Abstract
Herpes simplex virus mutants lacking the vhs protein are severely attenuated in animal models of pathogenesis and exhibit reduced growth in primary cell culture. As a result of these properties, viruses with vhs deleted have been proposed as live-attenuated vaccines. Despite these findings and their implications for vaccines, the mechanisms by which vhs promotes infection in cell culture and in vivo are not understood. In this study we demonstrate that vhs-deficient viruses replicate to reduced levels in interferon (IFN)-primed cells and that this deficit has both IFN-dependent and IFN-independent components. Furthermore, vhs-defective viruses induce increased and physiologically active levels of IFN, increased amounts of IFN-stimulated transcripts, and more phosphorylated eIF2α. In addition, we demonstrate greater accumulation of viral RNAs following infection with a vhs-deficient virus. This leads to the hypothesis that attenuation of viruses lacking vhs may be attributed to increased levels of double-stranded RNA, a potent pathogen-associated molecular pattern. Together these data show that vhs likely functions to reduce innate immune responses and thereby acts as a critical determinant of viral pathogenesis.
Herpes simplex virus type (HSV) is a ubiquitous human pathogen responsible for a variety of conditions, including cold sores, genital sores, and keratitis in immunocompetent hosts. In neonates, or in hosts lacking a competent immune system, HSV can become disseminated and cause severe and life-threatening diseases, including hepatitis and encephalitis. The outcome of infection is determined largely by the status of the host's immune response and its interaction with the many viral genes that act to counter it. Of these, the virion host shutoff protein (vhs), while being nonessential for replication cell in culture, is a crucial factor for both HSV type 1 (HSV-1) and HSV-2 replication, virulence, and pathogenesis in vivo (35, 39). Viruses lacking vhs have an increased susceptibility to interferon (IFN), an increased ability to activate dendritic cells, and a decreased ability to down-regulate major histocompatibility complex class I in cell culture (31, 40, 43). These observations highlight the multifactorial impact of vhs on the innate and adaptive immune system (32). In addition, the reduced replication of vhs-deficient viruses relative to wild-type virus is observed rapidly, within 24 h of infection in the cornea or vaginal epithelium (35, 39), suggesting that an inability to counter innate immunity, rather than adaptive immunity, contributes most significantly to the attenuated phenotype. In addition, vhs of HSV-2 plays an important role early in HSV-2 pathogenesis in vivo by interfering with the IFN-mediated antiviral response (5). It has been proposed that HSVs with vhs deleted might be useful as live attenuated vaccines in both prophylactic and therapeutic settings and as such have proved effective in animal models (11, 15, 44, 45). Together these data point to a critical role for vhs in the counteraction of the innate immune system, but the precise mechanisms have yet to be determined.
Biochemically, the function of vhs has been well characterized. vhs, a 58-kDa phosphoprotein, is a virally encoded RNase with RNase activity that degrades both host and viral mRNA and thereby contributes to the marked decrease in host protein synthesis that follows HSV infection (16). As a component of the HSV tegument, vhs targets host mRNAs before de novo viral gene expression by association with cellular translation initiation factors (8, 9). vhs is believed to promote viral infection by depleting cellular mRNA, thereby decreasing its competition with viral mRNA for cellular translation machinery and perhaps by decreasing certain specific genes important for the host response to infection (7). vhs also contributes to infection by regulating the stability of viral RNA as infection progresses and through stabilization of the gE/gI complex, a complex necessary for cell-to-cell spread in vitro and in vivo (14).
In order to sense infection and to rapidly inhibit the growth and spread of pathogens, mammalian cells have evolved a number of innate defenses, many of which are triggered by the recognition of a variety of pathogen-associated molecular patterns (PAMPs). For viruses, these PAMPs include envelope proteins and nucleic acids, especially double-stranded RNA (dsRNA) and DNA (30). All three of these species of PAMP are implicated in the recognition of HSV and the initiation of the cellular antiviral response pathway. In brief, nuclear translocation and activation of NF-κB as well as interferon regulatory factors 3 and 7 (IRF-3 and IRF-7) result in the increased expression of hundreds of genes, including the type I beta interferon (IFN-β) (3, 4). IFN-β further amplifies the response and signals the infection to adjacent cells through JAK/STAT signaling. In the infected cell, the dsRNA-dependent protein kinase R (PKR) is activated by dsRNA and phosphorylates the translation initiation factor eIF2α, promoting the shutdown of viral and host protein synthesis. While PKR is recognized as a major kinase mediating the phosphorylation of eIF2α, other kinases also contribute to the phosphorylation of eIF2α (47).
To date, four HSV genes are known to play major roles in IFN resistance (vhs and the ICP0, ICP34.5, and US11 genes) (2, 6, 12, 18, 21, 23), but the mechanism of vhs in IFN resistance remains obscure. In addition, the profound in vivo attenuation of HSV strains lacking vhs remains poorly understood. In continuous cell lines, viruses lacking vhs grow with wild-type kinetics in one-step and multiple-step growth curves. In primary cells, however, a distinct pattern of multiple-step growth emerges. We have shown previously that HSV-1 vhs mutants show wild-type virus growth up to 24 h, but thereafter their growth is significantly reduced (37). For HSV-2 vhs mutants, the attenuation occurs even sooner under these culture and infection conditions (5). Therefore, we have hypothesized that vhs plays a crucial role in maintaining the susceptibility of cells to HSV beyond the initial round of infection via suppression of innate immune responses.
To test this hypothesis, we measured cellular events downstream of viral recognition, including host gene expression, eIF2α phosphorylation, and IFN-β expression, comparing the cellular response to a wild-type (KOS) virus to that of a vhs-null mutant (Δvhs1). We report herein that infection of mouse embryo fibroblasts (MEFs) with a virus lacking a functional vhs protein leads to an accumulation of viral RNAs, increased phosphorylation of eIF2α, and increased production of IFN-β. Taken together, these findings imply that accumulated viral RNAs induce expression of antiviral genes that attenuate vhs-deficient viruses.
MATERIALS AND METHODS
Cells and viruses.
Viral stocks were grown and titers were determined on Vero cells as previously described (27). The HSV-1 wild-type strain KOS was the background strain for most of this study (34). Construction of the KOS-derived vhs-null HSV-1 strain NHB (referred to in this paper as the Δvhs1 mutant) has been described previously (39). For studies with HSV-2, strain 333 was the background strain from which the vhs-null virus strain 333d41 was constructed, and this null strain is referred to as the Δvhs2 mutant (35). MEF cultures were generated from 129 Sv/Ev mice at embryonic day 15 and passaged once before being plated for infection. MEFs were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 0.1 mM sodium pyruvate, 250 U/ml penicillin, 250 μg/ml streptomycin, and 250 ng/ml amphotericin B. Isogenic IFN-αβγR−/− (AG129) (24) and PKR−/− (129PK) (48) MEFs were also utilized. For multiple-step growth curves, cells were pretreated overnight with 100 U/ml IFN-α (Sigma, St. Louis, MO) where appropriate, and cells were infected at a multiplicity of infection (MOI) of 0.01.
HSV-1 microarray production.
ArrayOligoSelector was used to select 182 70-mer oligonucleotides specific for HSV-1 transcripts (1). The oligonucleotides were compared to the HSV-1 strain 17 viral genome (GenBank accession no. NC_001806) and the mouse RefSeq transcriptome. When possible, three nonoverlapping oligonucleotides were chosen for each HSV-1 transcript. In addition, 68 HSV-1-specific oligonucleotides identical to those generated previously were synthesized (36). Selected cellular genes were also represented on the array. To facilitate normalization, Arabidopsis thaliana ready-to-spot oligonucleotides (Stratagene, La Jolla, CA) were also included. Each oligonucleotide probe was synthesized by Illumina, Inc. (San Diego, CA). Oligonucleotides were solubilized at 20 μM in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.75 M betaine for printing. The oligonucleotides were spotted in triplicate onto expoxide-coated glass slides (Corning, Corning, NY) by use of a linear servo arrayer (17). The printed slides were placed in an oven at room temperature under 70% humidity for 12 to 14 h. After incubation, the slides were cross-linked using the UV Stratalinker 2400 (Stratagene, La Jolla, CA) at 150 μJ/cm2.
RNA preparation.
Cultures of 2.5 × 106 MEFs in 100-mm dishes were infected at an MOI of 5 with KOS or the Δvhs1 mutant for 30 min followed by removal of the inoculum and addition of complete medium. At the times postinfection indicated in the figures, RNA was harvested as previously described (26). RNA was purified and DNase treated with the RNeasy minikit (Qiagen, Valencia, CA). Total RNA quality was determined with an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA) according to the manufacturer's recommendations.
cDNA synthesis and labeling.
First-strand cDNA was generated by random and oligo(dT)-primed reverse transcription (Superscript II; Invitrogen, Carlsbad, CA) utilizing the 3DNA Array 900MPX kit (Genisphere, Hatfield, PA), according to the manufacturer's protocol. The cDNAs were quantified via spectrophotometry to confirm recovery. Samples were paired and balanced by mass.
Hybridization.
Each sample pair was suspended in sodium dodecyl sulfate-based hybridization buffer (Genisphere) and Array 50dT blocker (Genisphere). Two hybridizations were carried out in a sequential manner. The primary hybridization was performed under a supported glass coverslip (Erie Scientific, Portsmouth, NH) at 62°C for 16 to 20 h. Prior to the secondary hybridization, the slides were washed as described in the manufacturer's protocol. Secondary hybridization was carried out using the 3DNA Array 350 kit (Genisphere) according to the manufacturer's protocol, washed, and treated with Dyesaver (Genisphere) before scanning.
Data analysis.
Slides were scanned immediately following hybridization on a ScanArray Express HT scanner (PerkinElmer, Waltham, MA). Laser power was kept constant, and photomultiplier tube values were set for optimal intensity with minimal background. Gridding and analysis of images were performed with Scanarray software express v3.0 (PerkinElmer). The resulting median pixel values were imported into GeneSpring v7.3 software (Agilent), and local background intensities were subtracted from individual spot intensities. To account for dye swap, the “signal” channel and “control” channel measurements were reversed in such samples so that signal derived from Δvhs1 mutant-infected MEF RNA occupied the signal channel and that of the KOS-infected MEF RNA occupied the control channel. The mean signal and control intensities of the on-slide duplicate spots were calculated, and the control values were normalized with GeneSpring's “normalize to positive control genes” function using four SpotReport oligonucleotide array validation system spike-in controls. If the control channel signal was lower than 10 relative fluorescence units (rfu), then 10 rfu was used instead. Signal-to-normalized-control ratios were calculated, and the cross-chip averages were derived from the antilog of the mean of the natural log ratios across all biologic replicates. Oligonucleotide elements that received a detectable call (intensity of >200 rfu or local signal-to-background ratio of >2 in at least one channel) by Scanarray software in either the Cy3 or Cy5 in at least two of four samples of at least one time point were identified, and all others were excluded from the analysis. Genes were not filtered based on significance in order to be able to visualize global trends. Differences in dye incorporation and quantum yield were normalized by the addition of A. thaliana mRNA spikes into the reverse transcription reaction in equal molar amounts along with the experimental mRNA. Data represent two independent biological experiments with dye swaps such that two technical experiments were performed for each independent biological sample.
Real-time reverse transcription (RT)-PCR.
Total RNA was harvested from MEFs in 35-mm wells that were mock treated or infected at an MOI of 5 utilizing the Aurum total RNA minikit as described in the kit protocol (Bio-Rad, Hercules, CA). cDNA was generated using the iScript cDNA synthesis kit as described in the kit protocol (Bio-Rad). PCRs were prepared with iQ SYBR green supermix (Bio-Rad), 5% acetamide, primers (IDT, Coralville, IA), and 2 μl of cDNA. All PCRs were performed in duplicate with the Bio-Rad iCycler. Primers specific for 18S rRNA were used as a reference gene for normalization between samples.
IFN-β ELISA.
Confluent MEFs plated in 35-mm wells were mock treated or infected at an MOI of 5 for 3, 6, 9, or 12 h with KOS or the Δvhs1 mutant and cultured in 1 ml of medium. Culture supernatants were harvested at the indicated times and stored at −20°C until assayed. IFN-β in the medium was measured using 50 μl of harvested medium in a mouse IFN-β enzyme-linked immunosorbent assay (ELISA) as described in the kit protocol (PBL Biomedical Laboratories, Piscataway, NJ).
VSV and HSV challenge assay.
MEF monolayers were mock treated or infected at MOI of 5 with KOS or the Δvhs1 mutant for 12 h, at which time the medium (1 ml) was collected and clarified by low-speed centrifugation (300 rpm, 5 min) to remove cellular debris. To neutralize infectious HSV, the medium was divided and treated with either 50 μl of Dulbecco's modified Eagle's medium or HSV-1 neutralizing antibody (Sigma, St. Louis, MO) for 1 h at 37°C. Treated medium was used as a pretreatment for fresh secondary MEF monolayers, with 150 μl of medium used per 35-mm well. After 18 h of pretreatment, the medium was removed and the MEFs were infected with either vesicular stomatitis virus (VSV) or HSV at an MOI of 0.01. Some pretreated wells were left uninfected in order to assay the effectiveness of the neutralizing antibody treatment. After 2 days postinfection (VSV) or 3 days postinfection (HSV), the medium and cells were harvested, and the titers of infectious particles were determined on Vero cells.
Western blot analysis.
Protein lysates were prepared, processed, and analyzed as previously described (26). The antibodies used in this study included total eIF2α (FL-315) (Santa Cruz Biotechnology, Santa Cruz, CA), phospho-specific eIF2α (pS52) (BioSource International Inc, Camarillo, CA), and the secondary antibody goat anti-rabbit-horseradish peroxidase conjugate (Bio-Rad).
RESULTS
Growth of the Δvhs1 mutant is reduced in IFN-treated MEFs.
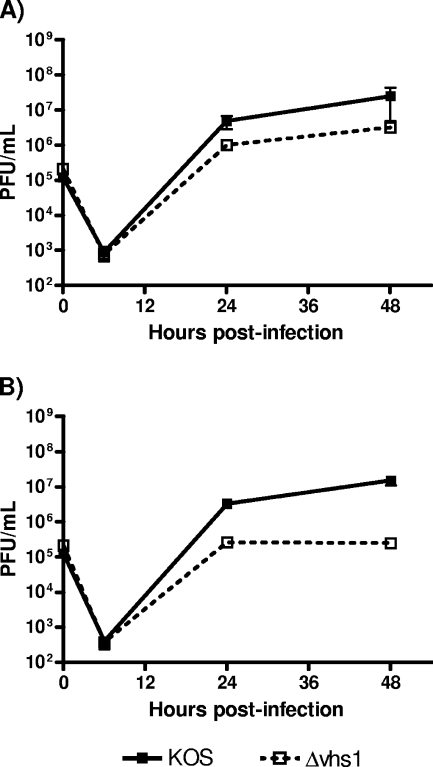
vhs-null viruses exhibit near-normal growth in continuous cell lines such as Vero and Hep-2 (28), with only slight growth defects in terms of plaque size and burst size (28, 29, 39). Studies of primary explant cultures of human and murine corneal buttons, however, showed that late in infection (≥28 to 72 h), growth differences between a vhs-null virus and KOS became apparent (37). No differences, however, were apparent at high MOIs. In this study we compared the growth of the Δvhs1 mutant with that of KOS in primary MEF cultures infected at a low MOI that were mock treated or pretreated with recombinant alpha IFN (rIFN-α). As previously reported, the vhs-null virus showed about 1-log lower growth than the wild-type KOS (Fig. 1A). When cells were pretreated overnight with rIFN-α, significant differences between the growth of KOS and the Δvhs1 mutant were apparent at 24 h (>10-fold, P < 0.01), with the difference continuing to increase up to 48 h (80 to 100-fold, P < 0.01) (Fig. 1B). These data were consistent with previous data (5) and with the hypothesis that the vhs function is important for maintaining the susceptibility of IFN-primed cells to HSV infection.
FIG. 1.
In vitro replication in MEFs. Confluent primary MEF monolayers were infected with KOS or the Δvhs1 mutant at an MOI of 0.01. (A) Multiple-step growth curve analysis of untreated cells. (B) Multiple-step growth curve analysis of cells pretreated overnight with 100 U/ml of rIFN-α. At the indicated times postinfection, cells and supernatants were harvested and viral titers were measured on Vero cells. Results shown are the mean titers of the results for three independent experiments.
Δvhs1 virus-infected MEFs produce increased IFN-β and ISG RNA.
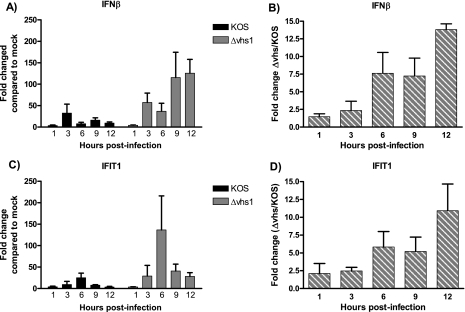
IFN-β transcript expression and IFN protein synthesis and secretion are induced in response to virus infection and recognition of pathogen-associated molecular patterns (PAMPs). In turn, IFN-stimulated genes (ISGs) are up-regulated and activated. We therefore assessed whether infection of primary cultured cells with the Δvhs1 mutant resulted in an increase in IFN or ISG synthesis relative to KOS-infected cells. Analysis of IFN-β RNA by real-time RT-PCR revealed that cells infected with both KOS and the Δvhs1 mutant expressed increased amounts of IFN-β RNA relative to mock-infected cells, although the increase in induction for the Δvhs1 mutant-infected cells relative to the mock-treated cells was higher than that for KOS-infected cells (Fig. 2A). When comparing the ratios of IFN-β expression (Δvhs1 to KOS), Δvhs1 mutant-infected cells expressed 8- to 13-fold more IFN-β RNA than KOS-infected cells from 6 to 12 h postinfection (hpi) (Fig. 2B). To assess the downstream effect of this induction, we subsequently examined the expression of the IFIT1 (interferon-induced protein with tetratricopeptide repeats 1, also known as ISG56) gene, which is rapidly induced by type 1 IFN (33, 41). Real-time RT-PCR revealed that as for IFN-β expression, IFIT expression was induced by both the Δvhs1 mutant and KOS relative to mock-infected cells, but the induction was higher for the Δvhs1 mutant (Fig. 2C). When the ratios of IFIT1 expression (Δvhs1 to KOS) are compared, the Δvhs1 virus-infected cells express 5- to 10-fold more IFIT1 RNA than KOS-infected cells from 6 to 12 hpi (Fig. 2D).
FIG. 2.
Real-time RT-PCR analysis of IFN-β and IFIT1 mRNA expression. Confluent MEF monolayers were mock treated or infected with KOS or the Δvhs1 mutant at an MOI of 5. At the indicated times postinfection, RNA was harvested and used for real-time RT-PCR. (A) Change in expression for IFN-β RNA relative to expression in mock-treated cells. (B) IFN-β RNA expression as a ratio of the Δvhs1 mutant to KOS at each time point indicated. (C) Change in expression for IFIT1 RNA relative to mock-treated cells. (D) IFIT1 RNA expression as a ratio of the Δvhs1 mutant to KOS at each time point indicated. All data shown are the mean changes from the results for three independent experiments.
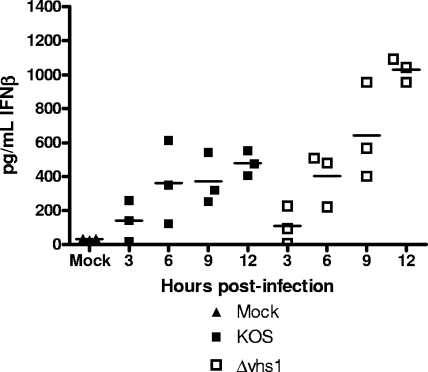
Having shown increases in the levels of IFN-β and IFIT1 RNA expressed by Δvhs1 mutant-infected cells compared to those expressed by KOS, we next wanted to assess the levels of IFN-β protein produced in these infected cells. We therefore measured IFN-β secreted into the culture medium over the first 12 h of infection by use of an IFN-β ELISA (Fig. 3). We found that while the levels of IFN-β secreted by KOS-infected cells increased from 0 to 6 hpi, they subsequently leveled off. Cells infected with the Δvhs1 mutant also showed an increase in IFN-β synthesis 0 to 6 hpi, but in contrast to KOS-infected cells, the secretion of IFN-β did not level off but rather continued to increase up to 12 h. In general, the data for the IFN-β protein expression mirrored that for its RNA expression.
FIG. 3.
IFN-β secretion by infected cells. Confluent MEF monolayers were mock treated or infected with KOS or the Δvhs1 mutant at an MOI of 5. At the indicated times postinfection, the supernatants were harvested and IFN-β secretion was measured by ELISA. Shown are the results from three independent experiments.
Δvhs1 virus infection in MEFs produces functional IFN.
The real-time RT-PCR and ELISA results described above indicated higher IFN-β production from MEFs infected with the Δvhs1 mutant than from MEFs infected with KOS. To test the physiological relevance of the observed difference and whether the levels of secreted IFN were antiviral, culture medium was collected from MEFs treated with mock lysate or infected with KOS or the Δvhs1 mutant and used to pretreat secondary wild-type and IFN-αβγR−/− MEF cultures. These MEFs were then challenged with VSV, KOS, or the Δvhs1 mutant and the titers of the resultant viruses were determined. Pretreatment with medium from KOS-infected cells reduced the VSV titer on MEFs an average of 100-fold, and medium from Δvhs1 mutant-infected cells reduced the VSV titer 400-fold. Changes are relative to titers on IFN-αβγR−/− MEFs and are from two experiments. It should also be noted that the VSV titers on IFN-αβγR−/− MEFs were comparable for KOS-infected supernatant- and Δvhs1 mutant-infected supernatant-treated groups.
Pretreatment with medium from KOS-infected cells reduced the KOS titer on MEFs an average of 6-fold and reduced Δvhs1 mutant titers an average of 50-fold, reflecting the higher sensitivity of the Δvhs1 mutant to IFN. Pretreatment with medium from Δvhs1 mutant-infected cells reduced the KOS titer on MEFs an average of 10-fold and reduced Δvhs1 mutant titers an average of 100-fold. As described above for the VSV challenge assays, changes are relative to titers on IFN-αβγR−/− MEFs and are from two experiments, and HSV titers on IFN-αβγR−/− MEFs were comparable for KOS-infected supernatant-treated and Δvhs1 mutant-infected supernatant-treated groups. Taken together, these data show that the released IFN is antiviral and that cells infected with an HSV-1 mutant that lacks vhs secrete more antiviral IFN than a KOS-infected cell.
HSV-1 Δvhs1 virus-infected MEFs have increased levels of viral transcripts.
The open reading frames of HSV, especially those of the late gene class, are located on opposing DNA strands and produce large amount of mRNAs with regions of partial complementarity. Previous studies have identified the presence of dsRNA in HSV-infected cells (13, 46). HSV-1 vhs degrades viral mRNAs during infection, and in its absence, viral immediate-early gene expression is prolonged, while early and late gene expression is delayed, compared to a wild-type virus (28). That said, the precise kinetics and preference of this degradation activity have not been examined. We therefore wished to compare viral RNA expression levels in Δvhs1 mutant- and KOS-infected MEFs, in order to evaluate the potential for overexpression of dsRNA in vhs-deficient virus infections. The assessment was performed using two concomitant approaches, gene array and real-time RT-PCR.
Viral gene array analysis.
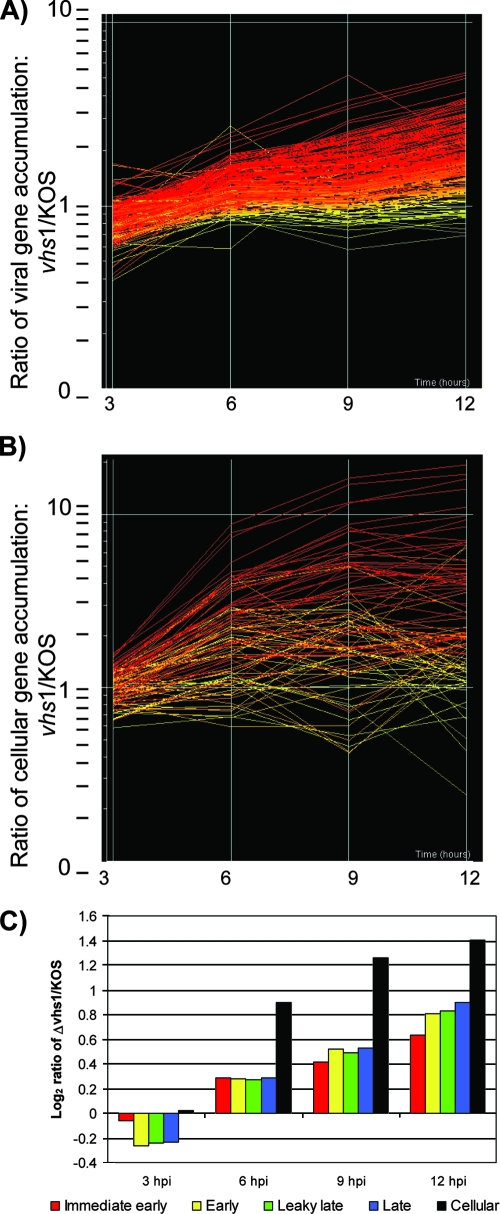
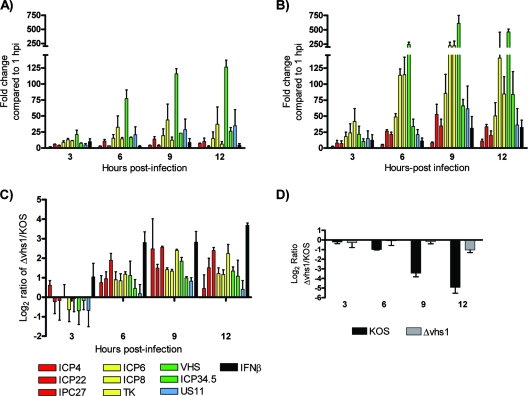
Our custom viral microarray as described in Materials and Methods was designed to examine all known HSV genes and selected cellular genes. The results are shown by a line graph, with each line representing the ratio (Δvhs1 mutant to KOS) of gene expression for each viral (Fig. 4A) and cellular (Fig. 4B) gene. The line graphs also indicate the significance of the expression ratio at 12 hpi. A y-axis value of 1 indicates no difference between the KOS and Δvhs1 mutant results. The data showed the same trends over two independent experiments. At the 3-h time point, there was expression of most classes of viral genes by both viruses but a generalized underexpression of many genes by the Δvhs1 mutant relative to KOS (Fig. 4C). At the 6-, 9-, and 12-h time points, however, there was overexpression of most genes in all classes for the Δvhs1 mutant relative to KOS (Fig. 4C). Similarly, there was overexpression of cellular genes at these time points in Δvhs1 mutant-infected cells. Together these data suggest that there is global degradation of viral and cellular RNAs mediated by vhs that occurs between 3 and 12 hpi. Further details of these data are available in Table S1 in the supplemental material.
FIG. 4.
Microarray analysis of viral and cellular mRNA expression. (A) Line graph depicting the expression of detectable signal from 231 oligonucleotide spots designed to detect all known viral genes. The approximately 90 viral genes were represented in a redundant fashion by at least three oligonucleotides. (B) Line graph depicting the expression of the 92 detectable cellular genes. The time in hours is represented on the x axis, and the change in expression is represented on the y axis (log scale). Each line represents the mean of the ratio of intensity signals for a particular transcript from Δvhs1 mutant-infected MEFs to that from KOS-infected MEFs. The lines are colored to indicate the significance of the under- or overexpression at 12 hpi, by standard error of measurement, ranging from red (+3 standard deviations) to yellow (0 standard deviations) to blue (−3 standard deviations). (C) Within each viral gene kinetic class and cellular genes, the ratio (Δvhs1 mutant to KOS) of the gene expression levels was averaged and converted to a log2 ratio to visualize trends in gene class expression over time.
Real-time RT-PCR analysis.
Validation of the viral gene array data was performed via real-time RT-PCR for the mRNAs encoding ICP4, -22, and -27 (immediate-early genes); ICP6, ICP8, and thymidine kinase (early genes); and vhs, ICP34.5, US11, and gC (leaky-late and true-late genes). The accumulation of these transcripts in KOS- and Δvhs1 mutant-infected MEFs over time is shown relative to their levels at 1 hpi. As expected, viral transcripts in KOS-infected cells accumulated over the 12-h time course, broadly according to their kinetic classes (Fig. 5A). Similarly, mRNA levels in Δvhs1 mutant-infected cells increased over time, although to a greater extent than seen in KOS-infected MEFs (Fig. 5B). For both KOS and the Δvhs1 mutant, the greatest changes were seen in early and late genes. When the levels of viral mRNA were compared as a ratio (Δvhs1 mutant to KOS), there was a notable underexpression of almost all genes analyzed at 3 hpi by the Δvhs1 mutant (Fig. 5C), consistent with the gene arrays (Fig. 4). In contrast, from 6 to 12 hpi, the Δvhs1 mutant-infected cells exhibited a marked accumulation of all viral genes examined, compared to KOS-infected cells. To summarize, when absolute values of viral mRNA were assessed, both viruses accumulated viral transcripts throughout infection, but Δvhs1 mutant-infected cells accumulated them at a greater rate than did KOS-infected cells. When the expression levels of actin mRNA for the two viruses were compared (Fig. 5D), we observed evidence of strong RNA degradation mediated by KOS, but not by the Δvhs1 mutant, between 6 and 12 hpi. Taken together, these data are consistent with the gene array studies and also with the hypothesis that a vhs-deficient virus can express more dsRNA than a wild-type virus.
FIG. 5.
Real-time RT-PCR analysis of selected viral and cellular genes. Confluent MEF monolayers were mock treated or infected with KOS or the Δvhs1 mutant at an MOI of 5, and RNA was extracted for RT-PCR analysis. Viral genes are color coded by kinetic class, and genes are shown in the graphs in the same order as they appear in the key at the bottom of the figure. (A) Change in mRNA expression for selected viral genes relative to that for KOS at the 1-h time point. (B) Change in mRNA expression for selected viral genes relative to that for the Δvhs1 mutant at the 1-h time point. (C) Ratio of gene expression for each time point for selected genes reported as a log2 ratio of Δvhs1 mutant to KOS. (D) Actin mRNA expression reported as a log2 ratio of Δvhs1 mutant to KOS. Results shown are the means for two independent experiments.
Phosphorylation of eIF2α is increased in HSV Δvhs mutant-infected MEFs.
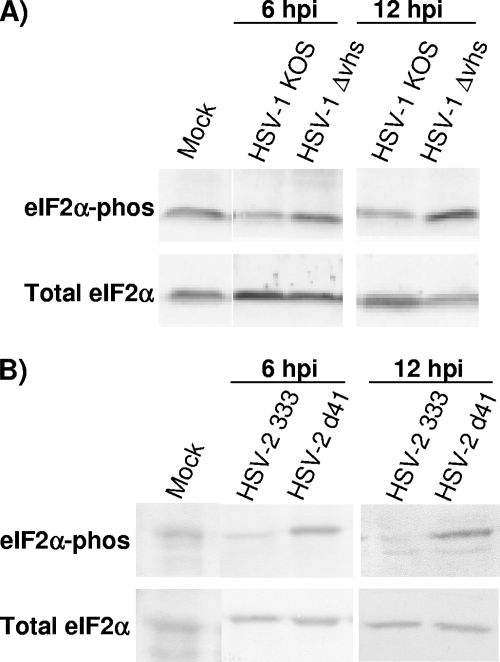
Activation of the PKR pathway heralds the presence of dsRNA, stimulates IFN expression, and is a pivotal marker of the cellular antiviral state. Once activated by dsRNA binding, PKR phosphorylates the translation initiation factor eIF2α on serine 51. In this state, eIF2α is unable to recycle GDP for GTP and translation initiation is shut down. The HSV-1 gene products ICP34.5 and US11 are known to directly counter this translational shutoff response by inactivating or reversing the effects of PKR, thereby allowing viral replication to continue. No other HSV proteins have been shown to have activity in this regard, but given the role for vhs in countering IFN production (Fig. 3) (5) and controlling viral mRNA levels (Fig. 4 and 5), we hypothesized that the presence of vhs activity might impact PKR-dependent eIF2α phosphorylation. Because of the demonstrated ability of HSV-2 vhs to strongly counter IFN production and responses in vitro and in vivo (5), we also wanted to compare the impacts of HSV-2 vhs on the phosphorylation of eIF2α. We therefore assayed for phosphorylated eIF2α by Western blotting lysates of MEFs that were mock treated or infected with KOS, Δvhs1 mutant, HSV-2 333, or Δvhs2 mutant (333d41) viruses (Fig. 6A and B). At 6 and 12 hpi, increases were seen in phosphorylated eIF2α in Δvhs1 and Δvhs2 mutant-infected cells compared with the respective wild-type-infected MEFs. The increases, however, in eIF2α phosphorylation for the Δvhs2 mutant-infected cells (eightfold at 6 hpi) relative to those infected with HSV-2 333 were greater than those seen for the Δvhs1 mutant (twofold at 6 hpi) relative to KOS.
FIG. 6.
Western blot analysis of eIF2α phosphorylation in infected cells. Confluent MEF monolayers were mock treated or infected at an MOI of 5. At the indicated times postinfection, cell lysates were prepared and eIF2α phosphorylation was assessed by Western blotting. (A) Total and phosphorylated eIF2α assessed over time following infection with KOS or the Δvhs1 mutant. (B) Total and phosphorylated eIF2α assessed over time following infection with HSV-2 333 or the Δvhs2 mutant.
Partial restoration of Δvhs1 mutant growth in IFN-αβγR−/− MEFs but not PKR−/− MEFs.
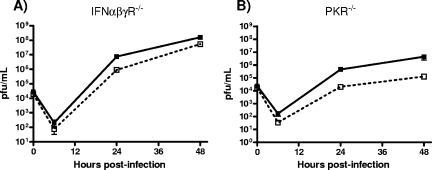
In order to further determine the role of IFNs and PKR in controlling Δvhs1 virus replication, we performed a multiple-step in vitro growth analysis of primary MEFs derived from either IFN-αβγR−/− or PKR−/− mice (Fig. 7). Compared to the result with control MEFs, in IFN signaling-deficient IFN-αβγR−/− MEFs, we found that the growth of the Δvhs1 virus was only partially restored and still slightly and reproducibly attenuated relative to that of KOS (Fig. 1). As expected, pretreatment with rIFN-α did not attenuate Δvhs1 virus replication (data not shown), in contrast to what was observed in the control MEFs (Fig. 1B). These results indicated that IFN-dependent signaling was mostly, but not wholly, responsible for the attenuation of Δvhs1 virus observed in control MEFs. In PKR−/− MEFs, Δvhs1 virus replication was reduced 10-fold compared to replication of KOS (Fig. 7B), comparable to the attenuation observed for control MEFs (Fig. 1). Furthermore, the phosphorylation of eIF2α induced by Δvhs1 virus was not significantly increased over levels for mock infection in PKR−/− MEFs (data not shown). These data show that despite the induction of increased eIF2α phosphorylation, the loss of PKR does not restore the IFN-dependent growth defect to vhs-deficient HSV-1.
FIG. 7.
In vitro replication in IFN-αβγR−/− and PKR−/− MEFs. Confluent primary MEF monolayers were infected with KOS (solid lines) or the Δvhs1 (dashed lines) mutant at an MOI of 0.01. (A) Multiple-step growth curve analysis of IFN-αβγR−/− MEFs. (B) Multiple-step growth curve analysis of PKR−/− MEFs. At the indicated times postinfection, cells and supernatants were harvested and viral titers were measured on Vero cells.
DISCUSSION
The vhs function of HSV has been extensively studied in vitro and in vivo, and a clear dichotomy has emerged. In most continuous cell culture systems, lack of vhs activity has little impact on viral growth at any MOI. In primary cells, however, loss of vhs has a far greater impact, especially at a low MOI, and especially for HSV-2 (5). For HSV-2, vhs is a major general mediator of viral resistance to IFN-αβ and to the antiviral effects of PKR, and the virulence of a vhs-deficient HSV-2 is largely restored in IFN-αβR−/− mice (5, 25). Perhaps the most different result between HSV-1 and HSV-2 is that while vhs deletion from both HSV types induces higher levels of phosphorylated eIF2α, only HSV-2 Δvhs virus replication is restored in the absence of PKRs. The IFN-dependent attenuation of an HSV-1 Δvhs mutant is therefore independent of PKR and likely independent of eIF2α phosphorylation. This may explain, at least in part, the greater impact of vhs on the IFN resistance of HSV-2 than on that of HSV-1. While ICP0, ICP34.5, and US11 all play roles in modulating aspects of the IFN response to HSV-1, viruses lacking vhs are not hypersensitive to IFN in culture, and virulence is not restored to vhs-deficient viruses in IFN-αβγR−/− mice (19, 23). While vhs of HSV-2 is more critical than its HSV-1 counterpart for resistance to the IFN response, paradoxically, the in vivo attenuation of HSV-1 mutants lacking vhs is far greater than that of an equivalent vhs-deficient HSV-2 strain (35, 38, 39). The rapidity with which HSV-1 vhs mutants are so attenuated, coupled with the inability of mouse strains lacking adaptive immunity to restore any growth or virulence to vhs-negative HSV-1 strains (25), nevertheless underscores the role for vhs in countering the IFN response.
In this study we have shown that IFN significantly reduces the replication of the Δvhs1 mutant but that the replication of KOS is almost completely unaffected. Moreover, in the absence of exogenous rIFN-α, the growth defect in primary cells of a virus lacking vhs is more apparent following a secondary round of replication. Together these data are consistent with the idea that vhs is important for maintaining cellular permissiveness to HSV infection following priming with IFN. These data for mouse cells are consistent with those seen previously for human cells, showing that this is not a mouse-specific phenomenon (37). The results of other studies have differed with this work and with each other regarding the IFN sensitivity of viruses lacking vhs (5, 23, 37, 40). It seems very likely that these differences arise from the use of differing cell lines, and it is notable that results obtained with primary cells of either human or mouse origin are consistent with each other.
We show herein that vhs significantly affects IFN-β expression. The pattern of IFN-β RNA and protein induction by KOS was consistent with previous studies, with a rapid induction of an antiviral response followed by its shutoff (22). In contrast, Δvhs1 virus infection caused an induction that was followed by an inexorable increase in IFN-β expression over time. This too is consistent with previous studies of HSV-1 and HSV-2 (5, 40). Moreover, the VSV and HSV challenge assays showed that the IFN secreted into supernatants of infected cells is actively antiviral, despite the fact that the IFN produced is significantly diluted by the culture media and is likely unstable in culture. Some contrasts between these studies and others, however, are seen when the expression of the downstream IFIT1 (ISG56) gene is examined. IFIT1 inhibits the initiation of translation by binding to various subunits of the translation initiation factor eIF3 and has been suggested to be a major antiviral mechanism for blocking viral replication (10). Our current work shows strong induction of IFIT1 by KOS but significantly stronger induction in the absence of vhs. Previous work has shown IFIT1 to be undetectable following infection with KOS or with a vhs-null mutant (20) and that IFIT1 was detectable only when ICP0, a major factor in the shutoff of IFN expression and of ISGs by a proteasome-dependent mechanism, was mutated (6). Consistent with our work, however, was that IFIT1 induction by an ICP0 gene-null virus was further enhanced following deletion of vhs. Yet both vhs-null viruses in both studies were ICP0 competent, used the KOS strain, employed similar MOIs, and were similar otherwise (37). It seems unlikely therefore that different viruses or infection methods can account for this discrepancy. It is possible that technical variances such as primer optimization, assay sensitivity, and quantification methods for RT-PCR contribute to the difference in IFIT1 results. In addition, the use of primary cells (MEFs) in this study, compared with continuous cells (human embryonic lung fibroblasts) in the previous studies, is likely to have an impact, especially in light of recent data showing that expression of IFIT1 in vivo is tissue specific (42). Ongoing studies in our laboratory investigating the expression profiles and phenotypes of vhs-deficient viruses in vivo are likely to further define the impact of vhs on IFN and ISG expression.
A key issue in this study is the mechanism by which vhs dampens the innate immune response to HSV. This mechanism has been difficult to define in part because the pathways by which HSV infection is recognized that lead to the antiviral state are also poorly understood. Many potential mechanisms exist, but it is known that efficient recognition of viral dsRNA and signal transduction are required to induce host gene expression. Data in this study suggest that vhs may be operating on both of these requirements. First, vhs may serve to minimize the accumulation of viral RNA throughout infection to levels optimal for efficient viral replication but below the levels detected by RNA sensing molecules such as RIG-I and below those that trigger dsRNA-inducible antiviral pathways such as PKR and RNase L. Second, vhs may serve to reduce the accumulation of RNA expressed by pivotal genes involved in the antiviral state, such as IFN genes and ISGs, as evidenced by the altered expression of IFN-β and IFIT1 in this study. vhs also likely enhances, quantitatively or temporally, the expression of the many viral genes involved in the countering of the IFN response. This may occur in a selective or a nonselective fashion, but either way represents a powerful general mechanism by which HSV can serve to dampen many host response pathways simultaneously. Together, all these activities of vhs serve as a versatile and powerful counter to the various arms of the host's innate immune response. Current in vivo functional genomic analysis ongoing in our laboratory using vhs mutants will likely elucidate this possibility further.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to David A. Leib (EY10707), to Lynda A. Morrison (AI57573), and to the Department of Ophthalmology and Visual Sciences (P30EY02687). Support for the Department from Research to Prevent Blindness and a Lew Wasserman Scholarship to David A. Leib are gratefully acknowledged.
We acknowledge David Wang for help with selection of oligonucleotides and Michael Heinz and Eric Tycksen of the Washington University Microarray Core for helpful design, construction, and implementation of microarray technology.
Footnotes
Published ahead of print on 26 March 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, G., K. Yang, and B. He. 2003. Dephosphorylation of eIF-2α mediated by the γ134.5 protein of herpes simplex virus type 1 is required for viral response to interferon but is not sufficient for efficient viral replication. J. Virol. 7710154-10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9515623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69912-920. [PubMed] [Google Scholar]
- 5.Duerst, R. J., and L. A. Morrison. 2004. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology 322158-167. [DOI] [PubMed] [Google Scholar]
- 6.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 762180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esclatine, A., B. Taddeo, L. Evans, and B. Roizman. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci. USA 1013603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, P., D. N. Everly, Jr., and G. S. Read. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 799651-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, P., D. N. Everly, Jr., and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 7510272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436939-945. [DOI] [PubMed] [Google Scholar]
- 11.Geiss, B. J., T. J. Smith, D. A. Leib, and L. A. Morrison. 2000. Disruption of virion host shutoff activity improves the immunogenicity and protective capacity of a replication-incompetent herpes simplex virus type 1 vaccine strain. J. Virol. 7411137-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harle, P., B. Sainz, Jr., D. J. Carr, and W. P. Halford. 2002. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293295-304. [DOI] [PubMed] [Google Scholar]
- 13.Jacquemont, B., and B. Roizman. 1975. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J. Virol. 15707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalamvoki, M., J. Qu, and B. Roizman. 2008. Translocation and colocalization of ICP4 and ICP0 in cells infected with herpes simplex virus 1 mutants lacking glycoprotein E, glycoprotein I, or the virion host shutoff product of the UL41 gene. J. Virol. 821701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keadle, T. L., K. A. Laycock, J. L. Morris, D. A. Leib, L. A. Morrison, J. S. Pepose, and P. M. Stuart. 2002. Therapeutic vaccination with vhs(-) herpes simplex virus reduces the severity of recurrent herpetic stromal keratitis in mice. J. Gen. Virol. 832361-2365. [DOI] [PubMed] [Google Scholar]
- 16.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 841926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lashkari, D. A., J. L. DeRisi, J. H. McCusker, A. F. Namath, C. Gentile, S. Y. Hwang, P. O. Brown, and R. W. Davis. 1997. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. USA 9413057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leib, D. A. 2002. Counteraction of interferon-induced antiviral responses by herpes simplex viruses. Curr. Top. Microbiol. Immunol. 269171-185. [DOI] [PubMed] [Google Scholar]
- 19.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 781675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melroe, G. T., N. A. DeLuca, and D. M. Knipe. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 788411-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 742052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 2641918-1921. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, J. A., R. J. Duerst, T. J. Smith, and L. A. Morrison. 2003. Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J. Virol. 779337-9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasieka, T. J., T. Baas, V. S. Carter, S. C. Proll, M. G. Katze, and D. A. Leib. 2006. Functional genomic analysis of herpes simplex virus type 1 counteraction of the host innate response. J. Virol. 807600-7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rader, K. A., C. E. Ackland-Berglund, J. K. Miller, J. S. Pepose, and D. A. Leib. 1993. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. J. Gen. Virol. 741859-1869. [DOI] [PubMed] [Google Scholar]
- 28.Read, G. S., and N. Frenkel. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate early) viral polypeptides. J. Virol. 46498-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 677149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito, T., and M. Gale, Jr. 2007. Principles of intracellular viral recognition. Curr. Opin. Immunol. 1917-23. [DOI] [PubMed] [Google Scholar]
- 31.Samady, L., E. Costigliola, L. MacCormac, Y. McGrath, S. Cleverley, C. E. Lilley, J. Smith, D. S. Latchman, B. Chain, and R. S. Coffin. 2003. Deletion of the virion host shutoff protein (vhs) from herpes simplex virus (HSV) relieves the viral block to dendritic cell activation: potential of vhs− HSV vectors for dendritic cell-mediated immunotherapy. J. Virol. 773768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smiley, J. R. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 781063-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, J. B., and H. R. Herschman. 1996. The glucocorticoid attenuated response genes GARG-16, GARG-39, and GARG-49/IRG2 encode inducible proteins containing multiple tetratricopeptide repeat domains. Arch. Biochem. Biophys. 330290-300. [DOI] [PubMed] [Google Scholar]
- 34.Smith, K. O. 1964. Relationship between the envelope and the infectivity of herpes simplex virus. Proc. Soc Exp. Biol. Med. 115814-816. [DOI] [PubMed] [Google Scholar]
- 35.Smith, T. J., L. A. Morrison, and D. A. Leib. 2002. Pathogenesis of herpes simplex virus type 2 virion host shutoff (vhs) mutants. J. Virol. 762054-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 749916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strelow, L., T. Smith, and D. Leib. 1997. The virion host shutoff function of herpes simplex virus type 1 plays a role in corneal invasion and functions independently of the cell cycle. Virology 23128-34. [DOI] [PubMed] [Google Scholar]
- 38.Strelow, L. I., and D. A. Leib. 1996. Analysis of conserved domains of UL41 of herpes simplex virus type 1 in virion host shutoff and pathogenesis. J. Virol. 705665-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 696779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyama, and M. Azuma. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 811763-1771. [DOI] [PubMed] [Google Scholar]
- 41.Terenzi, F., S. Pal, and G. C. Sen. 2005. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340116-124. [DOI] [PubMed] [Google Scholar]
- 42.Terenzi, F., C. White, S. Pal, B. R. Williams, and G. C. Sen. 2007. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J. Virol. 818656-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tigges, M. A., S. Leng, D. C. Johnson, and R. L. Burke. 1996. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J. Immunol. 1563901-3910. [PubMed] [Google Scholar]
- 44.Walker, J., K. A. Laycock, J. S. Pepose, and D. A. Leib. 1998. Postexposure vaccination with a virion host shutoff defective mutant reduces UV-B radiation-induced ocular herpes simplex virus shedding in mice. Vaccine 166-8. [DOI] [PubMed] [Google Scholar]
- 45.Walker, J., and D. A. Leib. 1998. Protection from primary infection and establishment of latency by vaccination with a herpes simplex virus type 1 recombinant deficient in the virion host shutoff (vhs) function. Vaccine 161-5. [DOI] [PubMed] [Google Scholar]
- 46.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 805059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wek, R. C., H. Y. Jiang, and T. G. Anthony. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc Trans. 347-11. [DOI] [PubMed] [Google Scholar]
- 48.Yang, Y. L., L. F. Reis, J. Pavlovic, A. Aguzzi, R. Schafer, A. Kumar, B. R. Williams, M. Aguet, and C. Weissmann. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 146095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.