Abstract
Following interruption of antiretroviral therapy among individuals with acquired drug resistance, preexisting drug-sensitive virus emerges relatively rapidly. In contrast, wild-type virus is not archived in individuals infected with drug-resistant human immunodeficiency virus (HIV) and thus cannot emerge rapidly in the absence of selective drug pressure. Fourteen recently HIV-infected patients with transmitted drug-resistant virus were followed for a median of 2.1 years after the estimated date of infection (EDI) without receiving antiretroviral therapy. HIV drug resistance and pol replication capacity (RC) in longitudinal plasma samples were assayed. Resistance mutations were characterized as pure populations or mixtures. The mean time to first detection of a mixture of wild-type and drug-resistant viruses was 96 weeks (1.8 years) (95% confidence interval, 48 to 192 weeks) after the EDI. The median time to loss of detectable drug resistance using population-based assays ranged from 4.1 years (conservative estimate) to longer than the lifetime of the individual (less conservative estimate). The transmission of drug-resistant virus was not associated with virus with reduced RC. Sexual transmission of HIV selects for highly fit drug-resistant variants that persist for years. The prolonged persistence of transmitted drug resistance strongly supports the routine use of HIV resistance genotyping for all newly diagnosed individuals.
Primary human immunodeficiency virus (HIV) infection is typically initiated with a monoclonal or oligoclonal viral population (48) from which viral dissemination occurs with the establishment of a latent reservoir (42). Latent virus persists within resting memory CD4+ T cells, precluding eradication with available antiretroviral therapy (36). Drug-resistant (DR) virus is transmitted by sexual exposure (6, 15), parenteral exposure (44), and perinatal exposure (4). Initial antiretroviral treatment options may be limited and the virologic response compromised in those with transmitted drug resistance (22). Routine drug resistance testing has recently been recommended for all recently diagnosed HIV-infected individuals, with the assumption that transmitted DR virus will persist for an unknown period of time after infection (9, 14).
DR virus is transmitted only about 20% to 35% as readily as drug-sensitive virus, perhaps because selective pressures during transmission may restrict transmission to only the more “fit” of the viral strains within the complex mixture of genetic variants replicating in the source subject (21, 49). Since transmission represents a monoclonal or oligoclonal infection, transmitted drug resistance differs fundamentally from drug resistance acquired during treatment of chronic infection. This acquired DR virus exists as a complex mixture of genetic variants that includes the original wild-type (WT) virus. The interruption of failing antiretroviral therapy in patients with acquired drug resistance results in the relatively rapid reemergence, over 12 to 16 weeks, of more “fit” WT variants with the loss of detectable drug resistance as measured with standard tests (7). In contrast, transmitted DR variants have been shown to persist for up to 3 years in antiretroviral-naive subjects (28). The characterization of transmitted drug resistance has previously been limited to 26 subjects followed for 1 to 3 years following HIV infection without antiretroviral therapy (1, 2, 8, 11, 28). We characterized the natural history of transmitted drug resistance among 14 individuals with primary HIV infection who elected to defer antiretroviral therapy for as long as 4.1 years.
MATERIALS AND METHODS
Study subjects.
Subjects with an acute retroviral syndrome or recent HIV infection presenting to the UCSD Antiviral Research Center in San Diego, CA, between June 1996 and June 2003 were evaluated for study entry. Acute HIV type 1 infection was defined by detectable HIV RNA (>5,000 copies/ml) with a negative HIV enzyme immunoassay (EIA), followed by HIV seroconversion. Early HIV infection was defined with a positive HIV EIA, together with a less sensitive “detuned” HIV EIA standardized optical density of <1.0 by Vironostika (31) or a standardized optical density of <1.5 by Abbott (16). Subjects who elected to defer antiretroviral therapy were eligible for study participation. Patients were categorized as followed: A1 (acute), infection date defined as the date 21 days prior to the first positive plasma HIV RNA while all other HIV test results were negative; A2 (acute), infection date defined as the date 28 days prior to the first positive plasma HIV RNA with a simultaneous indeterminate Western blot; E1 (early), infection date defined as the date 85 days prior to the date of the first positive HIV EIA with a simultaneous negative less sensitive (detuned) EIA; and E2 (early), infection date defined as the midpoint date between the last negative EIA and the first positive EIA. All subjects were followed as part of a larger observational study (AIEDRP CORE01-ACTG 5228) and were provided access to their drug resistance test results. These studies were conducted with appropriate written subject consent and were approved by the Human Research Protections Program at the University of California, San Diego.
Sequence analysis.
Population-based nucleotide sequence analysis of the whole protease sequence (codons 1 to 99) and codons 1 to 305 of the reverse transcriptase gene was performed locally (Viroseq v.2.0; Celera Diagnostics, Alameda, CA) or centrally (GeneSeq HIV; Monogram Biosciences, Inc., South San Francisco, CA) for each of the 14 baseline and a total of 2 to 19 sequential plasma samples. Minority species can be detected at a threshold of approximately 30% (Viroseq) (13) or 10 to 20% (GeneSeq), depending on the mutation. Drug resistance mutations were defined according to the International AIDS Society-USA 2005 Drug Resistance Group guidelines (18). Specifically, we considered the following mutations (using the reference virus NL4-3 as the WT): for a protease inhibitor (PI), D30N, L33F/I, M46I/L, G48V, I50L/V, V82A/F/L/S/T, I84A/C/V, and L90M; for a nucleoside reverse transcriptase inhibitor (NRTI), M41L, A62V, K65R, D67N, K70R, L74V, V75I, F77L, Y115F, F116Y, Q151M, M184V, L210W, T215Y/F/C/D/E/S/I/V, and 215A/G/H/L/N (41) and K219E/Q/R*; and for a nonnucleoside reverse transcriptase inhibitor (NNRTI), L100I, K103N, V106A/M, V108I, Y181C/I, Y188C/H/L, G190A/S, P225H, M230L, and P236L (219R was not considered as a transmitted resistance mutation but rather as a mutation that emerged during follow-up). Mixtures of WT and mutant sequences were considered DR. To determine whether mutations occurred at cytotoxic T lymphocyte (CTL) epitopes, we downloaded all known CTL epitopes from the Los Alamos HIV immunology database and determined whether each epitope was present or absent in each sequence. Evolution within CTL epitopes was measured in terms of numbers of mutations that resulted in the gain or loss of these epitopes, compared to the baseline sequence for each individual. In addition, clonal sequences spanning codons 54 to 231 (HXB2 numbering) of reverse transcriptase were isolated from patient 01-0180 at three time points (n = 21, 14, and 13 clones, corresponding to 134, 148, and 180 weeks after the estimated date of infection [EDI]). Briefly, HIV RNA was isolated from blood plasma using the QIAamp viral RNA extraction kit (Qiagen, Chatsworth, CA). Reverse transcriptase PCR performed using the Finnzyme Robust reverse transcriptase PCR kit (MJ Research, Waltham, MA), per previously published protocols (19). PCR products were cloned into TOPO cloning vectors according to the manufacturer's protocol, and 13 to 20 clones were analyzed using dye terminator sequencing with the ABI Prism 3100 genetic analyzer.
Phenotypic analysis.
Drug susceptibility testing was performed at baseline and during follow-up for each of the 14 patients using a recombinant virus assay (PhenoSense HIV; Monogram Biosciences, Inc., South San Francisco, CA) (30). Reduced drug susceptibility was defined as a fold change (FC) in the 50% inhibitory concentration over the biological cutoff (99th percentile of the FC distributions among WT viruses) (29). HIV pol replication capacity (RC) was assessed using a modification of the phenotypic susceptibility assay (30). Determinations of phenotypic susceptibility and pol RC were performed at the same frequency (see Table 2; resistance testing was performed at the designated weeks after EDI), though pol RC results were recovered slightly less frequently (a mean of 7 measures/patient; range, 2 to 18) during follow-up due to assay failure. pol RC was determined as the ability of the patient-derived virus to replicate in the absence of drug, relative to a WT strain, by measuring luciferase activity in infected cells after a single round of replication. The median RC of WT viruses was set at 100% based upon measurement of a large number of WT viruses (3). Based on assay reproducibility experiments, RC measurements are accurate within ±0.25 log10 95% of the time. RC results were compared to those from the entire San Diego primary infection cohort infected with WT virus (n = 140) or with transmitted DR virus (n = 33) but not included in the current analysis because of their subsequent initiation of antiretroviral treatment. In addition, RC data from samples submitted for routine drug resistance testing in the Monogram Biosciences database were analyzed using the following group definitions: (i) samples with no recognized resistance-associated mutations in PR or RT (n = 962), (ii) samples with one or two major NNRTI mutations (n = 1,522 and 236, respectively), (iii) samples with PI resistance mutations only (n = 459), and (iv) samples with NRTI and PI but no NNRTI resistance mutations (n = 437).
TABLE 2.
Persistence of drug resistance mutations over time in study subjects
| Patient | Wk after EDI | Mutations associated with resistance to:
|
||
|---|---|---|---|---|
| NRTI | NNRTI | PI | ||
| 01-0125 | 12, 15 | None | K103N | None |
| 01-0143 | 6, 10, 18, 22, 26, 28 | None | K103N | None |
| 01-0180 | 12, 18, 22, 25 | None | K103N | None |
| 29, 33, 38, 42, 46, 59, 74, 85, 102, 119, 134 | None | K103K/N | None | |
| 148, 160, 172, 185 | None | None | None | |
| 01-0182 | 12, 19, 21, 33 | None | K103N, Y181C | None |
| 38, 46 | None | K103K/N, Y181Y/C, G190G/A | None | |
| 01-0183 | 12, 18, 22 | None | K103N | None |
| 01-0449 | 12, 18, 26, 35, 44, 52, 68, 101 | M41L, D67N, K70R, T215Y, K219Q | None | D30N, L33I, M46I, I84V, L90M |
| 60, 86, 135, 189 | M41L, D67N, K70K/R, T215Y, K219Q | None | D30N, L33I, M46I, I84V, L90M | |
| 118, 152, 162, 201 | M41L, D67N, T215Y, K219Q | None | D30N, L33I, M46I, I84V, L90M | |
| 213, 226 | M41L, D67N, T215C/Y, K219Q | None | D30N, L33I, M46I, I84V, L90M | |
| 01-0483 | 12, 15 | M41L, V75I, T215L, K219E | Y188L | D30N |
| 01-0503 | 4, 7, 15, 24, 38, 46, 59 | None | K103N | None |
| 01-0507 | 6, 10, 16, 24 | M41L, D67N, T215Y | K103N | I84V, L90M |
| 30, 34 | M41ML, D67N, T215C/Y | K103N | I84V, L90M | |
| 42 | M41L, D67N, T215C/Y | K103N | I84V, L90M | |
| 61 | M41ML, D67D/N, T215C/D/G/Y | K103N | I84V, L90M | |
| 69 | M41M/L, D67D/N, T215C/D/G/Y | K103N | I84V, L90M | |
| 01-0512 | 8, 11, 21, 29, 37, 48, 65, 79, 91, 111, 123, 157 | None | K103N | None |
| 136, 147, 167, 181 | None | K103N, P225P/Ha | None | |
| 212 | None | K103K/N | None | |
| 01-0559 | 4, 15, 27, 44, 50, 60, 78, 97, 111, 151 | None | K103N, P225H | None |
| 132 | K219K/Ra | K103N, P225H | None | |
| 171 | K103N, P225P/H | None | ||
| 01-0566 | 12, 26, 40, 56, 71, 95, 111, 118, 128, 147 | None | K103N | None |
| 01-0575 | 12, 26, 38 | M41L, L74V, M184V, L210W, T215Y | None | M46I, V82A |
| 50 | M41L, L74V, M184MV, L210W, T215Y | None | L33L/Ia, M46I, V82A | |
| 66, 80 | M41L, L74V, L210W, T215Y, K219K/Ra | None | M46I, V82A | |
| 98 | M41L, L74V, L210W, T215C/S | None | M46I, V82A | |
| 111, 122 | M41L, L74V, L210W, T215C/S | None | M46I, V82V/A | |
| 133 | M41L, L74V, L210W, T215C/S | Y181Y/Ca | L33L/I, M46I, V82A | |
| 143, 153 | M41L, L74V, L210W, T215C/S | Y181Y/Ca | L33L/I, M46I, V82V/A | |
| 170 | M41L, L74V, L210W, T215S | Y181Y/Ca | L33L/I, M46I, V82V/A | |
| 01-0629 | 6 | D67N, M184M/V, T215S, K219E | K103K/N | None |
| 21, 33, 42, 56, 71, 91, 114, 126, 137, 161, 173 | D67N, T215S, K219E | None | None | |
Emergence of a mutation not detected in earlier samples.
Statistical analysis.
The mean time to the first mixture of WT and resistant virus or to complete replacement was estimated using a parametric survival model, based on an exponential distribution of replacement (DR-to-WT substitution) times. As the precise time at which the mixture became detectable was not observed, the model used interval-censored times based upon the times at which the sequences were sampled. We estimated the median time to the first mixture of WT and mutant virus or to complete replacement using the Kaplan-Meier method. This method does not accept interval-censored data as input. We imputed times as the midpoint between the sampling times before and after the appearance of a WT variant at the codon in question, either as a pure population or as a mixture. In order to be as conservative as possible, we used a modified method to calculate lower confidence intervals (CIs), in which the CI becomes wider at each censored observation, in order to obtain the most accurate estimates of CIs given the heavy censoring of the data. The relationship between the duration over which a mixture persisted and the fitness advantage of the WT virus over the DR virus was modeled using a simple population genetics model, which assumes that the frequency of the WT virus, p, changes according to the ordinary differential equation dp/dt = (s/g)p(1 − p), where s is the selective coefficient and g is the generation time of productively infected cells, which is assumed to be 1 day. From this, we can calculate the selective advantage of WT virus from the duration of time in which there is a mixture of WT and DR virus. Assuming that there are only two major variants (DR and WT) and assuming that the outgrowth of WT virus is deterministic, the length of time, t, taken for the WT to increase from a frequency p1 (assumed to be 25%) to a frequency p2 (assumed to be 75%) is related to the selective coefficient, s, and the generation time, g, by s = (g/t){log[p2/(1 − p2)] − log[p1/(1 − p1)]}. This model can also be used to estimate the selective advantage of WT virus from phenotypic data. Assuming that the log-transformed FC of the WT virus is f1 [which is close to log(1) but may not be exactly log(1) due to differences between the primary virus and the reference strain] and that of the resistant virus is f2, the dynamics of the log-transformed FC, r, is given by the equation dr/dt = (s/g)(r − f2)(f1 − r)/(f1 − f2). We fitted this model to phenotype data taken over time from patient 01-0180 using nonlinear least squares, fixing f1 and f2 to the first and last measurements for reasons of numerical stability. To estimate 95% CIs of the frequency of K103N in samples of clonal sequences, we performed exact binomial tests. The number of substitutions at CTL epitopes at each time point compared to the baseline sequence was fitted using a generalized estimating equation approach, to correct for the repeated sampling of individuals, using a Poisson family and a log link. To estimate variation in RC over time, we fitted a general linear model, with individual-level intercepts and slopes, with variation between individuals in the slope of RC tested using analysis of variance. RCs of different groups of viruses were compared using t tests and the Bonferroni posttest correction for multiple comparisons (Prism 4.0; GraphPad Inc.).
Phylogenetic analysis.
The clustering of pol sequences by patient was confirmed by phylogenetic analysis. A neighbor-joining tree was reconstructed using a distance matrix based upon synonymous changes per synonymous site, estimated using the Nei-Gojobori (26) distance, to avoid potentially erroneous clusters due to convergent DR mutations. One thousand nonparametric bootstrap samples were generated to assess the statistical significance of the clustering by patient. Phylogenetic analyses were performed using HyPhy v. 0.99 beta (20).
Nucleotide sequence accession numbers.
The sequences analyzed in this study have been deposited in GenBank under accession numbers EU636241 to EU636382.
RESULTS
Fourteen men with acute or recent HIV infection were identified between August 1999 and May 2003. All participants remained treatment naive for the duration of the study. All subjects reported sex with men as their risk for HIV infection. Subjects were predominantly white (11 of 14) and presented a mean of 66 days (range, 27 to 85 days) after their EDI with a median baseline CD4 count of 537 cells/mm3 and 5.15 log10 RNA copies/ml (Table 1). The mean baseline RC was 87% of WT. Study participants were selected from a cohort of 210 recently HIV-infected subjects who had baseline (study entry) and follow-up pol sequence data available for analysis, had transmitted DR virus, and chose to defer antiretroviral therapy for at least 2 weeks after study entry (mean, 20 months).
TABLE 1.
Baseline characteristics
| Subject | Age (yr) | Days since EDI | CD4 cells/mm3 | CD4 (%) | Log10 RNA | RC (%)a |
|---|---|---|---|---|---|---|
| 01-0125 | 45 | 85 | 476 | 10 | 6.22 | 35 |
| 01-0143 | 33 | 45 | 383 | 22 | 4.88 | 60 |
| 01-0180 | 22 | 85 | 547 | 22 | 5.26 | 91 |
| 01-0182 | 36 | 85 | 573 | 20 | 6.05 | 208 |
| 01-0183 | 46 | 85 | 398 | 27 | 5.88 | 21 |
| 01-0449 | 41 | 85 | 480 | 41 | 3.88 | 72 |
| 01-0483 | 59 | 85 | 472 | 25 | 4.57 | 85 |
| 01-0503 | 33 | 30 | 263 | 24 | 7.33 | 172 |
| 01-0507 | 36 | 45 | 527 | 31 | 4.22 | 110 |
| 01-0512 | 34 | 55 | 431 | 25 | 7.76 | 58 |
| 01-0559 | 47 | 27 | 930 | 26 | 7.19 | 48 |
| 01-0566 | 30 | 85 | 819 | 31 | 4.59 | 162 |
| 01-0575 | 24 | 85 | 813 | 33 | 3.86 | 16 |
| 01-0629 | 22 | 45 | 959 | 41 | 5.04 | 71 |
One hundred percent equals the median RC of wild-type viruses.
Genotypic analysis.
Drug resistance mutations were identified at baseline for each of the 14 subjects (Table 2). Virus from 5 subjects contained NRTI mutations, that from 12 contained NNRTI mutations, and that from 4 contained major PI mutations. Three subjects (01-0449, 01-0575, and 01-0629) were infected with two-class multi-DR virus, and two subjects (01-0483 and 01-0507) were infected with three-class multi-DR HIV. Longitudinal plasma samples were collected for a median of 108 weeks (range, 15 to 226 weeks) after infection and analyzed for the persistence of transmitted DR variants.
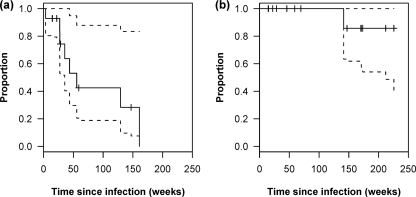
For each individual, at each time point, and at each codon, we determined whether a resistance mutation was present and, if so, whether it was present as a mixture or as a pure population. Resistance mutations that persisted without replacement were censored by the last day of follow-up. To estimate the rate of replacement of resistance mutations under conservative assumptions, we used the time at which the first mixture appeared (although other mutations often persisted as pure populations in the same patient). Mixtures or complete replacements of DR by WT sequences were detected at resistance-associated positions, which were previously unmixed mutant, in virus from 6 of 14 individuals (01-0180, 01-0182, 01-0449, 01-0507, 01-0575, and 01-0629) during the follow-up period. Of these, two individuals (01-0180 and 01-0629) exhibited replacement of at least a single mutation to WT, as assessed by bulk sequencing (Table 2). Using a parametric survival model, the mean time to first detection of a WT/DR mixture based on all 14 patients, considering any resistance mutation, was 96 weeks (1.8 years) (95% CI, 48 to 192 weeks) after the EDI. The median time to the first detection of a mixture was lower, at 56 weeks (95% CI, 36 weeks to infinity) (Fig. 1a).
FIG. 1.
(a) Kaplan-Meier plot of the time to first detection of the WT virus as a mixture (WT/DR) in the population. (b) Kaplan-Meier plot of the time to complete replacement (i.e., last detection of any DR virus). Dashed lines indicate upper and lower 95% CIs.
Over a median follow-up period of 108 weeks (2.1 years), one or more resistance mutations, either as pure populations or as mixtures, persisted for the entire period of observation in 13 of the 14 patients (only patient 01-0180 demonstrated complete replacement to WT by population sequencing during follow-up). It would require an unknown period of longer follow-up to observe complete replacement of transmitted drug resistance (as detected by bulk sequencing) in all patients, especially as resistance is comprised of multiple codons in many patients (>1 codon involved in 7 of 14 patients in this study). Based upon these data, a conservative estimate of 212 weeks (4.1 years) can be made for the median time to complete replacement of DR by WT variants by bulk sequencing, based on the lower 95% CI of the Kaplan-Meier analysis (Fig. 1b). The lower CI of the mean time to complete replacement as calculated using an exponential survival model was similar (210 weeks).
Susceptibility analysis.
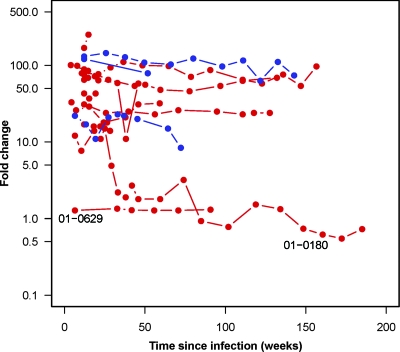
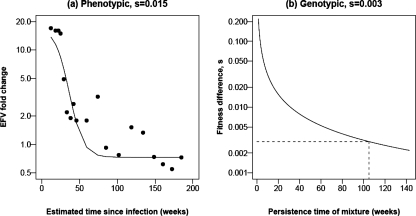
All subjects had reduced drug susceptibility consistent with the drug resistance mutations identified at baseline except when mutations were present as mixtures (i.e., subject 01-0629). Longitudinal susceptibility results for a representative drug for each subject (Fig. 2) demonstrate that the virus from all but subject 01-0629 had 10- to >100-fold-reduced susceptibility to either efavirenz (EFV) or lopinavir at baseline. Among the nine subjects with over 50-fold-reduced NNRTI susceptibility at baseline, only one (01-0180) showed waning of the resistant phenotype. The relative proportion of resistant virus at each time point for subject 01-0180 was derived using drug susceptibility as a marker of the proportion of DR virus. In vitro mixing of 103N or 181C site-directed mutants with WT strain NL4-3 demonstrate that as the proportion of resistant virus is increased, there is a log-linear increase in FC for each NNRTI (C. Petropoulos, personal communication). By measuring the rate at which FC decreases, over the generation time of HIV, we estimate that the WT virus has a 1.5% fitness advantage over the 103N mutant, using a simple population genetic model, given a stable population size and assuming a generation time of 1 day (Fig. 3 and 4). Under this same model, we can also estimate the fitness advantage of the WT virus from the time during which the WT and the 103N mutant persist together as a mixture. Assuming that a mixture is detected at between 25% and 75%, a generation time of 1 day, and a conservative time of 105 weeks during which the K103K/N mixture persisted, we estimated a fitness advantage of 0.3% (Fig. 3). Although it is unclear why this genotypic estimate is lower than the estimate from phenotypic data, it highlights the ability of genotypic testing to detect transmitted resistance for longer periods of time following infection than phenotypic testing. We also obtained clonal sequences of reverse transcriptase from this individual at three time points in order to quantify the proportion of K103N over time. At 134 weeks, the last time point at which a K103K/N mixture was detected, 9/21 clones had the K103N mutation (43%; 95% CI, 22 to 66%). At 148 weeks, the first time point with a pure K103 in the bulk sequence, the frequency of K103N had decreased to 1/14 clones (7%; 95% CI, 0.2 to 34%). By 180 weeks after the EDI, we did not detect any K103N mutants, based on 13 clones (0%; 95% CI, 0 to 25%).
FIG. 2.
Longitudinal FC susceptibility results for a representative drug for each subject. Red circles indicate EFV susceptibility. Blue circles indicate lopinavir susceptibility. Subject 01-0629, infected with a K103K/N mixture at baseline, did not show reduced EFV susceptibility at baseline or during follow-up. Subject 01-0180 showed a waning of reduced EFV susceptibility over time.
FIG. 3.
Estimation of the relative fitness advantage of WT virus over virus harboring K103N in patient 01-0180. (a) FC in susceptibility to EFV over time (points) and the fit of a model that assumes a log-linear relationship between FC and frequency of the WT variant (see Materials and Methods). (b) Relationship between the time during which a mixture persists and the selective advantage of WT.
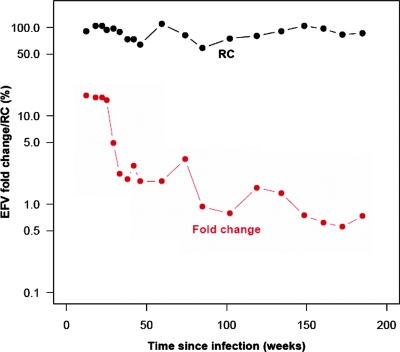
FIG. 4.
Dynamics of RC and FC in susceptibility to EFV in patient 01-0180. Despite a dramatic drop in FC, the RC remains relatively stable over the period of observation.
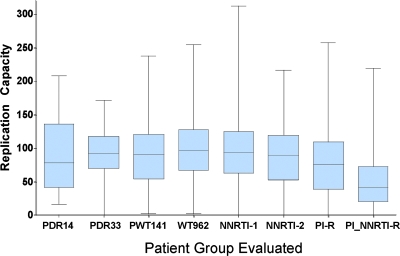
The transmission of DR virus was not associated with virus of reduced RC (Fig. 5). The mean baseline RC of the 14 transmitted viruses (RC = 87%) was not significantly different from that observed in either newly infected patients with transmitted WT virus (mean RC = 89%; P > 0.05) or a larger group of newly infected patients with DR virus (mean RC = 92%; P > 0.05). The mean baseline RC value for virus from the 14 study subjects was also not significantly different from that observed in individuals infected for unknown lengths of time with virus lacking resistance mutations (WT) (RC = 100%; P > 0.05), with isolated NNRTI resistance (RC = 96% and 88% for 1 and 2 mutations, respectively; P > 0.05), with PI resistance (RC = 78%; P > 0.05), or with a combination of NRTI and PI resistance (mean RC = 50%; P > 0.05). Virus from the newly infected patients with transmitted drug resistance (n = 33) had significantly higher RC values than that from subjects with acquired drug resistance with a combination of NRTI and PI mutations (mean RC of 92% versus 50%; P < 0.001). RC remained relatively stable during a mean follow-up of 88 weeks (1.7 years) following EDI in the 14 study participants. Due to the low rate of replacement of resistance mutations by WT sequences, we were unable to correlate within-individual changes in RC with replacement of drug resistance. However, in the single individual who did demonstrate complete replacement of resistance, the RC remained relatively stable despite a decrease of over an order of magnitude in susceptibility to NNRTIs (Fig. 4). Given that the RC of WT viruses is similar to that of NNRTI-resistant viruses, the lack of detectable changes in RC as resistance wanes is not surprising.
FIG. 5.
Box plots showing the median RC among eight different groups, with 25th and 75th percentiles shown by the box and the complete range indicated by whiskers. PDR14, study patients with transmitted DR virus; PDR33, 33 primary infection patients infected with DR virus but not included in present study due to antiretroviral treatment choices after entry; PWT141, 141 primary patients infected with WT virus; WT962, unrelated WT samples with infection of undetermined duration (n = 962); NNRTI-1, samples with an isolated single NNRTI major drug resistance mutation (n = 1,522); NNRTI-2, samples with only two major NNRTI drug resistance mutation (n = 236); PI-R, samples with ≥1 major PI mutation (no NRTI or NNRTI mutations) (n = 459); and PI_NRTI-R, samples with ≥1 major PI mutation and ≥1 major NRTI mutation (without NNRTI mutations).
Analysis of viral evolution.
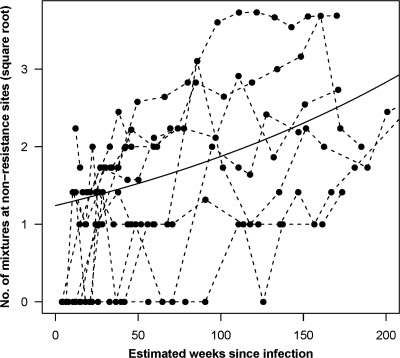
Despite the low rate of replacement of resistance mutations, evidence of continued viral evolution was present. Mixtures involving synonymous substitutions, i.e., those that do not affect the amino acid sequence, emerged in many subjects at resistance-conferring sites, despite the lack of DR-to-WT replacement at the amino acid level. Furthermore, multiple nonsynonymous (i.e., amino acid-changing) substitutions within known CTL epitopes were observed over time (Fig. 6). Although we could not confirm that these changes were actually due to CTL selection pressure, these results are consistent with ongoing genetic evolution and increasing viral diversity over time and in contrast with the apparent low rate of change from resistant variants to drug-sensitive forms. It is possible that compensatory mutations could explain the lower rate of reversion among the patients with transmitted resistance mutations. However, such compensatory mutations would have to act on the transmissibility of the virus in order for it to be possible to observe lower rates of reversion compared to those with acquired resistance. We were unable to detect any sites in pol that discriminated between the transmitted resistant strains and resistant strains downloaded from the Stanford HIV resistance database. This may simply reflect the small number of study participants or the involvement of sites outside the region of protease and reverse transcriptase sequenced. Although none of the 14 patients showed evidence of superinfection using previously published screening methods (39) or received any antiretroviral therapy, novel drug resistance mutations were observed in 6 of 14 subjects during follow-up (Table 2).
FIG. 6.
The total number of nonsynonymous mutations in predicted CTL epitopes between the baseline sequence and subsequent sequences was determined for all study participants over time. Overall, there is a large variation in the number of substitutions in each individual subject, though the mean number (represented by the solid line) shows an increase over time, demonstrating that viral evolution is ongoing despite the persistence of drug resistance mutations.
DISCUSSION
High levels of transmitted drug resistance persisted in 13 of 14 subjects. In only one person did a transmitted DR K103N variant become completely replaced by a WT virus such that it was not detectable by conventional population sequencing nearly 3 years after infection. Among all 14 subjects, the first detection of a mixture of WT and DR virus by population sequencing occurred at a mean of 96 weeks (1.8 years) after the EDI. Given the very low rate of resistance replacement, it was not possible to measure directly the time to complete loss of detectable drug resistance by bulk sequencing; rather, we imputed the time to this event using the lower 95% CIs (Fig. 1b). Despite a conservative estimate of a median time to complete replacement of 4.1 years, the transmitted resistant variant is archived for life, even when below the threshold of detection (34). A less conservative estimate allows the lifelong persistence of detectable transmitted drug resistance in many people. The rate of replacement of the resistant mutant is likely to vary directly with the fitness cost associated with the resistance mutation and perhaps other host selective pressures not yet recognized. Although a significant in vitro fitness impairment has been observed for several of the major PI resistance mutations relative to WT virus (23), we did not observe an increased rate of replacement of PI resistance mutations compared to that observed for NNRTI resistance mutations. Overall, replacement of resistance is gradual and usually incomplete, resulting in the persistence of mixtures of WT and resistant variants in plasma HIV RNA for years following initial infection.
The number of years after initial HIV infection during which detection of transmitted drug resistance is routinely feasible has not yet been defined, but patients identified many years, perhaps more than 10 years, after their EDI may not be properly identified as having been initially infected with DR virus (38). The prevalence of transmitted DR HIV measured within the first year following HIV infection is between 20% and 23% in some North American populations and between 10% and 14% in some European and Canadian populations (12, 22, 25, 37, 47, 49) More recent estimates suggest that the prevalence of transmitted resistance may be declining in both North America and Europe (S. Little, S. Frost, D. Smith, S. May, N. Parkin, and D. Richman, abstr. 60, presented at the 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA, 25 to 28 February 2007). Among 900 recently diagnosed treatment-naive individuals with established infection from 10 U.S. cities, the prevalence of detectable resistance at 7.4% remains lower than even the most recent estimates among patients with primary infection (46). Resistance prevalence estimates among untreated patients with chronic infection from Europe are quite similar, with 8.7% of patients with chronic infection (n = 607) or unknown durations of infection (n = 824) demonstrating resistance between 1996 and 2002 (47). These data suggest that the prevalence of resistance declines with time after infection, resulting from the gradual replacement of DR virus by drug-sensitive virus until ultimately resistance testing fails to detect the resistant variant. Patients with chronic infection and older historical infection dates may predate the transmission of significant drug resistance or, if infected more recently (i.e., <10 years), may maintain some level of detectable drug resistance following transmission.
Transmitted DR variants were not associated with low RC values (mean, 87%). The mean RC was 89% for recently infected participants (PWT141) lacking any well-characterized drug resistance mutations (Fig. 5). There was no difference in the mean RCs of the patients with transmitted DR virus who remained treatment naive, those who received antiretroviral treatment, and those who were infected with WT virus. Previous studies have demonstrated that DR virus is transmitted only about 20% as readily as drug-sensitive virus (21). Among the subset of DR viruses that are transmitted, however, RC values are equivalent to those measured among newly infected individuals infected with WT virus, suggesting that the more fit DR viruses are more likely to be transmitted. This is consistent with the observation that the initial levels of viremia are comparable following acute infection with either DR or WT virus (12). The fitness cost of individual RT resistance mutations is well established, particularly for 215Y, 184V, and 65R (5). The transmission of any of the resistant variants more frequently associated with a fitness impairment might be associated with a more rapid reversion to a more “fit,” WT genotype that might fail to be detected by conventional sequencing (17). Further studies are needed to determine whether these traditionally less fit variants are uniformly replaced at a significantly higher rate than other variants associated with higher RC values. Following transmission, the relatively high RC values associated with the majority of the transmitted resistant variants favor their persistence in the context of viral fitness conferred by the complete genome. This is in contrast to the observation in chronic infection where antiretroviral selective pressure selects for drug resistance mutations, which often reduce viral RC (23, 24).
Chronic HIV infection is characterized by a complex mixture of genetic variants, with approximately 75% of chronically infected individuals harboring DR forms (32, 33, 49). The interruption of failing treatment has been associated with rapid emergence of more “fit” WT variants, typically over 12 to 16 weeks (7). Since subjects infected with DR virus do not harbor WT variants at the time of infection, loss of detectable resistance requires mutational replacement of the DR virus by the WT virus rather than reemergence of a preexisting WT virus. The detection of complete shift from DR virus to drug-sensitive virus by bulk sequencing was demonstrated in only 1 of 14 patients, who were followed for a median of 2.1 years after the EDI. Using drug susceptibility as a marker of the proportion of resistant virus within the viral population in this patient, and based on the persistence of the K103K/N mixture, we estimated that the WT virus had less than a 1.5% fitness advantage compared to the K103N NNRTI-resistant variant. Given the relatively trivial fitness advantage of the transmitted WT virus compared to the transmitted DR virus, it is expected that most DR variants will persist, though the rate of replacement of mutations associated with larger changes in viral fitness (i.e., M184V) may be higher (43). The persistence of 103K/N mixtures for over 2.6 years despite the short mutational distance between these mutants suggests that the mutation confers a small fitness cost consistent with high RC values. Given the low rate of replacement, it is impossible to estimate to what extent replacement times may vary between individuals due to factors such as host genetic background; however, if the few individuals in whom replacement of resistance occurred have higher rates of replacement, then the persistence of drug resistance may be even longer.
The persistence of drug resistance, however, does not imply the absence of ongoing genetic evolution. Substitutions occurred at sites not involved in drug resistance, presumably because of HIV-specific immune responses (10, 27). The rate of evolution at the amino acid level appeared to be independent of steady-state viremia (data not shown). The spontaneous appearance of drug resistance mutations in 3 of 14 patients (01-0512, 01-0550, and 01-0575) in the absence of selective drug pressure was most likely related to the presence of these particular resistant variants as relatively minor populations at the time of transmission, below the threshold of assay detection. More sensitive real-time PCR methods for detecting low-frequency minor variants among treatment-naive individuals have shown that resistant variants identified by real-time PCR and missed by conventional sequencing may represent 0.7% to 11% of the population by clonal sequencing (17). Potential selective advantages may have resulted in the emergence of these resistant isolates over time to detectable levels despite the absence of selective drug pressure.
The persistence of DR variants provides a prolonged “window of opportunity” for secondary transmission of DR variants at a time when plasma viral load measures are routinely very high. Two subjects (study subject 01-0559 and a nonstudy subject) both acquired 103N as a secondary transmission from an untreated source subject (01-0512) 4 to 5 months after his EDI at a time when his viral load ranged from 5.5 to 6.3 log10 copies/ml. The same source subject, 3.2 years after his EDI and with a plasma viral load that remains stable at 5.0 log10 copies/ml, recently transmitted the same DR variant to yet another index subject. Of additional concern, the lower rates of viral turnover in the male genital tract result in even slower decay of HIV drug resistance in semen than in plasma (40). The relative stability of drug resistance mutations within both source and recipient partners suggests that it is unlikely that we missed many replacement of resistance mutations by WT in the brief time between infection and study entry.
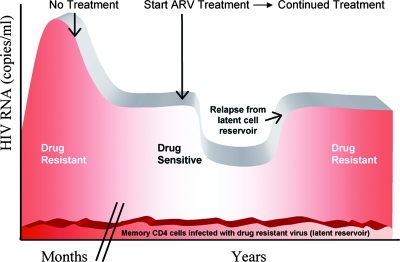
The persistence of transmitted drug resistance for years, prevalence estimates of transmitted drug resistance in newly infected individuals, and estimates of drug resistance among treatment-naive individuals with established infection all support the implementation of routine screening for primary drug resistance in all newly HIV-diagnosed, treatment-naive individuals. Primary resistance testing is estimated to be cost-effective in areas where the prevalence of resistance is greater than 4 to 5% (35, 45). The greater sensitivity of pol genotype testing compared to phenotype testing in detecting drug resistance, coupled with its reduced cost, suggests that sequence-based resistance testing should be the resistance test of choice for newly diagnosed patients (38). The health care implications of missing transmitted drug resistance are potentially significant. There are the costs of early treatment failures, the potential selection of additional DR variants in the setting of suboptimal treatment due to unrecognized resistance, and secondary transmissions that may continue for years. The relatively rapid emergence of drug resistance in the setting of treatment in a previously treatment-naive patient (infected for more than 5 to 7 years) should prompt the question as to whether initial infection with a DR variant may have been missed (Fig. 7). Despite the apparent fitness “cost” of acquired drug resistance mutations in many treated patients, sexual transmission of HIV appears to select for the most fit DR variants when resistance is transmitted such that persistence of a highly replication-competent, DR variant is ensured for years in the new host. If proper records are maintained, a single genotype done for patients initiating care may alter the course of antiretroviral therapy initiated years later.
FIG. 7.
Schematic illustration of the first decade (approximately) of HIV infection following infection with a resistant strain of virus. Patients initially infected with a DR variant will typically demonstrate a transient high-titer viremia, followed by a spontaneous decline to a steady-state or “set point” viremia. During these first 6 months, bulk sequencing of plasma virus will typically detect only resistant virus in patients with transmitted DR virus. Years may pass during which the gradual process of random and potentially selective mutations results in the appearance of a mixture at the site(s) of a previous drug-resistant mutation(s) followed by ultimate “reversion” or, more accurately, replacement by WT virus. The resistant variant, however, will persist for the life of the patient, harbored within the reservoir of long-lived memory CD4 cells. ARV, antiretroviral.
Acknowledgments
This work was supported by National Institutes of Health grants AI27670, AI69432, AI38858, AI68636, AI43638, AI47745, AI57167, AI29164, the UCSD Centers for AIDS Research (AI36214), and the San Diego Veterans Affairs Healthcare System. We received additional institutional support from the California HIV/AIDS Research Program (CHRP [formerly UARP]) (IS02-SD-701).
Douglas D. Richman, Susan Little, and Simon D. W. Frost are consultants for Monogram Biosciences. Christos J. Petropoulos and Neil T. Parkin are employees and stockholders of Monogram Biosciences, which provided some of the diagnostic assay services used to generate data for this paper. The other authors have declared that no competing interests exist.
We thank Laureen Copfer for her excellent administrative assistance with the preparation of the manuscript and Deya Collier, Prentice Higley, Nancy Keating, and Sherry Rostami for their technical assistance. We are grateful to the AIEDRP study participants for their unwavering generosity and to the University of California, San Diego, AVRC staff (Heidi Aiem, Tari L. Gilbert, Jill Kunkel, Paula Potter, and Joanne Santangelo) for their many contributions.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Barbour, J. D., F. M. Hecht, T. Wrin, T. J. Liegler, C. A. Ramstead, M. P. Busch, M. R. Segal, C. J. Petropoulos, and R. M. Grant. 2004. Persistence of primary drug resistance among recently HIV-1 infected adults. AIDS 181683-1689. [DOI] [PubMed] [Google Scholar]
- 2.Brenner, B. G., J. P. Routy, M. Petrella, D. Moisi, M. Oliveira, M. Detorio, B. Spira, V. Essabag, B. Conway, R. Lalonde, R. P. Sekaly, and M. A. Wainberg. 2002. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J. Virol. 761753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, T. B., K. Schneider, T. Wrin, C. J. Petropoulos, and E. Connick. 2003. Relationship between in vitro human immunodeficiency virus type 1 replication rate and virus load in plasma. J. Virol. 7712105-12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colgrove, R. C., J. Pitt, P. H. Chung, S. L. Welles, and A. J. Japour. 1998. Selective vertical transmission of HIV-1 antiretroviral resistance mutations. AIDS 122281-2288. [DOI] [PubMed] [Google Scholar]
- 5.Cong, M. E., W. Heneine, and J. G. Garcia-Lerma. 2007. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J. Virol. 813037-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlon, C. P., P. Klenerman, A. Edwards, B. A. Larder, and R. E. Phillips. 1994. Heterosexual transmission of human immunodeficiency virus type 1 variants associated with zidovudine resistance. J. Infect. Dis. 169411-415. [DOI] [PubMed] [Google Scholar]
- 7.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344472-480. [DOI] [PubMed] [Google Scholar]
- 8.Delaugerre, C., L. Morand-Joubert, M. L. Chaix, O. Picard, A. G. Marcelin, V. Schneider, A. Krivine, A. Compagnucci, C. Katlama, P. M. Girard, and V. Calvez. 2004. Persistence of multidrug-resistant HIV-1 without antiretroviral treatment 2 years after sexual transmission. Antiviral Ther. 9415-421. [PubMed] [Google Scholar]
- 9.DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents. 10 October 2006. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. U.S. Department of Health and Human Services, Washington, DC. www.aidsinfo.nih.gov.
- 10.Frost, S. D., Y. Liu, S. L. Pond, C. Chappey, T. Wrin, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J. Virol. 796523-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi, R. T., A. Wurcel, E. S. Rosenberg, M. N. Johnston, N. Hellmann, M. Bates, M. S. Hirsch, and B. D. Walker. 2003. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin. Infect. Dis. 371693-1698. [DOI] [PubMed] [Google Scholar]
- 12.Grant, R. M., F. M. Hecht, M. Warmerdam, L. Liu, T. Liegler, C. J. Petropoulos, N. S. Hellmann, M. Chesney, M. P. Busch, and J. O. Kahn. 2002. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA 288181-188. [DOI] [PubMed] [Google Scholar]
- 13.Gunthard, H. F., J. K. Wong, C. C. Ignacio, D. V. Havlir, and D. D. Richman. 1998. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res. Hum. Retroviruses 14869-876. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch, M. S., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, R. T. D'Aquila, L. M. Demeter, S. M. Hammer, V. A. Johnson, C. Loveday, J. W. Mellors, D. M. Jacobsen, and D. D. Richman. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 37113-128. [DOI] [PubMed] [Google Scholar]
- 15.Imrie, A., A. Beveridge, W. Genn, J. Vizzard, and D. A. Cooper. 1997. Transmission of human immunodeficiency virus type 1 resistant to nevirapine and zidovudine. J. Infect. Dis. 1751502-1506. [DOI] [PubMed] [Google Scholar]
- 16.Janssen, R., G. Satten, S. Stramer, B. Rawal, T. O'Brien, B. Weiblen, F. Hecht, N. Jack, F. Cleghorn, J. Kahn, M. Chesney, and M. Busch. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 28042-48. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. A., J. F. Li, X. Wei, J. Lipscomb, D. Bennett, A. Brant, M. E. Cong, T. Spira, R. W. Shafer, and W. Heneine. 2007. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS ONE 2e638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, V. A., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, D. Pillay, J. Schapiro, A. Telenti, and D. Richman. 2005. Update of the drug resistance mutations in HIV-1: 2005. Top. HIV Med. 1351-57. [PubMed] [Google Scholar]
- 19.Koelsch, K. K., D. M. Smith, S. J. Little, C. C. Ignacio, T. R. Macaranas, A. J. Brown, C. J. Petropoulos, D. D. Richman, and J. K. Wong. 2003. Clade B HIV-1 superinfection with wild-type virus after primary infection with drug-resistant clade B virus. AIDS 17F11-F16. [DOI] [PubMed] [Google Scholar]
- 20.Kosakovsky Pond, S. L., S. D. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21676-679. [DOI] [PubMed] [Google Scholar]
- 21.Leigh Brown, A. J., S. D. Frost, W. C. Mathews, K. Dawson, N. S. Hellmann, E. S. Daar, D. D. Richman, and S. J. Little. 2003. Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population. J. Infect. Dis. 187683-686. [DOI] [PubMed] [Google Scholar]
- 22.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347385-394. [DOI] [PubMed] [Google Scholar]
- 23.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 748524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maree, A. F., W. Keulen, C. A. Boucher, and R. J. De Boer. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 7411067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masquelier, B., K. Bhaskaran, D. Pillay, R. Gifford, E. Balestre, L. B. Jorgensen, C. Pedersen, L. van der Hoek, M. Prins, C. Balotta, B. Longo, C. Kucherer, G. Poggensee, M. Ortiz, C. de Mendoza, J. Gill, H. Fleury, and K. Porter. 2005. Prevalence of transmitted HIV-1 drug resistance and the role of resistance algorithms: data from seroconverters in the CASCADE collaboration from 1987 to 2003. J. Acquir. Immune Defic. Syndr. 40505-511. [DOI] [PubMed] [Google Scholar]
- 26.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3418-426. [DOI] [PubMed] [Google Scholar]
- 27.Ogg, G. S., S. Kostense, M. R. Klein, S. Jurriaans, D. Hamann, A. J. McMichael, and F. Miedema. 1999. Longitudinal phenotypic analysis of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes: correlation with disease progression. J. Virol. 739153-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pao, D., U. Andrady, J. Clarke, G. Dean, S. Drake, M. Fisher, T. Green, S. Kumar, M. Murphy, A. Tang, S. Taylor, D. White, G. Underhill, D. Pillay, and P. Cane. 2004. Long-term persistence of primary genotypic resistance after HIV-1 seroconversion. J. Acquir. Immune Defic. Syndr. 371570-1573. [DOI] [PubMed] [Google Scholar]
- 29.Parkin, N. T., N. S. Hellmann, J. M. Whitcomb, L. Kiss, C. Chappey, and C. J. Petropoulos. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawal, B. D., A. Degula, L. Lebedeva, R. S. Janssen, F. M. Hecht, H. W. Sheppard, and M. P. Busch. 2003. Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J. Acquir. Immune Defic. Syndr. 33349-355. [DOI] [PubMed] [Google Scholar]
- 32.Recsky, M. A., Z. L. Brumme, K. J. Chan, B. Wynhoven, B. Yip, W. W. Dong, K. V. Heath, J. S. Montaner, A. R. Levy, R. S. Hogg, and P. R. Harrigan. 2004. Antiretroviral resistance among HIV-infected persons who have died in British Columbia, in the era of modern antiretroviral therapy. J. Infect. Dis. 190285-292. [DOI] [PubMed] [Google Scholar]
- 33.Richman, D. D., S. C. Morton, T. Wrin, N. Hellmann, S. Berry, M. F. Shapiro, and S. A. Bozzette. 2004. The prevalence of antiretroviral drug resistance in the United States. AIDS 181393-1401. [DOI] [PubMed] [Google Scholar]
- 34.Ruff, C. T., S. C. Ray, P. Kwon, R. Zinn, A. Pendleton, N. Hutton, R. Ashworth, S. Gange, T. C. Quinn, R. F. Siliciano, and D. Persaud. 2002. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J. Virol. 769481-9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sax, P. E., R. Islam, R. P. Walensky, E. Losina, M. C. Weinstein, S. J. Goldie, S. N. Sadownik, and K. A. Freedberg. 2005. Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin. Infect. Dis. 411316-1323. [DOI] [PubMed] [Google Scholar]
- 36.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9727-728. [DOI] [PubMed] [Google Scholar]
- 37.Simon, V., J. Vanderhoeven, A. Hurley, B. Ramratnam, M. Louie, K. Dawson, N. Parkin, D. Boden, and M. Markowitz. 2002. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. AIDS 161511-1519. [DOI] [PubMed] [Google Scholar]
- 38.Smith, D., N. Moini, R. Pesano, E. Cachay, H. Aiem, Y. Lie, D. Richman, and S. Little. 2007. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin. Infect. Dis. 44456-458. [DOI] [PubMed] [Google Scholar]
- 39.Smith, D. M., J. K. Wong, G. K. Hightower, C. C. Ignacio, K. K. Koelsch, E. S. Daar, D. D. Richman, and S. J. Little. 2004. Incidence of HIV superinfection following primary infection. JAMA 2921177-1178. [DOI] [PubMed] [Google Scholar]
- 40.Smith, D. M., J. K. Wong, H. Shao, G. K. Hightower, S. H. Mai, J. M. Moreno, C. C. Ignacio, S. D. Frost, D. D. Richman, and S. J. Little. 2007. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: implications for secondary transmission. J. Infect. Dis. 196356-360. [DOI] [PubMed] [Google Scholar]
- 41.Stanford HIVDB Team. 2007. HIV drug resistance database. Stanford University, Stanford, CA. hivdb.stanford.edu.
- 42.Strain, M. C., S. J. Little, E. S. Daar, D. V. Havlir, H. F. Gunthard, R. Y. Lam, O. A. Daly, J. Nguyen, C. C. Ignacio, C. A. Spina, D. D. Richman, and J. K. Wong. 2005. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J. Infect. Dis. 1911410-1418. [DOI] [PubMed] [Google Scholar]
- 43.Turner, D., B. Brenner, J. P. Routy, D. Moisi, Z. Rosberger, M. Roger, and M. A. Wainberg. 2004. Diminished representation of HIV-1 variants containing select drug resistance-conferring mutations in primary HIV-1 infection. J. Acquir. Immune Defic. Syndr. 371627-1631. [DOI] [PubMed] [Google Scholar]
- 44.Veenstra, J., R. Schuurman, M. Cornelissen, A. B. van't Wout, C. A. Boucher, H. Schuitemaker, J. Goudsmit, and R. A. Coutinho. 1995. Transmission of zidovudine-resistant human immunodeficiency virus type 1 variants following deliberate injection of blood from a patient with AIDS: characteristics and natural history of the virus. Clin. Infect. Dis. 21556-560. [DOI] [PubMed] [Google Scholar]
- 45.Weinstein, M. C., S. J. Goldie, E. Losina, C. J. Cohen, J. D. Baxter, H. Zhang, A. D. Kimmel, and K. A. Freedberg. 2001. Use of genotypic resistance testing to guide HIV therapy: clinical impact and cost-effectiveness. Ann. Intern. Med. 134440-450. [DOI] [PubMed] [Google Scholar]
- 46.Weinstock, H. S., I. Zaidi, W. Heneine, D. Bennett, J. G. Garcia-Lerma, J. M. Douglas, Jr., M. LaLota, G. Dickinson, S. Schwarcz, L. Torian, D. Wendell, S. Paul, G. A. Goza, J. Ruiz, B. Boyett, and J. E. Kaplan. 2004. The epidemiology of antiretroviral drug resistance among drug-naive HIV-1-infected persons in 10 US cities. J. Infect. Dis. 1892174-2180. [DOI] [PubMed] [Google Scholar]
- 47.Wensing, A. M., D. A. van de Vijver, G. Angarano, B. Asjo, C. Balotta, E. Boeri, R. Camacho, M. L. Chaix, D. Costagliola, A. De Luca, I. Derdelinckx, Z. Grossman, O. Hamouda, A. Hatzakis, R. Hemmer, A. Hoepelman, A. Horban, K. Korn, C. Kucherer, T. Leitner, C. Loveday, E. MacRae, I. Maljkovic, C. de Mendoza, L. Meyer, C. Nielsen, E. L. Op de Coul, V. Ormaasen, D. Paraskevis, L. Perrin, E. Puchhammer-Stockl, L. Ruiz, M. Salminen, J. C. Schmit, F. Schneider, R. Schuurman, V. Soriano, G. Stanczak, M. Stanojevic, A. M. Vandamme, K. Van Laethem, M. Violin, K. Wilbe, S. Yerly, M. Zazzi, and C. A. Boucher. 2005. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J. Infect. Dis. 192958-966. [DOI] [PubMed] [Google Scholar]
- 48.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 2551134-1137. [DOI] [PubMed] [Google Scholar]
- 49.Yerly, S., S. Jost, A. Telenti, M. Flepp, L. Kaiser, J. P. Chave, P. Vernazza, M. Battegay, H. Furrer, B. Chanzy, P. Burgisser, M. Rickenbach, M. Gebhardt, M. C. Bernard, T. Perneger, B. Hirschel, and L. Perrin. 2004. Infrequent transmission of HIV-1 drug-resistant variants. Antiviral Ther. 9375-384. [PubMed] [Google Scholar]