans-Golgi network, is cleaved into the mature products by a member of the furin family of endoproteases (44). Mature Env proteins are transported to the plasma membrane, where they are rapidly endocytosed or incorporated into virions (1, 28, 42). Recent evidence suggests that endocytosis and intracellular trafficking of Env are required for its interaction with Gag precursors and for efficient assembly into virions (16).
The gp41 monomer has three subdomains, i.e., an ectodomain, a membrane-spanning domain (MSD), and a cytoplasmic domain (39). The ectodomain of gp41, which mediates membrane fusion, is composed of a fusion peptide, two heptad repeats, and a tryptophan-rich membrane-proximal external region. Following the binding of gp120 to the CD4 receptor and CCR5/CXCR4 coreceptor, conformational changes are induced in Env and result in the exposure of the gp41 fusion peptide (27). This peptide inserts into the target cell membrane, allowing gp41 to form a bridge between the viral and cellular membranes. Interaction of the heptad repeats to form a six-helix bundle then brings the target and viral membranes together, allowing membrane fusion to occur (17).
The MSD is one of the most conserved regions of the gp41 sequence. Currently, two models have been proposed to explain the intramembrane topology of the MSD from several genetic studies. In the initial model, 25 amino acid residues (from K681 to R705 in the NL4-3 sequence) were suggested to cross the viral membrane in the form of an alpha-helical structure (10, 12, 14), the length of which is approximately equal to the theoretical depth of a membrane bilayer. A major caveat of this model is that it places a basic amino acid residue (R694) into the hydrophobic center of the lipid bilayer without any known mechanism to neutralize the basic amino group. Point mutation studies have yielded variable results, but in general replacement of K681 is detrimental to fusion and infectivity while mutation of R694 or R705 has only a limited effect on these activities (12, 24). On the other hand, accumulating data argues for a different intramembrane structure of the HIV-1 MSD. Serial small deletions (3 amino acid residues) in the region between R694 and R705 showed normal cell-cell fusion, although larger deletions were detrimental (24), suggesting that, with respect to the biological functions of the Env glycoprotein, the length of this region is more important than its amino acid conservation. Moreover, previous C-terminal truncation studies of simian immunodeficiency virus (SIV) Env (15, 41) and more recently of HIV-1 Env (L. Yue et al., unpublished data) have demonstrated that the entire 25-amino-acid region is not required for the biological function of Env. In the case of SIV, only the 15 apolar amino acids flanked by K689 and R705 (equivalent to K681 and R694 in HIV) and 6 additional amino acids (for a total of 23 amino acids) were required for near-wild-type (WT) fusion (15, 41), and we have shown that in HIV 18 amino acid residues (from K681 to A698) are enough to anchor the truncated Env glycoprotein in the membrane and allow it to mediate cell-cell fusion. From these data, we have proposed a topology where the flanking K681 and R694 residues “snorkel” in the membrane (41). In this model for HIV-1 Env, 12 amino acid residues (from L682 to L693) form a hydrophobic core buried within the membrane, while the backbones of K681 and R694 residues, which are flanking the hydrophobic core region, are also buried within the membrane. However, their long side chains are proposed to extend outward to the surface and present the positively charged amino groups to the negatively charged head groups of the lipid bilayers—hence the term “snorkel.” Arginines 705 and 707 are located at the interface of membrane and cytoplasm in this model.
The high degree of conservation of the HIV-1 Env MSD, particularly within the core region, argues for a functional role in the biological activity of the HIV-1 Env complexes. Replacement of the HIV-1 MSD by equivalent regions from other transmembrane proteins has yielded variable results. Replacement of the HIV-1 MSD with a glycophospholipid anchor abrogated both cell-cell fusion and virus-cell fusion, though the chimeric protein could be normally expressed, processed, and incorporated into virions (30). Moreover, the replacement of the HIV-1 MSD with the MSDs of other transmembrane proteins, such as glycophorin A (19), vesicular stomatitis virus G protein (VSV-G) (19), and influenza virus hemagglutinin (40), resulted in an impaired ability of Env complexes to mediate membrane fusion. On the other hand, the replacement of the MSD of Env with that of CD22 (42) did not appear to block the replication of HIV in MT4 cells, and an Env chimera containing the transmembrane region of CD4 (26) has also been shown to induce normal membrane fusion.
According to the consensus sequence of the HIV-1 MSD, the 12 amino acid residues in the core region are generally more conserved than those C terminal to this region (from R694 to R705) (Fig. 1). Recent studies of the MSD core region have focused on the GXXXG motif, in which the glycine residues are the most conserved among all MSD residues. Statistical studies and bacterial models suggested that the GXXXG motif probably constructs a framework for the association of transmembrane helixes (29, 32, 33). It has been demonstrated that the GXXXG motif in the E1 glycoprotein of hepatitis C virus is important for the heterodimerization of its E1 and E2 envelope glycoproteins (5). The MSD of VSV-G also contains a GXXXG motif. Alanine and leucine scanning experiments showed that mutation of the two glycines reduced the fusion activity of VSV-G to half of the WT level, but simultaneous mutation of both residues almost completely blocked membrane fusion (6). Similar results were reported for the GXXXG motif in the HIV-1 MSD core, in which the fusogenicity of Env started decreasing when more than one glycine residue was substituted for alanine or leucine (18, 19).
FIG. 1.
Amino acid sequences of HIV-1 MSDs. The consensus sequence of the HIV-1 MSD was generated by the alignment of the Env MSD sequences of all M and N group HIV-1 isolates from the Los Alamos HIV sequence database (updated to June 2007). The percentages of isolates with the conserved amino acid residue are shown above the consensus sequence as a bar chart. The amino acid residues of the putative MSD region are shown as uppercase letters. The flanking sequences of the HIV-1 MSD are shown as lowercase letters. The position of the MSD in the sequence of HIV-1 NL4-3 is marked above the positively charged amino acid residues. The mutated portions of the HIV-1 MSD are underlined.
In order to further understand the biological functions of the more conserved HIV-1 MSD core structure, we initially changed all of the residues to leucine within the core, then sequentially reintroduced the GGXXG motif, and then introduced F683 with V687. We show here that the specific amino acid residues in the helical core of HIV-1 MSD are critical for the fusogenicity of Env complexes and infectivity of HIV-1. The loss of infectivity correlated with impaired virus-cell fusion. However, these mutations in the MSD core did not influence the biogenesis, intracellular transport, and incorporation of Env complexes. Truncation of the cytoplasmic domain of HIV-1 Env was not able to compensate for the fusion defects caused by MSD core mutations.
MATERIALS AND METHODS
Cells and antibodies.
COS-1, 293T, and JC53BL cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin (all from Gibco-BRL, Rockville, MD). Cells were grown at 37°C with 5% CO2 in humidified incubators. The anti-gp120 b12, anti-gp160 Chessie 13-39.1, and anti-p24 183-H12-5C monoclonal antibodies (MAbs) along with HIV-1 patient immunoglobulin were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The pooled HIV-1 patient sera were provided through the Emory CFAR Clinical Core. The sheep anti-HIV-1 gp120 polyclonal antibody and goat anti-human immunoglobulin G (heavy plus light chains) conjugated to horseradish peroxidase were purchased from Cliniqa Corp. (San Marcos, CA) and Pierce (Rockford, IL), respectively.
Glycoprotein and proviral expression constructs.
Construction of the HIV-1 Env expression vector pSRHS, which carries full-length env, tat, and rev genes from NL4-3, has been described previously (31). This simian virus 40 (SV40) (late promoter)-based vector contains a Mason Pfizer monkey virus-derived long terminal repeat providing a polyadenylation signal. A unique XbaI site (nucleotide 8213 in NL4-3) was previously introduced into the pNL4-3 env gene in both pSRHS and the proviral vector pNL4-3. In this study, we employed a two-step overlapping PCR approach to generate DNA fragments containing mutated MSDs. Briefly, forward and reverse oligonucleotides were designed to span the mutagenesis region between lysine 681 and arginine 694. The oligonucleotide sequences were as follows: L12FOR, 5′-TTACTACTGTTATTACTGCTCTTGTTACTGTTAAGAATAGTTTTTGCTG-3′; L12REV, 5′-CAGTAACAAGAGCAGTAATAACAGTAGTAATAATTTTATATACCACAGCC-3′; L12GFOR, 5′-TTACTACTGTTATTAGGAGGCTTGTTAGGTTTAAGAATAGTTTTT GCTG-3′; L12GREV, 5′-ACCTAACAAGCCTCCTAATAACAGTAGTAATAATTTTATATACCACAGCC-3′; L12G2FOR, 5′-TTCCTACTGTTAGTAGGAGGCTTGTTAGGTTTAAGAATAGTTTTTGCTG-3′; and L12G2REV, 5′-ACCTAACAAGCCTCCTAGTAACAGTAGGAATAATTTTATATACCA CAGCC-3′. We also designed a forward primer (PrimerXbaI) overlapping the unique XbaI site and a reverse primer (PrimerBamHI) overlapping a unique BamHI site downstream of the MSD sequence. In the first step of PCR, PrimerXbaI and reverse primers amplified sequence A of each MSD mutant, while PrimerBamHI and forward primers amplified sequence B of each MSD mutant, using WT NL4-3 env as the template in both reactions. The mixed A and B amplicons were then used as templates to amplify target sequences primed by PrimerXbaI and PrimerBamHI. The WT NL4-3 env region was replaced with mutated sequences by insertion into the XbaI and BamHI sites in both SV40-based expression vector pSRHS and proviral vector pNL4-3. We constructed at least three independent clones for the WT and each mutant. All mutations were confirmed by DNA sequencing using a primer approximately 100 bp upstream from the MSD sequence. In this study, nucleotides and amino acid residues were labeled according to the NL4-3 sequence (GenBank accession number AF324493) if not otherwise indicated.
Glycoprotein expression and immunoprecipitation.
The pSRHS Env expression vectors were transfected into COS-1 cells, and cytomegalovirus (CMV)-based Env expression vectors as well as proviral vectors were transfected into 293T cells, using Fugene6 (Roche, Indianapolis, IL) in six-well plates. The pSRHS vectors are not expressed in 293T cells because transcription from the SV40 late promoter is inhibited in these cells. At 36 to 48 h posttransfection, cells were starved for 15 min in methionine-free and cysteine-free DMEM and then labeled for 30 min in methionine-free and cysteine-free DMEM supplemented with [35S]methionine and [35S]cysteine (125 μCi/well). The labeled cells were then chased in complete DMEM for 1 h, 2 h, 3 h, and 5 h. All medium was filtered through a 0.45-μm membrane to remove cellular debris. For 293T cells transfected with proviral vectors, viral particles were pelleted through a 25% sucrose cushion by ultracentrifugation (100,000 × g, 2.5 h). Cells and viral pellets were lysed by a 10-min incubation on ice in lysis buffer (1% Triton X-100, 50 mM NaCl, and 0.1% sodium dodecyl sulfate [SDS] in 25 mM Tris-HCl [pH 8.0]). Cellular debris was removed by microcentrifugation at 13,200 rpm for 1 min at 4°C. HIV-1 proteins were immunoprecipitated from cell lysates, supernatants, and viral pellets by incubating overnight at 4°C with pooled HIV-1 patient sera. Immune complexes were incubated overnight at 4°C with fixed Staphylococcus aureus cells and pelleted in a microcentrifuge. The pellets were then washed three times in lysis buffer, and labeled proteins resolved by 8% SDS-polyacrylamide gel electrophoresis (PAGE) were visualized by autoradiography. The glycoprotein bands were quantified using a Cyclone phosphorimaging system (Packard, Meriden, CT) as previously described (41).
Cell-cell fusion assay.
COS-1 cells were transfected with pSRHS expression vectors, and 293T cells were transfected with pNL4-3 proviral vectors by using Fugene6. At 36 to 48 h after transfection, cells resuspended by trypsinization were combined with JC53BL indicator cells at a 5:1 ratio. Cell mixtures were incubated for 6 h, 12 h, and 24 h and then were lysed in luciferase reporter lysis buffer (Promega, Madison, WI) using two freeze-thaw cycles. Cellular debris was removed by centrifugation at 13,200 rpm for 5 min at 4°C in a microcentrifuge (Beckman, Palo Alto, CA). Luciferase substrate (100 μl) (Promega, Madison, WI) was added to 10 μl of each cell lysate, and light emission was quantified using a Synergy multidetector microplate reader (Biotek, Winooski VT).
Cell surface expression of Env glycoprotein.
COS-1 cells and 293T cells were transfected with expression vectors and proviral vectors, respectively, cultured for 36 to 48 h, and then, after removal from the plate, fixed for 20 min at 4°C in 4% paraformaldehyde (in phosphate-buffered saline [PBS] [pH 7.2]). Fixed cells were stained for 1 h at room temperature with 5 μg/ml of Alexa 647-conjugated anti-gp120 MAb b12. Then cell membranes were permeabilized using the Cytofix/Cytoperm fixation/permeabilization kit (BD Biosciences, San Diego, CA). Permeabilized COS-1 and 293T cells were further stained with Alexa 488-conjugated anti-gp160 Chessie13-39.1 or Alexa 488-conjugated anti-p24 183-H12-5C MAb, respectively, in order to normalize transfection efficiency. For some experiments a relative mean fluorescence index was calculated as percent transfected cells × mean fluorescence intensity. Both Alexa 647 and Alexa 488 conjugation kits were obtained from Invitrogen (Carlsbad, CA). Double-stained cells were subjected to flow cytometry analysis using the FACSCalibur system.
Infectivity of HIV-1 in JC53BL indicator cells.
Medium from 293T cells transfected with proviral vectors was harvested at 72 h posttransfection and filtered through a 0.45-μm membrane to remove cellular debris, and total virions were quantified using a p24 enzyme-linked immunosorbent assay (ELISA). The p24-normalized virus-containing supernatant was added to JC53BL indicator cells cultured in DMEM containing 1% fetal bovine serum and 80 μg/ml DEAE-dextran. Complete DMEM was added following a 2-h incubation, and cells were analyzed for luciferase activity at 48 h postinfection.
Single-round infection.
293T cells were cotransfected with pSRHS expression vectors and the pSG3Δenv proviral vector. Medium was collected at 72 h posttransfection and then was subjected to p24 ELISAs. The p24-normalized virus-containing supernatants were used to infected JC53BL indicator cells. Luciferase activity was measured at 48 h after infection.
Incorporation of Env into viral particles.
Two methods were used to measure the efficiency of incorporation of Env glycoprotein into viral particles. (i) Medium collected from 293T cells at 72 h after transfection with proviral vectors was filtered, and viral particles were pelleted through a 25% sucrose cushion by ultracentrifugation (100,000 × g, 2.5 h). The supernatant including the sucrose solution was collected, and viral pellets were resuspended in PBS (pH 7.2). The amounts of p24 and gp120 in both supernatants and viral pellets were measured using a p24 ELISA and a gp120 ELISA (11). Efficiencies of incorporation in the viral pellets were compared using the ratio of gp120 to p24 for each mutant. The gp120/p24 ratio in the supernatant reflected the relative amount of shed gp120. (ii) Transfected 293T cells were starved for 15 min in methionine-free and cysteine-free DMEM and then labeled for 30 min in methionine-free and cysteine-free DMEM supplemented with [35S]methionine and [35S]cysteine (125μCi/well). The labeled cells were then chased for 24 h. Viral particles in the supernatant were pelleted by ultracentrifugation (100,000 × g, 2.5 h) through a 25% sucrose cushion, followed by lysis in lysis buffer. HIV-1 proteins were immunoprecipitated by incubation overnight at 4°C with pooled HIV-1 patient sera. Immune complexes were incubated overnight at 4°C with fixed Staphylococcus aureus cells and pelleted in a microcentrifuge (13,200 rpm, 1 min). Labeled proteins resolved by 8% SDS-PAGE were visualized by autoradiography and quantified using a phosphorimager, as described above. Incorporation efficiencies were calculated as the ratio of the digital light units of gp120 bands to those of p24 bands.
Virus-cell fusion.
A virion-based virus-cell fusion assay was performed as described previously (2-4). Briefly, 293T cells were cotransfected with proviral vector pNL4-3, pCMV-BlaM-Vpr construct (kindly provided by W. Greene, UCSF), and pAdvantage construct (Promega, Madison, WI) using calcium phosphate precipitation. The pCMV-BlaM-Vpr vector contains a CMV promoter-driven β-lactamase-coding region fused to the N terminus of the HIV-1 vpr gene. At 48 h posttransfection, medium filtered through a 0.45-μm membrane was loaded onto a 25% sucrose cushion (in PBS [pH 7.2]) and centrifuged at 100,000 × g for 2.5 h at 4°C. Pelleted virus was resuspended in serum-free DMEM, the titer was determined using a p24 ELISA, and then 200 ng p24 equivalents was added to 3 × 105 JC53BL cells cultured in CO2-independent medium (Gibco-BRL, Rockville, MD) supplemented with 1% fetal bovine serum. After a 6-h incubation at 37°C, free viruses were washed away in serum-free CO2-independent medium. The fluorescent dye, CCF2-AM, was then loaded into these cells during a 2-h incubation at room temperature according to the instructions for the β-lactamase loading kit (Invitrogen, Carlsbad, CA). The extracellular dye was then washed away in serum-free CO2-independent medium, and cells were resuspended in CO2-independent medium supplemented with 10% fetal bovine serum and 2.5 mM probenecid. The cells were then incubated for 16 h at room temperature in the dark, fixed with 4% paraformaldehyde for 20 min at 4°C, and analyzed by flow cytometry in a Beckman Dickinson LSR2 cytometer.
RESULTS
In this study, we used a recovery-of-function mutagenesis strategy to define the roles of specific amino acid residues within the 12-amino-acid MSD core in HIV-1 assembly and entry. The amino acid sequences and nomenclature of the MSD core mutants are shown in Fig. 1. In the L12 mutant, the 12 amino acid residues between K681 and R694 were replaced by a polyleucine peptide of identical length. Leucine is the most hydrophobic amino acid, and a polyleucine peptide would be expected to form a homogenous hydrophobic, helical structure in the lipid bilayer. Subsequently, in the L12G mutant we reintroduced the conserved GGXXG motif into the polyleucine backbone. Finally, V687, which is immediately upstream of the GGXXG motif and has been postulated to contribute to the helix-helix interactions mediated by the GXXXG motif, as well as the highly conserved bulky amino acid F683, were reintroduced into the MSD core of the L12G mutant.
MSD core mutants exhibit normal Env biosynthesis and processing.
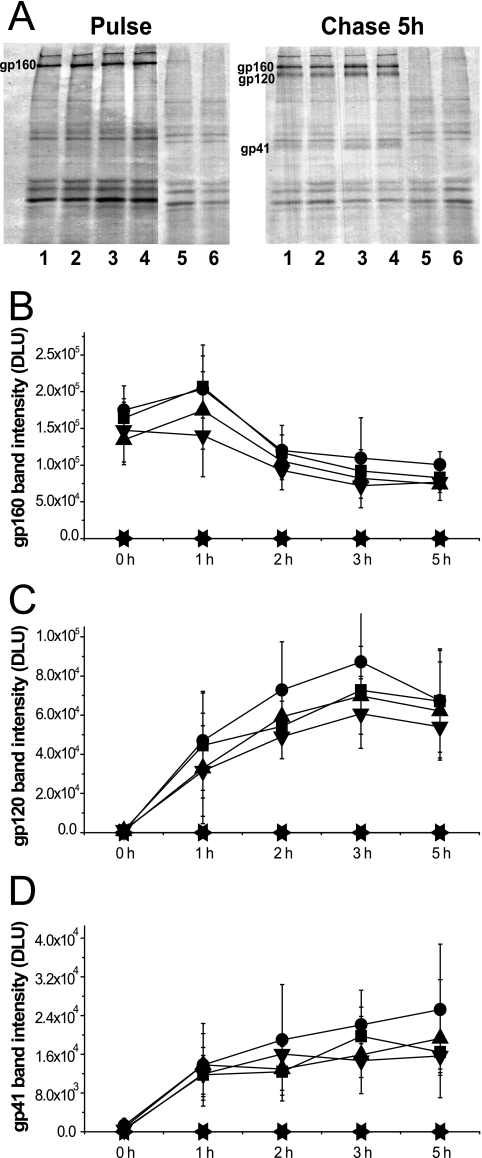
WT and mutant env sequences carrying, in addition, full-length tat and rev genes were cloned into the vector pSRHS, in which expression of Env is under the control of the late promoter from SV40 and a polyadenylation signal from Mason Pfizer monkey virus (7, 13). Mutant Env glycoproteins expressed in COS-1 cells were metabolically labeled in a 30-min pulse and chased in complete medium over the course of 5 h. Viral glycoproteins were subsequently immunoprecipitated with pooled HIV-1 patient sera and analyzed by SDS-PAGE followed by autoradiography. Quantification of protein bands was carried out using a phosphorimager (Packard, Meriden, CT). As shown in Fig. 2A, we observed similar profiles for the cell-associated Env glycoproteins in COS-1 cells expressing either WT or mutant Envs after a 5-h chase. In a time course experiment (Fig. 2B to D), the MSD core mutants had processing kinetics similar to those of WT Env, with the gp160 precursors decreasing and gp120 and gp41 subunits increasing at similar rates. These data suggested that the replacement of specific amino acid residues in the MSD core did not alter the biosynthesis of the glycoprotein precursor or its transport to the Golgi network for completion of carbohydrate processing and cleavage to the SU and TM subunits.
FIG. 2.
Expression profiles of HIV-1 Env glycoprotein. (A) COS-1 cells transiently transfected with SV40-based Env expression vectors were metabolically labeled and chased over 5 h. Env glycoprotein was immunoprecipitated, followed by 8% SDS-PAGE analysis and autoradiography (lane 1, pSRHS WT; lane 2, pSRHS L12; lane 3, pSRHS L12G; lane 4, pSRHS L12G2; lane 5, empty vector; lane 6, mock). The bands of gp160 precursors (B), gp120 (C), and gp41 (D) in cell lysates at each time point were quantified by phosphorimager calculation, and the intensity of bands is shown as digital light units (DLU) (squares, pSRHS WT; circles, pSRHS L12; upward triangles, pSRHS L12G; downward triangles, pSRHS L12G2; leftward triangles, empty vector; rightward triangles, mock). Results are from four independent experiments; error bars represent standard deviations from the means.
MSD core mutants exhibit normal cell surface expression of Env glycoprotein.
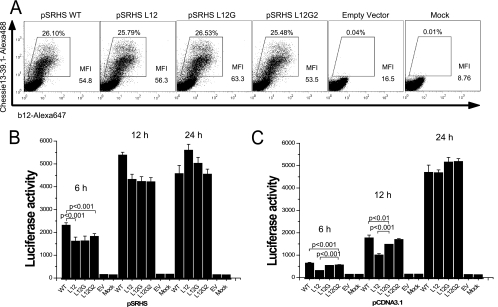
Since synthesis and processing of MSD-mutated HIV-1 Env glycoproteins appeared to be normal, we examined the surface expression of these mutant proteins to determine whether they were transported to, and retained normally in, the plasma membrane. Transfected COS-1 cells were fixed in 4% paraformaldehyde and stained with Alexa 647-conjugated anti-gp120 MAb b12, and then cells were subjected to flow cytometry analysis. Transfection efficiency was monitored by intracellular staining following permeabilization using Alexa 488-conjugated anti-gp160 MAb Chessie13-39.1. As shown in Fig. 3A, the MSD-mutated Env glycoproteins were presented on the cell surface at levels equivalent to that of the WT glycoprotein, with mean fluorescence intensities varying from 54.8 for the WT to 63.5 for L12G, indicating that the GGXXG motif and specific amino acid residues in the MSD core region are not required for stable surface expression of the Env glycoprotein.
FIG. 3.
Cell surface level of Env glycoprotein and cell-cell fusion examined using SV40-based Env expression vectors. (A) COS-1 cells transfected with pSRHS expression vectors were fixed and labeled with Alexa 647-conjugated anti-gp120 MAb b12. Cells were then permeabilized and stained with Alexa 488-conjugated anti-gp160 MAb Chessie 13-39.1 as a transfection efficiency control. (B) Cell-cell fusion assay. COS-1 cells transfected with the pSRHS Env expression vectors were cocultured with JC53BL indicator cells. Cell mixtures were lysed, and luciferase activity was measured after 6 h, 12 h, and 24 h of incubation. (C) Cell-cell fusion assay. 293T cells transfected with pCDNA3.1 Env expression vectors were cocultured with JC53BL indicator cells. Cell mixtures were lysed, and luciferase activity was measured after 6 h, 12 h, and 24 h of incubation. Results are from three independent experiments; error bars represent standard deviations from the means. EV, empty vector.
MSD core mutants exhibit minor defects in cell-cell fusion when expressed at high levels.
Because all of the MSD core mutants and the WT were expressed at similar levels on the surface of COS-1 cells following transfection with the pSRHS vectors, we determined whether these mutant Env glycoproteins had an equivalent capacity to mediate cell-cell fusion. Transiently transfected COS-1 cells were cocultured with JC53BL indicator cells, which are susceptible to infection by both R5 and X4 HIV-1 isolates and contain a luciferase reporter gene under the control of the HIV-1 long terminal repeat promoter and which can be activated by the Tat protein (38). Cell-cell fusion was measured by calculating the relative luciferase enzyme activity in cells expressing the mutant Envs compared to those expressing WT Env at 6 h, 12 h, and 24 h after coculture. Each of the mutants exhibited reduced fusion relative to the WT at 6 h, with L12 inducing 70% the fusion observed with the WT. This effect was reduced at 12 h and lost at 24 h (Fig. 3B). Similar results were observed when the same env sequences were cloned into the pCDNA3.1 vector (CMV promoter) and transfected 293T cells were used as the effector cells (Fig. 3C). In this case, fusion induced by the L12 mutant was only 44% of that induced by the WT at 6 h (L12G, 83%; L12G2, 86%), but it increased to 56% at 12 h and was equivalent to the WT level at 24 h after mixing. The small differences in fusion defects observed in the two expression vectors could reflect differences in surface expression levels, since in general the mean fluorescence intensities observed with the pSRHS vectors were double that with the pCDNA3.1 vector. Moreover, transfection with 300 ng (10-fold reduction) of the pCDNA3.1 vector yielded surface expression levels approximately fivefold less than that observed with the pSRHS vector, and under these conditions we observed a 50% decrease in fusion by the L12 mutant relative to the WT at 24 h postcoculture (data not shown).
MSD core mutant viruses show defects in infectivity.
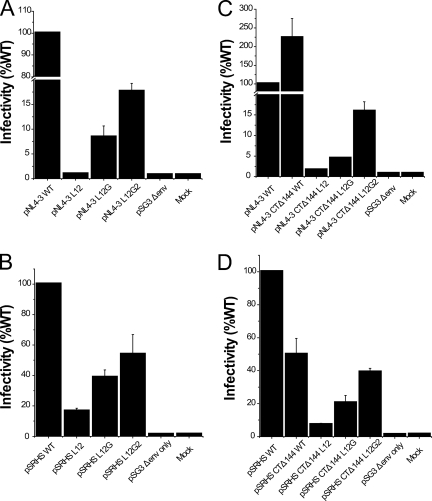
In order to analyze whether the MSD core mutations affected virus infectivity, each mutated env gene was substituted into the proviral vector pNL4-3, and virions were produced by transfection of 293T cells. Following normalization based on p24 content, these virions were used to infect JC53BL indicator cells. The substitution of a polyleucine peptide for the MSD core in the L12 mutant reduced infectivity on JC53 cells more than 100-fold (Fig. 4A). The single reintroduction of the GGXXG motif in the L12G mutant increased infectivity to 8.4% of that of the WT, and the further addition of conserved amino acid residues, F683 and V687, into the L12G2 mutants resulted in another twofold increase in infectivity to 17.6% of that of the WT. An env deletion mutant of HIV-1 (pSG3Δenv) exhibited only background infectivity (0.75% of that of the WT) in this assay. These data suggest that the specific amino acid residues and conserved motifs in the MSD core region are critical for Env-mediated infectivity in the context of the virion.
FIG. 4.
Infectivity of HIV-1 Env mutants. (A and C) HIV-1 viruses containing the Env MSD core mutations (A) and viruses containing truncated Env plus the MSD core mutations (C) were produced by transfecting proviral DNA into 293T cells. The p24-normalized cell culture supernatant was used to infect JC53BL indicator cells. Cells were lysed and luciferase enzyme activity was assayed after 48 h of incubation. (B and D) Single-round infectivity assay. Env-defective proviral construct pSG3Δenv was cotransfected into 293T cells with WT and mutant Env expression vectors (B) and cytoplasmic domain-truncated Envs (D). Pseudotyped viruses in the cell culture supernatant were quantitated using a p24 ELISA assay. The p24-normalized supernatant was then used to infect JC53BL indicator cells. Infectivity of MSD mutants is shown as relative luciferase activity compared to that of the WT. Results are from three independent experiments; error bars represent standard deviations from the means.
MSD core mutant Envs expressed in the context of virus are processed normally but show defects in cell-cell fusion.
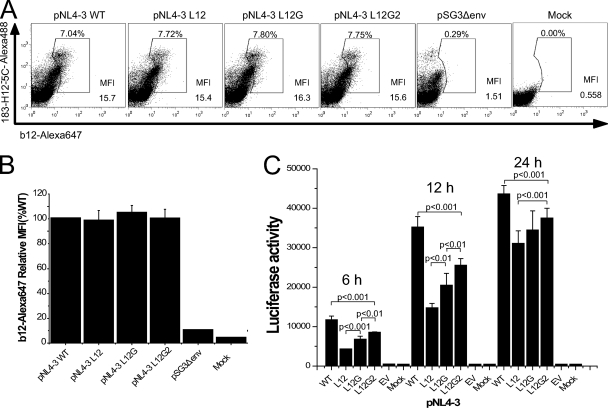
In order to understand the mechanism of the defect in infectivity caused by the MSD core mutations, we examined the biosynthesis, surface expression, and induction of cell-cell fusion by these mutants in the context of virus. In pulse-chase experiments similar to those shown for the pSRHS expression vector (Fig. 2), we observed that in the context of the pNL4-3 genome the MSD core mutant Env precursor gp160 was expressed and proteolytically processed to gp120 and gp41 with kinetics similar to those of the WT gp160 (data not shown). Flow cytometric analyses of cells stained with Alexa 647-conjugated anti-gp120 MAb b12 showed that while the WT and MSD mutants had equivalent levels of surface Env (Fig. 5A and B), the mean fluorescence index for the virus-based vector was fourfold lower than that for the pSRHS vector.
FIG. 5.
Cell surface level of Env glycoprotein and cell-cell fusion examined in the context of provirus expression. (A) 293T cells transfected with pNL4-3 proviral vectors were fixed and were labeled with Alexa 647-conjugated anti-gp120 MAb b12. Cells were then permeabilized and stained with Alexa 488-conjugated anti-p24 MAb 183-H12-5C as a transfection efficiency control. (B) The surface levels of Env glycoprotein are shown as relative mean fluorescence index (MFI) compared to that of WT. (C) Cell-cell fusion in the context of viruses. 293T cells transfected with proviral DNA were mixed with JC53BL indicator cells and cocultured for 6 h, 12 h, and 24 h before the measurement of luciferase activity. P values were calculated using Student's t test. Results are from three independent experiments; error bars represent standard deviations from the means.
In order to determine the ability of virus-producing cells to mediate cell-cell fusion, 293T cells transfected with proviral DNA were cocultured with JC53BL indicator cells for 6 h, 12 h, and 24 h in the presence of 10 μM zidovudine to prevent virus replication and then assayed for luciferase activity. In the context of virus, we found significant differences (P < 0.01) in the abilities of the MSD mutants to mediate cell-cell fusion (Fig. 5C). Mutant NL4-3 L12 induced only 35% of the cell-cell fusion observed with WT NL4-3. When the GGXXG motif was reintroduced into the mutated MSD core, fusion increased to 56% of the WT level, while the further addition of V687 and F683 increased fusion to 72% of that of the WT. The difference in cell fusion observed in the context of Env expression vectors and the NL4-3 provirus is most likely explained by differences in the amount of Env expressed at the cell surface. Therefore, under conditions where cell surface expression levels were reduced to that characteristic of virus-infected cells, the mutants did exhibit a reduced ability to mediate cell-cell fusion, although the extent of this defect was less than that observed in infectivity assays.
MSD core mutant Envs are incorporated efficiently into viral particles.
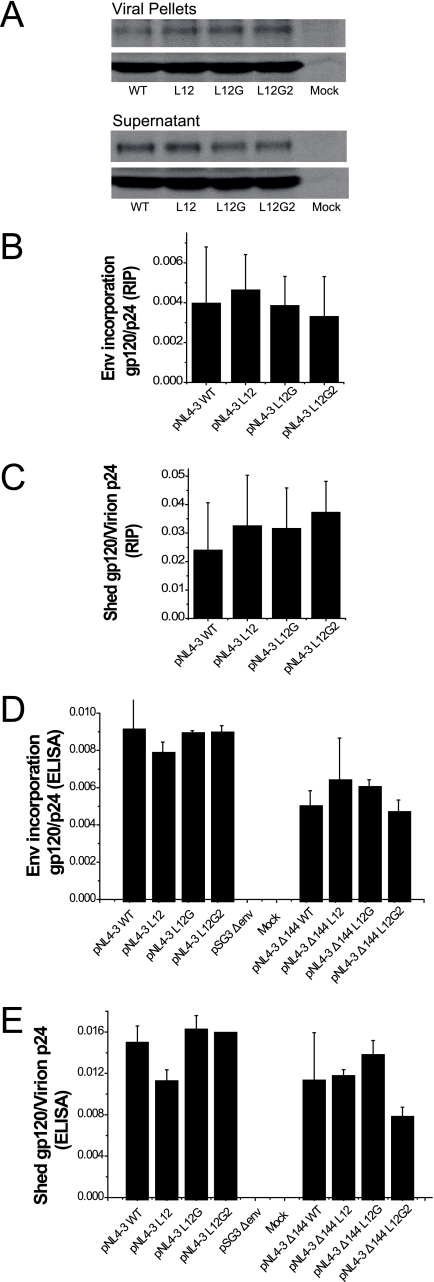
In order to determine whether a defect in Env incorporation might also contribute to the MSD core mutant impaired infectivity, we utilized two different approaches to measure the efficiency of incorporation of mutated Envs into viral particles. In the first, virions released from provirus-transfected 293T cells that were pulse-labeled and chased for 24 h were pelleted through a sucrose cushion and analyzed by SDS-PAGE (Fig. 6A). The ratios of gp120 to p24 for WT and mutant proviruses were calculated using a phosphorimager, and, as can be observed in Fig. 6B, similar levels of glycoprotein incorporation were observed for each construct. To confirm this result, the amounts of gp120 and p24 in viral pellets and supernatants were analyzed using gp120 and p24 ELISAs after centrifugation of unlabeled transfected 293T cell culture fluids. We observed that the WT and mutant virions had similar gp120/p24 ratios (Fig. 6D), confirming that the MSD core mutations did not alter the incorporation of Env complexes into viruses. Moreover, the gp120 glycoprotein of the MSD mutants appeared to be as stable as the WT, because equivalent amount of gp120 were found in medium after centrifugation (Fig. 6C and E).
FIG. 6.
Incorporation of Env into virions. (A) 293T cells transfected with proviral DNA were pulse-labeled with [35S]methionine and [35S]cysteine and chased over 24 h. Viral particles in medium were pelleted through a 25% sucrose cushion by ultracentrifugation. Viral proteins were immunoprecipitated with pooled AIDS patient sera and analyzed by 8% SDS-PAGE followed by autoradiography. (B) The viral gp120 and p24 bands in viral pellets and supernatants after ultracentrifugation were quantified by phosphorimaging analysis. The efficiency of incorporation of Env into viral particles is shown as the ratio of the intensities of gp120 bands in virions to those of p24 in virions. (C) The relative amount of gp120 shed into medium is shown as the ratio of the intensities of gp120 bands in supernatant after ultracentrifugation to those of p24 in virions. (D) Unlabeled viral pellets and supernatants were subjected to gp120 ELISA and p24 ELISA. The incorporation efficiency is shown as the ratio of the concentration of gp120 to that of p24 in virions for both full-length (left) and truncated (right) Envs. (E) The degree of shedding of gp120 is shown as the ratio of the amount of gp120 in supernatant after centrifugation to that of p24 in virions for both full-length (left) and truncated (right) Envs. The standard deviations of three repeats for each experiment are shown as error bars.
Pseudotyped virions containing MSD core mutant Envs are defective for entry and virus-cell fusion.
In order to determine if the MSD core mutants were defective at the stage of entry, and specifically at the stage of virus-cell fusion, we utilized a single-round infection pseudotyped virus assay. In order to compare infectivities of these viruses with the NL4-3 constructs, pSG3Δenv was pseudotyped with WT and mutant Envs expressed from the pSRHS vector, and then the pseudotyped virions were used to infect JC53BL indicator cells. A pattern of defective infectivity similar to that observed in the NL4-3 infectivity assay (Fig. 4A) was seen in this assay, although the extent of the defect was reduced (Fig. 4B). The L12 mutant induced only 17% of the luciferase activity observed for WT Env pseudotyped virus, but reintroduction of conserved amino acid residues increased it to 39% in mutant L12G and 54% in mutant L12G2.
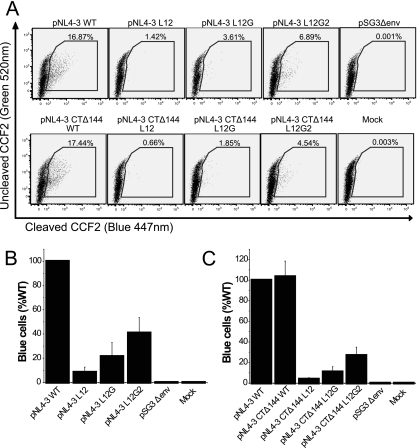
Because the expression of luciferase in JC53BL cells depends on the expression of the tat gene, a late event after viral entry, it was possible that the defect of the MSD core mutants in single-round infection could be at a postentry step. Therefore, we utilized WT and mutant NL4-3 viruses that contained a β-lactamase-Vpr fusion protein (BlaM-Vpr) to pinpoint the stage at which virus infection was blocked. JC53BL target cells infected with the BlaM-Vpr viruses and then loaded with the fluorescent dye CCF2-AM (green), which can be cleaved by β-lactamase into CCF2 (blue) and AM immediately after viral entry. This method is a sensitive indicator of the membrane fusion between the viral envelope and cell plasma membrane. As shown in Fig. 7A and B, the L12 mutation decreased virus-cell fusion by approximately 92% compared to WT, but reintroduction of GGXXG motif increased fusion almost threefold to 21% of that of the WT. Further addition of F683 and V687 increased virus-cell fusion to 41% of that of the WT. Virus attachment assays carried out at 4°C, 10°C, and 25°C showed that WT and mutant viruses bound to the target cells with equivalent efficiency (data not shown). These data confirm that the MSD core mutants are blocked at the stage of virus-cell fusion during the entry process and that conserved amino acid residues play a key role in this process.
FIG. 7.
Virus-cell fusion assay. (A) Proviral constructs were cotransfected into 293T cells with pCMV-BlaM-Vpr vectors. Viruses containing the BlaM-Vpr fusion protein were pelleted by ultracentrifugation through 25% sucrose. Viral pellets normalized by p24 ELISA were used to infect JC53BL indicator cells. Then cells were loaded with fluorescent dye CCF2-AM. Blue cells were counted by flow cytometry analysis after 16 h of incubation. (B and C) The ability of the Env MSD mutants (B) and the truncation mutants (C) to mediate virus-cell fusion is shown as relative numbers of blue cells compared to those of the WT. The experiment was repeated at least three times; standard deviations are shown as error bars.
C-terminally truncated MSD core mutants.
SIV Env and HIV Env have remarkably long cytoplasmic domains. It has been recently reported that the second-site reversion mutants of SIV and HIV-1 that were resistant to amphotericin B methyl ester (AME), a cholesterol-binding compound, had truncated Env cytoplasmic domains (36). It was suggested that the truncated protein had a higher potential for movement in the lipid bilayer, which resulted in the escape from inhibition by AME. Since it was possible that the loss of conserved amino acid residues in the MSD core region altered the mobility of the Env glycoprotein in the membrane at the time of fusion pore formation, we determined whether truncation of the cytoplasmic domain of HIV-1 Env could relieve the defect observed in the absence of the glycine motif. Each mutant and the WT were therefore reconstructed into the pNL4-3 Δ144 proviral vector, in which a stop codon was engineered into the gp41 cytoplasmic domain of the WT and the MSD core mutants at Y710 (Y712 in HXB2). Incorporation of truncated Env complexes was reduced for both the MSD WT and core mutants (Fig. 6D). In addition, the amount of gp120 shed into the supernatant was also decreased (Fig. 6E). Moreover, we did not observe any reversal of the infectivity defect in either NL4-3-based provirus assays (Fig. 4C) or pseudotyped virus single-cycle assays (Fig. 4D). While a pNL4-3 provirus encoding a truncated Env was approximately twice as infectious as WT pNL4-3, this was not the case for virus pseudotyped with the truncated Env. Indeed, in general viruses encoding the truncated mutant Envs were less infectious than their nontruncated counterparts (compare Fig. 4A and B with Fig. 4C and D), with virus pseudotyped with the truncated WT Env exhibiting a 50% decrease relative to its nontruncated counterpart. Even taking into account this reduced WT infectivity, the L12 and L12G mutants showed defects in infectivity equivalent to that observed with their nontruncated versions. Similar results were also observed in virus-cell fusion assays based on viruses containing the BlaM-Vpr fusion protein (Fig. 7A and C). Thus, truncation of the gp41 cytoplasmic domain is not able to complement the fusion and infectivity defects observed with the MSD core mutants.
DISCUSSION
The primary function of the HIV-1 MSD is as a membrane anchor of the Env glycoprotein in both viral and cellular membranes; deletion of this region results in secretion of soluble gp160 molecules (10, 34). However, studies have suggested that this domain is involved in other important biological functions during the viral life cycle: (i) sequence alignments of the MSDs from more than 900 HIV isolates (in the Los Alamos HIV database) showed that the amino acid sequence of this domain is more highly conserved than might be expected to merely anchor the protein in the membrane; (ii) replacement of the HIV-1 MSD with the equivalent regions of glycophorin A, VSV-G, or the influenza virus hemagglutinin leads to significant defects in cell-cell fusion mediated by Env glycoprotein (19, 40); and (iii) synthetic peptides corresponding to the HIV-1 MSD region are able to mediate phospholipid mixing and then fusion between two lipid vesicles (22).
In the absence of any crystallographic or nuclear magnetic resonance-derived structural information on the HIV-1 MSD, the exact length of this region remains unresolved. Two topological models have been proposed to explain the intramembrane structure of the HIV-1 MSD, and they differ in the location of the three basic residues (K681, R694, and R705) with respect to the lipid bilayer (20, 41). In order to better understand the biological functions of this highly conserved MSD and the roles played by conserved motifs in this region, we defined an MSD core region (K681 to R694), which is more conserved than the rest of the HIV-1 MSD and is buried within the lipid bilayer in both models. Employing a recovery-of-function approach by first replacing the entire MSD core with leucine residues, we then determined the effect of reintroducing conserved residues and motifs into this leucine backbone.
A highly conserved GGXXG motif is present in the HIV-1 MSD core region, and it has been postulated that this motif on an intramembrane helix forms a framework for transmembrane helix-helix association. The absence of long side chains on the two glycine residues that are present on the same face of the helix is postulated to provide a flat platform to mediate helix-helix interactions (33). For hepatitis C virus Env, replacement of the glycine residues in the GXXXG motif of the E1 protein by tryptophan residues impaired the heterodimerization of E1E2 protein and the following fusion steps (5). The results of the studies presented here, however, show that specific amino acid residues between K681 and R694 are not required for the normal expression, carbohydrate modification, and proteolysis of Env glycoprotein precursor. There was no evidence that loss of the glycine-rich GGXXG motif or aromatic residues in the MSD interfered with the intracellular trafficking of Env from the endoplasmic reticulum to the trans-Golgi network, where the gp160 precursor is cleaved into gp120 and gp41 subunits. This argues that trimerization of Env monomers was unaffected by loss of these residues, since the transport of Env glycoprotein from the endoplasmic reticulum to the Golgi apparatus has been shown to require the oligomerization of gp160 monomers (23). This conclusion is consistent with the results observed when the membrane-spanning regions of other transmembrane proteins were used to replace the MSD of the HIV-1 Env (19, 40, 42). In all these cases, the chimeric Env glycoproteins were synthesized and cleaved as efficiently as WT Env, arguing that the HIV-1 MSD does not play a critical role in intracellular trafficking of Env. We also observed WT surface levels of the mutated Env glycoproteins on cells transfected with either an SV40 promoter-based expression vector in the absence of other viral proteins or in the context of full-length proviral vectors, suggesting that specific amino acid residues within the HIV-1 MSD core are also dispensable for the intracellular transport of Env to the plasma membrane. Moreover, the fact that mutant Envs were incorporated into viral particles as efficiently as the WT argues that specific residues in the MSD are not required for this process either. The levels of shed gp120 were similar for the MSD mutants and the WT, indicating that the MSD mutant Envs have the same stability as the WT after reaching the plasma membrane.
In contrast, the amino acid composition of the MSD core clearly influences the fusogenicity of Env glycoprotein. Mutant L12 Env, in which all 12 amino acid residues of the MSD core were leucine, was defective in mediating virus infectivity and cell-cell fusion. The inability of mutant viruses to infect target cells was correlated with defective fusion between the viral envelope and the cellular membrane, which could be rescued at least in part when conserved amino acid motifs (GGXXG, F683, and V687) were reintroduced into the MSD core. These results argue that in addition to its membrane-anchoring function, the HIV-1 MSD also plays an active role in gp41-mediated fusion during viral entry. Although its structural role is still unclear, the GGXXG motif is critical to the membrane fusion mediated by the HIV-1 Env, since single reintroduction of this motif into the leucine core of the L12 mutant significantly increased the efficiency of cell-cell fusion and virus-cell fusion. Further improvement of Env fusogenicity was achieved by reintroducing additional F683 and V687, indicating that these two amino acid residues also contribute to membrane fusion. It is worth mentioning that the fusogenicity and infectivity of mutant L12G2 are still less than those of the WT, so the remaining mutated amino acid residues (I684, M685, I686, and V692) appear to also facilitate the fusion process. These results argue that all of the 12 amino acid residues in the HIV-1 MSD core region are involved in the efficiency of Env-mediated membrane fusion, which again is consistent with the high conservation of this region. In initial mutagenesis studies, single mutations within the GGXXG motif failed to show a significant role for this motif in membrane fusion (19), perhaps because the remaining unmodified residues were able to compensate for the single change. In this respect, the “recovery-of-function” approach described here provides added power to the analysis.
According to the current model of HIV Env-mediated fusion, merging of the two membranes after the formation of the gp41 six-helix bundles consists of two major events, membrane disruption and fusion pore formation/expansion. It has been reported that the expression of peptides corresponding to the HIV-1 MSD plus four adjacent amino acid residues significantly increased membrane permeability in bacteria (22). Moreover, the replacement of the HIV-1 MSD with those of other transmembrane proteins impaired the efficiency of fusion pore formation/expansion (19). Therefore, the highly conserved MSD core region may contribute to both processes in membrane fusion, particularly when one considers the fact that the specific amino acid residues in the rest of the HIV-1 MSD (from I694 to N704) are not required to mediate efficient cell-cell fusion (24).
Interestingly, the defect in cell-cell fusion appeared to be modulated by the level of Env expressed on the cell surface. Although each of the mutant Envs (L12, L12G, and L12G2) exhibited reduced levels of fusion 6 h after coculture when expressed from the SV40-based or CMV-based vectors, greater differences were observed when Env was expressed in the context of the proviral genome. While we cannot rule out a modifying effect of Env-Gag interactions (25), this difference most likely reflects the higher surface expression levels of the Env glycoprotein by the SV40 and CMV promoter-based expression vectors. Proviral expression resulted in cell surface levels of Env approximately fourfold lower than that observed with the pSRHS Env expression vector, and analogous reductions in cell fusion were observed with the pCDNA3.1 vector when reduced input DNA resulted in equivalently low surface expression levels. A similar dependency on surface expression levels has been reported for C-terminally truncated Env mutants, where cell-cell fusion ability also increased with Env expression levels (15), and it may reflect the increased chance of fusion when higher numbers of molecules are present at the cell surface.
Because it was possible that modifications of the MSD core sequence could alter its interaction with and mobility in the lipid bilayer, we hypothesized that this might be the basis for decreased fusion. Previous work by Waheed et al. (37) had shown that a defect in virus replication imposed by the cholesterol-binding compound AME could be relieved by truncating the cytoplasmic domains of Env, presumably because this increased its mobility in the membrane. In contrast, we observed that C-terminal truncations were not able to rescue the defects imposed by the loss of conserved residues in the MSD core. C-terminal truncation of gp41 has also been shown to induce conformational changes in the ectodomains of gp120 and gp41 (9, 35, 45), and in the case of the truncated WT Env we did observe an approximately twofold enhancement of infectivity in the pNL4-3 construct. Viruses encoding mutant Envs, on the other hand, for the most part exhibited lower infectivity and lower virus-cell fusion than their full-length counterparts, suggesting that a combination of the two mutations was detrimental to biological activity rather than compensatory. Indeed, such mutants appear to be incorporated less efficiently into virions.
In summary, the experimental approaches described here have demonstrated that despite its high sequence conservation, the native MSD core amino acid sequence of HIV Env is not required for normal biosynthesis and intracellular transport. It does play a critical role in mediating efficient membrane fusion during viral infection, and at this stage the almost invariant GGXXG motif appears to be most important. Since the GXXXG motif has been postulated to provide a potential flat interface through which intramembrane helix-helix associations can occur (32, 33), it is tempting to speculate that it may facilitate MSD-MSD interactions during formation of the higher-order fusion pore rather than function during trimer assembly in the endoplasmic reticulum. Nevertheless, each amino acid residue within this core region appears to contribute to optimal fusogenicity of the HIV-1 Env. Further clarification of the mechanisms by which this region facilitates membrane fusion may provide insights into the intramembrane topology of the HIV-1 MSD and protein-lipid interactions of the region during the process of membrane fusion.
Acknowledgments
We thank Cynthia Derdeyn for critical reading of the manuscript and Richard Haaland for technical assistance. We are grateful to Jason Hammonds for assistance in establishing the p24 and gp120 ELISAs. We give our sincerest thanks to Warner Greene for providing the pCMV-BlaM-Vpr construct. We also thank Jin Song for preparing the anti-p24 MAb. The pooled HIV-1 patient sera were kindly provided by Jeffery Lennox through the Clinical Core.
Flow cytometry was performed in the Immunology Core of the Emory Center for AIDS Research (P30 AI050409). This work was supported by grant R01 AI33319 (to E.H.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Byland, R., P. J. Vance, J. A. Hoxie, and M. Marsh. 2007. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol. Biol. Cell 18414-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 201151-1154. [DOI] [PubMed] [Google Scholar]
- 3.Cavrois, M., J. Neidleman, M. Bigos, and W. C. Greene. 2004. Fluorescence resonance energy transfer-based HIV-1 virion fusion assay. Methods Mol. Biol. 263333-344. [DOI] [PubMed] [Google Scholar]
- 4.Cavrois, M., J. Neidleman, W. Yonemoto, D. Fenard, and W. C. Greene. 2004. HIV-1 virion fusion assay: uncoating not required and no effect of Nef on fusion. Virology 32836-44. [DOI] [PubMed] [Google Scholar]
- 5.Ciczora, Y., N. Callens, F. Penin, E. I. Pecheur, and J. Dubuisson. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 812372-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 953425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 666616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70777-810. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 762683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabuzda, D., U. Olshevsky, P. Bertani, W. A. Haseltine, and J. Sodroski. 1991. Identification of membrane anchorage domains of the HIV-1 gp160 envelope glycoprotein precursor. J. Acquir. Immune Defic. Syndr. 434-40. [PubMed] [Google Scholar]
- 11.Hammonds, J., X. Chen, L. Ding, T. Fouts, A. De Vico, J. zur Megede, S. Barnett, and P. Spearman. 2003. Gp120 stability on HIV-1 virions and Gag-Env pseudovirions is enhanced by an uncleaved Gag core. Virology 314636-649. [DOI] [PubMed] [Google Scholar]
- 12.Helseth, E., U. Olshevsky, D. Gabuzda, B. Ardman, W. Haseltine, and J. Sodroski. 1990. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J. Virol. 646314-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 673077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 2371351-1355. [DOI] [PubMed] [Google Scholar]
- 15.Lin, X., C. A. Derdeyn, R. Blumenthal, J. West, and E. Hunter. 2003. Progressive truncations C terminal to the membrane-spanning domain of simian immunodeficiency virus Env reduce fusogenicity and increase concentration dependence of Env for fusion. J. Virol. 777067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Verges, S., G. Camus, G. Blot, R. Beauvoir, R. Benarous, and C. Berlioz-Torrent. 2006. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc. Natl. Acad. Sci. USA 10314947-14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markovic, I., and K. A. Clouse. 2004. Recent advances in understanding the molecular mechanisms of HIV-1 entry and fusion: revisiting current targets and considering new options for therapeutic intervention. Curr. HIV Res. 2223-234. [DOI] [PubMed] [Google Scholar]
- 18.Miyauchi, K., R. Curran, E. Matthews, J. Komano, T. Hoshino, D. M. Engelman, and Z. Matsuda. 2006. Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions. Jpn. J. Infect. Dis. 5977-84. [PubMed] [Google Scholar]
- 19.Miyauchi, K., J. Komano, Y. Yokomaku, W. Sugiura, N. Yamamoto, and Z. Matsuda. 2005. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J. Virol. 794720-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modrow, S., B. H. Hahn, G. M. Shaw, R. C. Gallo, F. Wong-Staal, and H. Wolf. 1987. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J. Virol. 61570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 2501139-1142. [DOI] [PubMed] [Google Scholar]
- 22.Moreno, M. R., M. Giudici, and J. Villalain. 2006. The membranotropic regions of the endo and ecto domains of HIV gp41 envelope glycoprotein. Biochim. Biophys. Acta 1758111-123. [DOI] [PubMed] [Google Scholar]
- 23.Otteken, A., P. L. Earl, and B. Moss. 1996. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J. Virol. 703407-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owens, R. J., C. Burke, and J. K. Rose. 1994. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J. Virol. 68570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens, R. J., J. W. Dubay, E. Hunter, and R. W. Compans. 1991. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc. Natl. Acad. Sci. USA 883987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raja, N. U., M. J. Vincent, and M. Abdul Jabbar. 1994. Vpu-mediated proteolysis of gp160/CD4 chimeric envelope glycoproteins in the endoplasmic reticulum: requirement of both the anchor and cytoplasmic domains of CD4. Virology 204357-366. [DOI] [PubMed] [Google Scholar]
- 27.Ray, N., and R. W. Doms. 2006. HIV-1 coreceptors and their inhibitors. Curr. Top. Microbiol. Immunol. 30397-120. [DOI] [PubMed] [Google Scholar]
- 28.Rowell, J. F., P. E. Stanhope, and R. F. Siliciano. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J. Immunol. 155473-488. [PubMed] [Google Scholar]
- 29.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296911-919. [DOI] [PubMed] [Google Scholar]
- 30.Salzwedel, K., P. B. Johnston, S. J. Roberts, J. W. Dubay, and E. Hunter. 1993. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 675279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 732469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senes, A., D. E. Engel, and W. F. DeGrado. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14465-479. [DOI] [PubMed] [Google Scholar]
- 33.Senes, A., M. Gerstein, and D. M. Engelman. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 296921-936. [DOI] [PubMed] [Google Scholar]
- 34.Vincent, N., C. Genin, and E. Malvoisin. 2002. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim. Biophys. Acta 1567157-164. [DOI] [PubMed] [Google Scholar]
- 35.Vzorov, A. N., K. M. Gernert, and R. W. Compans. 2005. Multiple domains of the SIV Env protein determine virus replication efficiency and neutralization sensitivity. Virology 33289-101. [DOI] [PubMed] [Google Scholar]
- 36.Waheed, A. A., S. D. Ablan, M. K. Mankowski, J. E. Cummins, R. G. Ptak, C. P. Schaffner, and E. O. Freed. 2006. Inhibition of HIV-1 replication by amphotericin B methyl ester: selection for resistant variants. J. Biol. Chem. 28128699-28711. [DOI] [PubMed] [Google Scholar]
- 37.Waheed, A. A., S. D. Ablan, J. D. Roser, R. C. Sowder, C. P. Schaffner, E. Chertova, and E. O. Freed. 2007. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc. Natl. Acad. Sci. USA 1048467-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss, C. D. 2003. HIV-1 gp41: mediator of fusion and target for inhibition. AIDS Rev. 5214-221. [PubMed] [Google Scholar]
- 40.Welman, M., G. Lemay, and E. A. Cohen. 2007. Role of envelope processing and gp41 membrane spanning domain in the formation of human immunodeficiency virus type 1 (HIV-1) fusion-competent envelope glycoprotein complex. Virus Res. 124103-112. [DOI] [PubMed] [Google Scholar]
- 41.West, J. T., P. B. Johnston, S. R. Dubay, and E. Hunter. 2001. Mutations within the putative membrane-spanning domain of the simian immunodeficiency virus transmembrane glycoprotein define the minimal requirements for fusion, incorporation, and infectivity. J. Virol. 759601-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilk, T., T. Pfeiffer, A. Bukovsky, G. Moldenhauer, and V. Bosch. 1996. Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology 218269-274. [DOI] [PubMed] [Google Scholar]
- 43.Willey, R. L., and M. A. Martin. 1993. Association of human immunodeficiency virus type 1 envelope glycoprotein with particles depends on interactions between the third variable and conserved regions of gp120. J. Virol. 673639-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 2801884-1888. [DOI] [PubMed] [Google Scholar]
- 45.Wyss, S., A. S. Dimitrov, F. Baribaud, T. G. Edwards, R. Blumenthal, and J. A. Hoxie. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 7912231-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]