Abstract
Although the mechanism of simian virus 40 (SV40) DNA replication has been extensively investigated with cell extracts, viral DNA replication in productively infected cells utilizes additional viral and host functions whose interplay remains poorly understood. We show here that in SV40-infected primate cells, the activated ataxia telangiectasia-mutated (ATM) damage-signaling kinase, γ-H2AX, and Mre11-Rad50-Nbs1 (MRN) assemble with T antigen and other viral DNA replication proteins in large nuclear foci. During infection, steady-state levels of MRN subunits decline, although the corresponding mRNA levels remain unchanged. A proteasome inhibitor stabilizes the MRN complex, suggesting that MRN may undergo proteasome-dependent degradation. Analysis of mutant T antigens with disrupted binding to the ubiquitin ligase CUL7 revealed that MRN subunits are stable in cells infected with mutant virus or transfected with mutant viral DNA, implicating CUL7 association with T antigen in MRN proteolysis. The mutant genomes produce fewer virus progeny than the wild type, suggesting that T antigen-CUL7-directed proteolysis facilitates virus propagation. Use of a specific ATM kinase inhibitor showed that ATM kinase signaling is a prerequisite for proteasome-dependent degradation of MRN subunits as well as for the localization of T antigen and damage-signaling proteins to viral replication foci and optimal viral DNA replication. Taken together, the results indicate that SV40 infection manipulates host DNA damage-signaling to reprogram the cell for viral replication, perhaps through mechanisms related to host recovery from DNA damage.
DNA tumor viruses have been very successfully deployed as simple but powerful model systems to gain molecular insight into the mechanisms of DNA replication and oncogenic cell transformation in mammalian cells. One of the simplest DNA tumor viruses, simian virus 40 (SV40), packs the genetic information for both viral DNA replication and cell transformation into a 2.7-kb region of its genome, most of which encodes the large tumor (T) antigen (Tag) (1). Two related proteins, small t and 17k T, are encoded in partially overlapping reading frames with Tag but are not essential for virus propagation in primate cell lines.
Tag is a highly multifunctional modular protein that localizes primarily in the nuclei of infected cells. The protein is composed of an N-terminal J domain (residues 1 to 102) connected through an extended linker to the origin-DNA binding domain (residues 131 to 250), the replicative helicase domain (residues 251 to 627), and a region that determines host tropism through an unknown mechanism (residues 627 to 708) (67, 68, 83). The structures of the J domain linker region, the origin-DNA binding domain, and the helicase domain have been determined (30, 43, 53, 56, 86). Tag function in infected cells relies heavily on specific associations with host proteins; for example, it interacts with replication protein A (RPA), DNA polymerase alpha-primase (Pol-prim), and topoisomerase I to replicate viral DNA (9, 26, 27, 78, 79, 87). Oncogenic cell transformation depends on Tag binding to p53 tumor suppressor, retinoblastoma family proteins, the Hsc70 chaperone protein, the mitotic spindle checkpoint protein Bub1, and the ubiquitin ligase CUL7 (1, 14, 31, 42, 89). The Mre11-Rad50-Nbs1 (MRN) damage signaling and repair complex has also been reported to bind Tag, but its role in cell transformation has not been explored (19, 49, 92).
Although the activities of Tag in viral DNA replication and cell transformation are genetically and biochemically separable, they are tightly coupled in productively infected primate cells. The molecular chaperone function of the Tag J domain is essential for both virus propagation and cell transformation (1, 81). Furthermore, the ability of Tag to induce quiescent cells to enter S phase, a prerequisite for viral DNA replication, depends on multiple functions required for cell transformation (18, 20, 35, 65). Binding of Tag to Nbs1 has been reported to promote viral DNA replication (92), but it is not clear whether Tag binding to CUL7 or Bub1 is required for productive infection. Despite the contribution of these additional activities of Tag to viral replication in infected cells, their mechanisms of action in productively infected cells are not well understood.
Importantly, it has recently been recognized that replication of SV40 and murine polyomavirus DNA in infected cells occurs under conditions in which the ataxia telangiectasia-mutated (ATM)-mediated DNA damage-signaling pathway is activated (15, 77). Checkpoint signaling ordinarily arrests ongoing host DNA replication and delays cell cycle transitions, but in infected cells, this regulation is perturbed and appears to facilitate viral DNA replication. Thus, SV40 DNA replication does not mimic host DNA replication but may instead represent a damage- or stress-adapted pathway that utilizes mechanisms related to host DNA repair or replication fork recovery after damage. Consistent with this possibility, the 10 host proteins identified as essential for reconstitution of SV40 DNA replication in a cell-free system function not only in host DNA replication but also in DNA repair pathways (5, 13, 64, 74, 87).
The additional Tag activities required for in vivo viral DNA replication, together with the evidence that Tag operates at the interface of replication with repair in infected cells, led us to ask how SV40 utilizes DNA damage signaling to promote viral DNA replication in infected cells. We report here that in primate cells productively infected with SV40, activated DNA damage-signaling proteins ATM, γ-H2AX, Mre11, Rad50, and Nbs1 localize in large nuclear foci together with Tag and host proteins essential for viral DNA replication. During infection, Rad50 and Nbs1 levels decline in a proteasome-dependent manner. Mutational disruption of Tag interaction with the host ubiquitin ligase CUL7 prevented the decline in Rad50 and Nbs1 levels and reduced viral propagation. Specific inhibition of ATM kinase activity in infected cells not only reduced replication of SV40 DNA in infected cells but also inhibited colocalization of Tag and damage-signaling proteins in nuclear foci and stabilized Rad50 and Nbs1. We conclude that SV40-infected primate cells utilize ATM signaling at least in part to reprogram the intracellular milieu to support viral DNA replication.
MATERIALS AND METHODS
Cell culture.
Human osteosarcoma U2OS cells and BSC40 monkey kidney cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM l-glutamine. Mouse NIH 3T3 cells were maintained in the same medium and passed at 80% confluence.
SV40 infection.
Culture medium was removed and replaced with just enough DMEM to cover the cells. The SV40 virus stocks had a concentration of 1 × 10e8 to 2 × 10e8 PFU per ml, as determined by plaque assays on BSC40 cells (84), and cells were infected at a multiplicity of infection of 5 (Table 1 and Fig. 4D) or 10 PFU per cell. The virus infection was allowed to proceed for 2 h with gentle rocking every 30 min to ensure the distribution of virus. The virus inoculum was then replaced with fresh warm supplemented DMEM, and the infected cells were further incubated until harvest.
TABLE 1.
Time course of SV40 wt and F98A mutant production in BSC40 monkey cellsa
| Time (hpi) | wt titer | F98A titer (PFU) |
|---|---|---|
| 6 | 2.3 × 10e3 | 3.0 × 10e3 |
| 24 | 2.6 × 10e3 | 2.8 × 10e3 |
| 48 | 1.4 × 10e5 | 4.1 × 10e4 |
| 72 | 1.4 × 10e7 | 2.2 × 10e6 |
| 96 | 8.3 × 10e7 | 2.0 × 10e7 |
Confluent monolayers of BSC40 cells were infected with wt SV40 (strain 776) or the F98A mutant at a multiplicity of infection of 5. The infected dishes were monitored daily for cytopathic effect. At various times postinfection, the infected dishes were treated to three freeze-thaw cycles and stored at −20°C. Virus titers were determined by plaque assay on BSC40 cells (84).
FIG. 4.
An F98A substitution in Tag reduces CUL7 binding but does not inhibit DNA damage signaling or assembly of Tag foci in infected cells. (A) Alignment of the DnaJ domains of polyomavirus Tag. Numbers refer to SV40 Tag. J domain helices 1 to 4 are shown in red. The SV40 LXCXE motif is highlighted in blue. (B) Extract (Ex) from uninfected (con) (lane 1) or mock-infected (lane 2), wt-infected (lane 3), or F98A mutant-infected (lane 4) BSC40 cells at 48 hpi was immunoprecipitated with anti-Tag Pab101 covalently coupled to Sepharose beads and analyzed by immunoblotting as indicated at the right. *, Cross-reacting protein. (C) F98A mutant- or wt-infected BSC40 cells were preextracted, fixed, and stained at 48 and 60 hpi. Cells were visualized by confocal microscopy (red, Tag; green, γ-H2AX; blue, DNA; yellow, merge). Bars, 10 μm. (D) BSC40 cells that had been mock infected or infected with wt SV40 or F98A mutant SV40 were harvested and lysed at the indicated times after infection. Viral DNA was purified and digested with BamHI, electrophoresed in a 0.7% agarose gel, and visualized with Gelstar staining. BamHI-digested cloned SV40 DNA (pSVB3) was used as a marker (M).
DNA transfection.
Cells were plated in 60-mm dishes containing 4 ml of supplemented DMEM and incubated for 24 h to reach confluence. Plasmid DNA purified by CsCl equilibrium density gradient centrifugation (8 μg per dish) was transfected into the cells by use of 20 μl per dish of Lipofectamine (Invitrogen) in 0.5 ml of Opti-MEM medium (Gibco) according the manufacturer's protocol. Five hours later, 2.5 ml of DMEM was added to each dish and incubation was continued until the cells were harvested for analysis.
DNA cloning.
Site-directed mutagenesis (QuikChange; Stratagene) was used to generate an F98A substitution in the Tag coding sequence cloned in pBSKS+ and verified by DNA sequencing (details available on request). The mutant coding sequence (BsrGI fragment) was used to replace the corresponding wild-type (wt) coding sequence in pSV-wt DNA, which contains the SV40 genome (strain 776) cloned in the BamHI site of vector pAT153 (28). To generate virus, BamHI-cleaved pSV-F98A DNA was transfected into BSC40 cells. Cell cultures displaying cytopathic effect were harvested by freeze-thaw and titers of lysates were determined by plaque assay on BSC40 cells (84). To generate pSV-MB, an original GMSV40 Hirt supernatant DNA (59), kindly provided by M. Botchan, was used as the template to amplify the mutant Tag coding sequence on an EcoNI fragment. The mutant sequence was then inserted in place of the corresponding fragment of the wt Tag coding sequence cloned in pBSKS+ and verified by sequencing, and then the BsrGI fragment carrying the mutant Tag coding sequence was substituted for the corresponding fragment in pSV-wt.
Mre11 and Nbs1 mRNA quantification.
Total RNA was collected from mock-infected or SV40-infected U2OS cells (RNeasy mini kit; Qiagen). Equal amounts of total RNA (1.5 μg) from each sample were reverse transcribed to cDNA by reverse transcriptase-PCR exactly as described previously (32), and equal volumes (1 μl) of cDNA were used as templates in quantitative, real-time PCR (32) with the Mre11 or Nbs1 primer set. The Mre11 primer set was designed to cross the exon 17/19 junction, and the Nbs1 primer set was designed to cross the exon 10/11 junction, in order to specifically amplify cDNA from spliced mRNA but not from possible traces of genomic DNA that retained the intervening intron. Sequences for primers were as follows: for Mre11-Forward primer, 5′-GCC TTC CCG AAA TGT CAC TA-3′; for Mre11-Reverse primer, 5′-TTC AAA ATC AAC CCC TTT CG-3′; for Nbs1-Forward primer, 5′-AGC AGC AGA CCA ACT CCA TCA GAA-3′; and for Nbs1-reverse primer, 5′-TCC ACA ATG AGG GTC TAG CAG GTT-3′. The predicted products of amplification with cloned Mre11 and Nbs1 cDNA templates were verified by agarose gel electrophoresis, the template concentrations were measured, and 10-fold dilutions were used to generate the standard calibration curves in experimental quantifications of Mre11 and Nbs1 mRNA levels.
Antibodies.
Tag was detected with polyclonal rabbit serum against purified recombinant Tag or Pab101 mouse monoclonal antibody (33), which reacts with an epitope at the extreme C terminus of Tag (88). Pol-prim was detected with mouse anti-p180 monoclonal SJK132-20 (6). The following commercial antibodies were used: rabbit anti-promyelocytic leukemia (anti-PML) (Chemicon), mouse monoclonal anti-tubulin (Santa Cruz Biotechnology), mouse monoclonal anti-phospho-H2AX (Ser139) (Upstate Cell Signaling), rabbit anti-Mcm10 (Calbiochem), rabbit polyclonal anti-phospho-p53 (Ser15) (Cell Signaling Technology), rabbit polyclonal antibodies against Nbs1 and Mre11 (Novus Biologicals), rabbit polyclonal anti-CUL7 (Bethyl), mouse monoclonal anti-RPA (RPA 34-19; Calbiochem), rabbit polyclonal anti-p53 (fl-393, SC-6243; Santa Cruz), and Hsp70 (SPA-810; Stressgen). Peroxidase AffiniPure goat anti-mouse immunoglobulin G (heavy plus light chains) and goat anti-rabbit immunoglobulin G (heavy plus light chains) were from Jackson Immuno (115-035-246, 111-035-144).
Fluorescence immunostaining.
Cells grown and infected or mock-infected on coverslips were washed three times with cold phosphate-buffered saline (PBS) solution and then with cytoskeleton buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 100 mM NaCl, 300 mM sucrose, 1 mM MgCl2, 1 mM EGTA]. Soluble proteins were preextracted for 3 min with cytoskeleton buffer containing 0.5% Triton X-100 and protease inhibitors (0.15 μM aprotinin, 1 mM phenylmethylsulfonyl fluoride, and 1 μM leupeptin) in an ice bath. The cells were then washed with PBS, fixed in 4% paraformaldehyde in PBS for 30 min, washed with PBS, and blocked with 10% FBS-PBS at 4°C for 24 h (or longer in time course experiments). The samples were then incubated with signal enhancer (Image-iT FX; Invitrogen) for 20 min with primary antibodies for 3 h, washed, and then incubated with secondary antibodies for 1 h. Nuclear DNA in the cells was counterstained with 0.33 μM TO-PRO-3-iodide (Invitrogen) for 20 min. Primary antibody dilutions for rabbit anti-Tag, anti-Nbs1, anti-Mre11, and anti-CUL7 were 1:5,000, 1:300, 1:500, and 1:100, respectively. Mouse monoclonal Pab101 anti-Tag, anti-PML, anti-phospho-H2AX, and anti-RPA were diluted 1:300, 1:300, 1:100, and 1:300, respectively. Secondary antibodies (Invitrogen) AlexaFluor 488 anti-mouse and AlexaFluor 555 anti-rabbit were diluted 1:100. All antibodies were diluted in 10% FBS-PBS. Coverslips were mounted in ProLong antifade reagent (Molecular Probes, Eugene, OR).
Confocal microscopy.
Samples were examined using a Zeiss LSM 510 confocal microscope equipped with three lasers giving excitation lines at 633, 543, and 488 nm. The data from the channels were collected sequentially using the appropriate band-pass filters built into the instrument. Data were collected at a resolution of 512 by 512 or 1,024 by 1,024 pixels, using optical slices of 1.1 μm. The microscope was a Zeiss Axioplan utilizing a 63× oil immersion objective, numerical aperture 1.4. Data sets were processed using the LSM image browser software and then exported to Adobe Photoshop.
Immunoblotting.
Cells were washed three times with cold PBS and lysed on ice in 0.2 ml per 6-cm dish of lysis buffer (50 mM HEPES-KOH, pH 7.3, 250 mM NaCl, 0.1% Nonidet P-40, 10 mM NaF, 0.3 mM Na3VO4) with protease inhibitors. The lysate was centrifuged at 12,500 rpm for 15 min to separate proteins from cell debris, yielding most of the nonstructural proteins (reference 49 and data not shown). The protein concentration in the supernatant was measured using a bicinchoninic acid kit (Pierce Chemical, Rockford, IL), and equal amounts of total protein from each sample were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (48). Proteins were electroblotted to nitrocellulose in carbonate buffer (22) for 2.5 h at 150 mA. The gel was blocked with 5% nonfat dry milk for 30 min to prevent nonspecific binding and then placed in the primary antibody overnight. The membrane was then washed three times with Tris-buffered saline-Tween 20 (TBST) solution and incubated with the secondary antibody. The proteins were then visualized by chemiluminescence (SuperSignal; Pierce) and exposed to film for 30 s to 1 h. To reprobe the membranes, they were washed once with TBST for 10 min, stripped with 5% acetic acid for 30 min, and then washed three times with TBST and reblocked with 5% nonfat milk.
IP.
Cells (10e6) were washed twice with cold PBS, lysed with 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 10% glycerol, and protease inhibitors (immunoprecipitation [IP] buffer) for 20 min on ice, and centrifuged. The supernatant was incubated with Pab101-coupled CNBr-activated Sepharose beads (GE Healthcare) at 4°C for 2 to 3 h with continuous rocking. After four washes with IP buffer, the precipitated proteins were analyzed by SDS-PAGE and Western blotting.
RESULTS
DNA damage-signaling proteins colocalize with viral replication sites in SV40-infected primate cells.
Activation of ATM signaling by SV40 infection was initially detected in CV1 monkey cells (77). To determine whether ATM activation is observed for other primate cells infected with SV40, we investigated the phosphorylation of ATM substrate proteins in BSC40 monkey kidney cells as a function of time after infection, both in soluble extracts and bound to chromatin. Initially, proteins released from the cells by detergent and 250 mM NaCl extraction were examined in immunoblots. Traces of the early viral protein Tag were detected at 24 h postinfection (hpi) (Fig. 1A, lane 5) and accumulated at later times (lanes 6 to 8). Low levels of p53 were found in mock-infected cells, whereas higher levels accumulated in infected cells, as expected due to its stabilization by Tag (references 1 and 78). ATM and its target protein Chk2 were maintained at the same level as in mock-infected cells. In contrast, the level of protein detected by anti-phospho-ATM Ser1981 rose dramatically with time after infection (Fig. 1A, lanes 6 to 8). Robust phosphorylation of p53 on Ser15 was observed even at 24 hpi and increased during infection. Activated Chk2 phosphorylated on Thr68 was detected at 36 hpi and maintained up to 60 hpi.
FIG. 1.
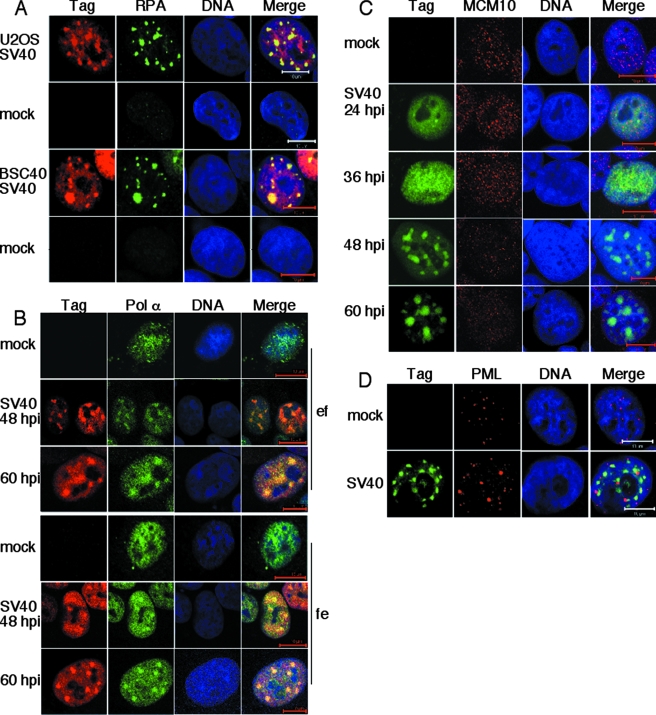
ATM-mediated damage signaling in SV40-infected primate cells localizes with Tag in prominent nuclear bodies. (A) Soluble extracts were prepared from BSC40 cells at the indicated times after SV40 infection or mock infection and analyzed by immunoblotting with the indicated antibodies. Tubulin was used as a loading control. (B) SV40-infected or mock-infected BSC40 cells were preextracted and fixed at different times after infection, stained, and visualized by confocal immunofluorescence microscopy (red, Tag; green, γ-H2AX). Bar, 10 μm. (C) BSC40 cells that had been mock infected, UV irradiated (40 J/m2), or SV40 infected for 48 h were preextracted and fixed (ef) or fixed and then permeabilized (fe) as indicated and then stained as described for panel B. (D) SV40-infected or mock-infected NIH 3T3 cells were examined at the indicated times after infection by immunofluorescence as for panel B.
In cells treated with ionizing radiation, ATM, its accessory factor MRN, and mediators accumulate on chromatin at sites of damage, where the activated kinase phosphorylates histone H2AX (γ-H2AX), MRN, and other substrates (50). To determine whether activated ATM phosphorylates H2AX on chromatin in SV40-infected cells, soluble proteins were preextracted and the residual chromatin-bound proteins were examined by immunofluorescence microscopy. Bright foci of γ-H2AX colocalized with Tag in infected cells, and their intensity increased as a function of time after infection (Fig. 1B). The size and intensity of the γ-H2AX foci in infected monkey cells were much greater than those in UV-irradiated cells (Fig. 1C). Foci in cells that had been preextracted prior to fixation were slightly less intense than those in cells that had been first fixed and then permeabilized (Fig. 1C). Antibody against Ser1981-phosphorylated ATM also detected prominent chromatin-bound nuclear foci in infected monkey cells (not shown). In contrast, γ-H2AX foci in SV40-infected nonpermissive murine NIH 3T3 cells were smaller, less intense, and dispersed through the nucleus (Fig. 1D). Tag staining was intense but diffuse throughout the nucleus and not colocalized with γ-H2AX foci (Fig. 1D). Occasionally, some less intense γ-H2AX foci were also observed in mock-infected cells, suggesting that these cells were in S phase (60). These observations demonstrate a correlation between host permissivity for viral DNA replication and Tag colocalization with damage-signaling proteins.
This correlation raised the question of whether the chromatin-bound Tag and colocalizing damage-signaling proteins might represent sites of SV40 DNA replication. In this case, host proteins required for reconstitution of cell-free viral DNA replication should also reside on chromatin at these sites. Indeed, RPA colocalized with Tag foci in infected human and monkey cells (Fig. 2A). Faint foci of chromatin-bound RPA were also barely detectable in nuclei of mock-infected cells (Fig. 2A) and presumably represent sites of DNA processing on host chromosomes. Similarly, Pol-prim, another protein required for both viral and host DNA replication, localized diffusely in nuclei of mock-infected monkey cells (Fig. 2B). However, in infected cells, large Pol-prim foci colocalized with Tag (Fig. 2B). Colocalizing Pol-prim and Tag foci were easily discernible even when the preextraction of soluble proteins was omitted (Fig. 2B, compare upper and lower sets of panels). In contrast, Mcm10, a protein that binds and stabilizes Pol-prim, recruits it to chromosomal origins prior to replication and migrates with progressing host replication forks (11, 63) localized in a dispersed punctate pattern that was not spatially related to Tag foci (Fig. 2C). The distinctive focal pattern of chromatin-bound viral replication factors, but not of a Pol-prim-associated host replication factor, in infected cells adds support to the notion that the Tag foci represent sites of viral DNA replication.
FIG. 2.
Tag and host replication proteins accumulate in chromatin-bound nuclear foci in SV40-infected primate cells. (A) SV40-infected or mock-infected U2OS and BSC40 cells were preextracted, fixed, and stained at 48 hpi and visualized by confocal immunofluorescence microscopy (red, Tag; green, RPA; DNA, blue). Bars, 10 μm. (B) SV40-infected or mock-infected BSC40 cells were preextracted and then fixed (top; ef) or fixed and then permeabilized (bottom; fe), stained with anti-Tag (red) or anti-Pol-prim (green), and examined by confocal fluorescence microscopy. (C) SV40-infected or mock-infected U2OS cells were preextracted, fixed, stained at the indicated times, and viewed by confocal microscopy (green, Tag; red, Mcm10; blue, DNA). Bars, 10 μm. (D) SV40-infected or mock-infected U2OS cells were preextracted, fixed, stained at 48 hpi, and viewed by confocal fluorescence microscopy (green, Tag; red, PML; blue, DNA). Bars, 10 μm.
By using in situ hybridization to detect newly synthesized viral DNA, SV40 DNA replication centers have been observed juxtaposed to host PML nuclear bodies, also known as nuclear domain 10 or promyelocytic oncogenic domains (38, 40, 82). PML bodies contain a variety of DNA damage-signaling and repair proteins and proteasomes, serve as sites of alternative telomere lengthening, and are targeted by many DNA viruses (7, 16, 25, 58, 72, 91). To determine the spatial relationship of the prominent Tag foci observed in infected cells to PML bodies, we stained SV40-infected and mock-infected human cells with antibody against the PML protein. At least half of the Tag foci in each confocal layer resided adjacent to PML bodies in the same layer (Fig. 2D). Consistent with the previously reported localization of SV40 replication centers (38, 40, 82), the Tag foci adjacent to PML bodies are likely to represent replication centers, while the other Tag foci may represent sites of transcription. The number and size of PML bodies were slightly greater in infected cells than in mock-infected cells (Fig. 2D). Taken together, the results strongly suggest that the prominent chromatin-bound Tag foci in infected cells represent viral replication centers.
MRN subunits localize in viral replication centers and undergo proteasome-dependent degradation.
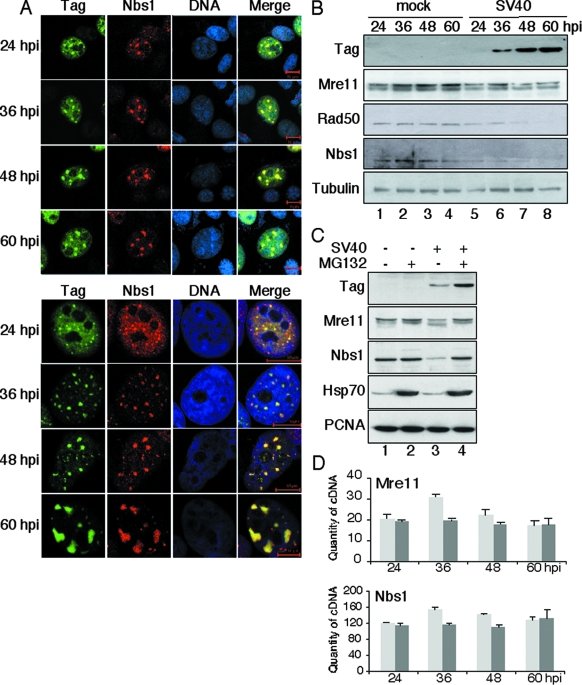
ATM signaling is tightly linked with the MRN complex. MRN recruits ATM to sites of damage in chromatin. It then undergoes phosphorylation by ATM and directly activates ATM to phosphorylate downstream substrate proteins (52, 90). In addition, MRN plays an important role in preventing chromosomal rereplication and in processing damage to facilitate DNA repair (51, 75). The functional coupling of ATM and MRN, as well as the physical interaction of Tag with MRN (92), suggested that MRN might also associate with Tag and DNA damage-signaling proteins in SV40-infected cells. Indeed, Nbs1 colocalized with Tag foci on chromatin in infected human and monkey cells, particularly during viral DNA replication in the late phase of the infection (Fig. 3A; data not shown). Similar foci of Mre11 and Tag were observed in infected human and monkey cells (data not shown). In contrast, diffuse Mre11 and Nbs1 staining was observed in nuclei of mock-infected monkey cells. SV40-infected NIH 3T3 mouse cells did not display prominent foci of Tag or MRN (data not shown).
FIG. 3.
MRN localization and stability are altered in SV40-infected primate cells. (A) At the indicated times after infection of BSC40 (top) or U2OS (bottom) cells with SV40, chromatin-associated Nbs1 and Tag were visualized by confocal immunofluorescence microscopy (green, Tag; red, Nbs1; blue, DNA; yellow, merge). Bars, 10 μm. (B) Soluble extracts of SV40-infected or mock-infected BSC40 cells were prepared at the indicated times after infection and equal amounts of protein from each sample were analyzed by SDS-PAGE and Western blotting. Tubulin was used as a loading control. (C) BSC40 cells mock infected (lanes 1 and 2) or infected with SV40 (lanes 3 and 4) were treated with 20 μM MG132 (+) or DMSO solvent control (−) at 44 hpi. Four hours later, cell lysates were prepared and analyzed by SDS-PAGE and Western blotting. PCNA was used as a loading control and Hsp70 as a positive control for inhibition of proteasome function. (D) Equal amounts of total RNA from SV40-infected (dark gray) or mock-infected (pale gray) U2OS cells were reverse transcribed to cDNA, and the Mre11- or Nbs1-specific cDNA in each sample was quantified by real-time PCR. The mean concentrations of specific cDNA determined from four independent RNA preparations at each time point are shown (fg/μl). Brackets indicate standard deviations of the mean.
The focal colocalization of Tag and MRN with activated ATM in infected primate cells suggested that ATM might phosphorylate not only Tag (77) but also MRN. To test this prediction, extracts of infected and mock-infected monkey cells were analyzed by Western blotting. Phosphorylation of Nbs1 Ser343 was observed for infected cells (not shown), but unexpectedly, we also found that the levels of Rad50 and Nbs1 subunits in soluble cell extracts dropped significantly over the course of the infection (Fig. 3B, lanes 5 to 8). Proteolytic clipping of Mre11 was also apparent in infected monkey cell extracts (Fig. 3B, lanes 7 and 8). In contrast, the level of Tag rose during infection as expected, and the tubulin loading control remained constant in each lane, suggesting the possibility of selective proteolysis of Rad50 and Nbs1 in infected cells.
To assess this possibility, we treated mock-infected and SV40-infected monkey cells with the reversible proteasome inhibitor MG132 (73) and examined MRN levels 4 hours later. Soluble extracts from cells treated with MG132 contained elevated amounts of Hsp70 heat shock protein, reflecting stress as expected (10) (Fig. 3C, lanes 2 and 4). The levels of Mre11 and Nbs1 in mock-infected cells with and without MG132 were quite similar (Fig. 3C, lanes 1 and 2). In SV40-infected cells, the levels of intact Mre11 and Nbs1 were clearly reduced in the absence of MG132 (Fig. 3C, compare lane 3 with lane 1). However, MG132 treatment restored the levels of intact Mre11 and Nbs1 to those in mock-infected cells (Fig. 3C, compare lane 4 with lane 1). We note that Tag was also stabilized by MG132 (Fig. 3C, compare lanes 3 and 4). These results indicate that the decrease in MRN levels in SV40-infected primate cells depended on proteasome activity. Consistent with this interpretation, the steady-state levels of Mre11 and Nbs1 mRNA, determined by quantitative reverse transcription-PCR, remained stable during SV40 infection (Fig. 3D).
Tag interaction with ubiquitin ligase CUL7 directs MRN degradation and facilitates virus propagation.
The colocalization of MRN and Tag in viral replication centers and the selective proteasome-dependent degradation of MRN subunits suggested that the Tag-associated ubiquitin ligase CUL7 might participate in directing MRN destruction. CUL7 is a novel member of the Skp1-Cullin-F-box (SCF) ligase family (17, 66) that was identified as a Tag-binding protein required for cell transformation by SV40 (2, 46, 85). CUL7 binds to the DnaJ chaperone domain of Tag, and specific J domain mutations can weaken or eliminate the interaction in cells overexpressing tagged CUL7 (2, 42, 46). Of note is that the unusually long loop between helices 3 and 4 of the SV40 Tag DnaJ domain is highly conserved among primate polyomaviruses (Fig. 4A) but not in murine polyomavirus Tag and other DnaJ homologs (34), suggesting a conserved function.
In order to determine whether endogenous CUL7 binding to Tag affects MRN subunit degradation in infected monkey cells, we first generated a virus stock with a mutation in Tag codon 98, substituting Ala for the wt Phe (F98A) (2). To examine the interaction of endogenous CUL7 with Tag in wt- and F98A mutant-infected monkey cells, Tag immunoprecipitates were analyzed by immunoblotting. Endogenous CUL7 coprecipitated with wt Tag, but only traces of CUL7 associated with F98A mutant Tag (Fig. 4B). Nevertheless, F98A mutant infection induced ATM-mediated DNA damage signaling similar to that in wt-infected cells, as analyzed by Western blotting (data not shown), and F98A mutant Tag colocalized with γ-H2AX in viral replication centers, although colocalization was less complete than in cells infected with wt SV40 (Fig. 4C). Consistent with these similarities between F98A mutant and wt infections, F98A mutant Tag replicated unit-length viral DNA at about 70% the level of that replicated by wt Tag (Fig. 4D).
If CUL7 interaction with Tag plays a crucial role in directing proteasome-dependent degradation of MRN subunits in infected cells, degradation should be absent from F98A mutant-infected cells. To test this prediction, soluble extracts of F98A mutant-infected monkey cells were prepared at different times after infection and MRN protein levels were monitored by Western blotting. Tag and phospho-p53 Ser15 accumulated as a function of time after infection. However, no decrease in the levels of MRN subunits was detected in infected cells (Fig. 5A, lanes 5 to 8). The results demonstrate that MRN degradation in infected cells depends, directly or indirectly, on CUL7 binding to Tag.
FIG. 5.
MRN degradation in SV40-infected and -transfected BSC40 cells depends on CUL7 interaction with Tag. (A) Extracts of BSC40 cells infected with F98A mutant virus or mock infected were prepared at 24, 36, 48, and 60 hpi and analyzed by SDS-PAGE and Western blotting. PCNA was used as a loading control. (B) BSC40 cells were transiently transfected with plasmid vector (lane 1), with pSV-wt plasmid (lane 2), or with MB SV40 plasmid (lane 3). Cell lysate was isolated 48 h after transfection for Western blot analysis. PCNA and tubulin served as loading controls.
To further substantiate this conclusion, we examined a mutant Tag with a deletion in the CUL7 binding region. This mutant viral genome (MB) was originally recovered as an episome replicating in an SV40-immortalized human fibroblast line, GM637 (59). A second mutant genome with the same deletion was recovered from an independently SV40-immortalized line of skin fibroblasts from a xeroderma pigmentosum patient (59). MB Tag supported robust SV40 DNA replication activity in cell-free assays, but its activity in monkey cells was severely attenuated and it did not produce viral progeny or transform rat cells (59). The MB Tag lacked residues 67 to 82 of wt Tag and Tyr-Lys replaced the wt Lys67-Tyr68, so that most of the loop connecting helix 3 and helix 4 of the Tag J domain was missing. A similar deletion led to the loss of ectopic CUL7 binding activity of Tag (42).
To compare the stabilities of MRN in cells expressing wt and MB Tag, it was not possible to infect cells as for Fig. 5A, as MB Tag does not support virus propagation (59). Thus, wt and MB SV40 plasmid DNA and vector DNA were transfected into monkey cells. Two days later, soluble cell extracts were analyzed by denaturing gel electrophoresis and Western blotting. The levels of wt and MB Tag were similar, and phosphorylation of p53 Ser15 was observed in both cell extracts (Fig. 5B, lanes 2 and 3), consistent with activation of ATM damage signaling. The level of MRN subunits was reduced in extracts of wt SV40-transfected cells (Fig. 5B, compare lanes 1 and 2), but little or no reduction in MRN subunit levels was observed in extracts of MB SV40-transfected cells (Fig. 5B, compare lanes 1 and 3). The results confirm that Tag interaction with CUL7 is a vital step in directing MRN degradation.
The inability of MB SV40 to support virus propagation (59) prompted us to ask whether the F98A mutant viral infection might also display a defect in virus propagation. Monkey cells infected with wt SV40 developed cytopathic effects with a typical time course, with all the cells lysed by 96 hpi. F98A mutant-infected cells also developed cytopathic effects, but with slower kinetics; full cell lysis did not develop until 144 hpi. F98A mutant produced less infectious progeny at all time points of the infection (Table 1). The final wt virus titer was 7.1 × 10e7 PFU/ml, corresponding to a total yield of 3.6 × 10e8 PFU, or 360 PFU/cell. The final F98A mutant titer was 2.3 × 10e7 PFU/ml, corresponding to 1.2 × 10e8 PFU, or 120 PFU/cell. The endpoint titer was repeated in three independent assays and in each case the yield of the F98A mutant was 20 to 30% of that of wt. These results indicate that although the F98A substitution did not prevent virus propagation in cultured BSC40 cells, it did significantly reduce virus yield.
ATM signaling promotes assembly of Tag and γ-H2AX in viral replication foci and proteolysis of MRN subunits.
The results above provide evidence that SV40 infection of primate cells induces ATM-mediated signaling, association of Tag and DNA damage-signaling proteins with viral replication centers, and proteasome-dependent destruction of MRN subunits. It is possible that these events take place in parallel but independently; alternatively, the events could be functionally coupled with one another. To begin to evaluate these alternatives, we examined the role of ATM signaling in greater detail.
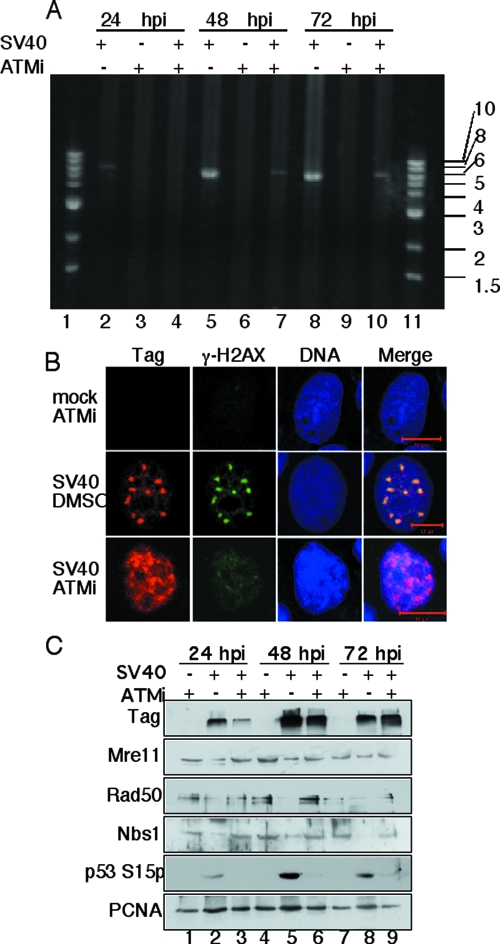
Silencing of ATM expression by small interfering RNA was recently shown to reduce SV40 DNA replication in infected CV1 cells by 50 to 90%, suggesting that either the ATM polypeptide or its signaling activity promotes viral DNA replication (77). To distinguish between these possibilities, we utilized a highly specific small molecule inhibitor of ATM kinase, KU-55933 (ATMi), to suppress ATM signaling without eliminating ATM protein (36). To determine the optimal concentration of ATMi, BSC40 cells were infected with SV40 for 48 h in the presence of increasing concentrations of ATMi or corresponding amounts of dimethyl sulfoxide (DMSO) solvent. The optimal concentration of ATMi for maximal inhibition of p53Ser15 phosphorylation with minimal cytotoxicity was determined to be 10 μM (data not shown), consistent with the literature (36, 39). To confirm the requirement for ATM signaling in viral DNA replication, BSC40 cells were infected with SV40 in the presence of ATMi or solvent. Supercoiled viral genomes were isolated 24, 48, and 72 h later and detected by agarose electrophoresis and staining. Viral DNA replication was detectable at 24 hpi in DMSO-treated cells but not in ATMi-treated cells (Fig. 6A, lanes 2 and 4). Larger amounts of viral DNA were produced at 48 and 72 hpi in DMSO-treated cells, but the levels in ATMi-treated cells were strongly reduced (Fig. 6A, compare lanes 5 and 8 with 7 and 10). The results confirm that ATM kinase activity is required to facilitate SV40 DNA replication in infected monkey cells.
FIG. 6.
Inhibition of ATM kinase activity in SV40-infected cells compromises SV40 DNA replication, foci of Tag and damage-signaling proteins, and proteolysis of Rad50 and Nbs1. The inhibitor KU-55933 (ATMi) or DMSO was added to BSC40 cells 30 min before infection with SV40 and maintained until the cells were harvested. (A) Hirt supernatant DNAs (93) were collected at 24, 48, and 72 hpi, digested with BamHI at 37°C, electrophoresed in a 0.8% agarose gel, and detected by SYBR green gold staining. DNA size markers are indicated in kilobase pairs. (B) At 48 h after infection, cells were preextracted, fixed, stained, and examined by confocal fluorescence microscopy (red, Tag; green, γ-H2AX; blue, DNA). Bars, 10 μm. (C) Cell extracts were collected at 24, 48, and 72 hpi and analyzed by SDS-PAGE and Western blotting. PCNA was used as a loading control.
To determine whether ATM signaling also promoted assembly of chromatin-bound replication foci in infected cells, BSC40 cells treated with ATMi or DMSO were infected with SV40 or mock infected. At different times after infection, cells were preextracted, fixed, stained for Tag and γ-H2AX, and examined by confocal fluorescence microscopy. As expected, Tag and γ-H2AX were not detected in mock-infected cells treated with ATMi (Fig. 6B, top row). DMSO-treated cells infected with SV40 expressed Tag in the familiar large replication centers as the infection progressed, and γ-H2AX colocalized with Tag (Fig. 6B, middle row). In contrast, chromatin-bound Tag expressed in infected cells treated with ATMi did not reside in prominent foci but rather dispersed throughout the nuclei (Fig. 6B, bottom row). In spite of the ATM kinase inhibitor, some chromatin-associated γ-H2AX was detected in a speckled pattern throughout the nuclei of infected cells (Fig. 6B, bottom row). These results indicate that ATM signaling promotes assembly of Tag and damage-signaling proteins in viral replication centers and suggest that SV40 infection induces DNA damage signaling not only through ATM kinase activity but perhaps also through other checkpoint kinases (29).
To assess whether ATM kinase activity influences proteasome-dependent degradation of MRN in SV40-infected cells, BSC40 cells treated with ATMi or DMSO were infected with SV40 or mock infected. Soluble cell extracts were prepared at 24, 48, and 72 hpi, separated by denaturing gel electrophoresis, and analyzed by Western blotting. Extracts from mock-infected cells treated with ATMi displayed constant levels of MRN subunits, no detectable Tag, and no phosphorylation on p53 Ser15 (Fig. 6C, lanes 1, 4, and 7). Extracts from SV40-infected cells treated with DMSO contained Tag, phospho-p53, and reduced levels of Rad50 and Nbs1 relative to the mock-infected extract (Fig. 6C, lanes 2, 5, and 8). The kinetics of Tag accumulation in infected cells treated with ATMi were slower than those in infected cells treated with DMSO (Fig. 6C, compare lanes 3, 6, and 9 with 2, 5, and 8). Importantly, however, in extracts from SV40-infected cells treated with the ATM kinase inhibitor, the levels of Rad50 or Nbs1 were essentially unchanged over the course of infection and comparable to those in extracts of mock-infected cells (Fig. 6C, compare lanes 3, 6, and 9 with 1, 4, and 7). These data demonstrate that ATM kinase activity facilitates the proteasome-dependent degradation of MRN subunits.
DISCUSSION
The SV40 proteins expressed early in infected primate cells prepare the host cell to support viral propagation, but understanding of these events remains incomplete. Here we have begun to explore these events more fully. We show that inhibition of ATM activity prevents or delays assembly of viral DNA replication centers in the nucleus, recruitment of other damage-signaling proteins to the centers, and proteasome-dependent degradation of selected host proteins, e.g., subunits of the MRN complex. Tag interaction with CUL7 ubiquitin ligase is also required for selective proteolysis of MRN subunits. These data confirm and significantly extend an earlier report that SV40 DNA replication is correlated with ATM activation and phosphorylation of Tag (77). Together, these findings imply that SV40 DNA replication in infected cells proceeds through a novel pathway induced by viral reprogramming of the host cell cycle and damage response pathways. This reprogramming is apparently more extensive and complex than previously appreciated, unveiling new puzzles to be solved, including those considered below.
ATM: master regulator for Tag-directed host reprogramming?
The observed requirement for ATM activity for phosphorylation of Tag Ser120 (76, 77), assembly of Tag in viral replication centers, recruitment of damage-signaling and repair proteins to the centers, and MRN proteolysis (Fig. 3 to 6) suggests that ATM activity lies at the top of a regulatory hierarchy. However, the time course of ATM activation, assembly of viral replication centers, accumulation of Tag Ser120p, and MRN proteolysis coincides with the latter phase of infection. This timing suggests that ATM may regulate primarily the late phase of infection and that ATM-independent damage signaling may arise early in infection, prior to the initiation of viral DNA replication. Consistent with this speculation, ATMi suppressed viral DNA replication by only 50 to 90% and chromatin-bound γ-H2AX was easily detected in cells treated with ATMi (Fig. 6A and B; also see reference 77). Moreover, checkpoint signaling by ATR-Chk1 has been reported for SV40-infected monkey cells (62). ATM-independent damage signaling early in infection may play a crucial role in viral reprogramming that facilitates ATM activation. Successive activation of ATR-Chk1, followed by ATM-Chk2, is elicited by some activated oncogenes and is proposed to induce cellular senescence or cell death as an anticancer barrier (4). Whether SV40 infection provokes a similar succession of checkpoint kinases is not known. Further studies will be needed to identify the nature of the ATM-independent signaling observed in Fig. 6B and the mechanism of its activation in SV40-infected cells.
Tag-CUL7-directed proteolysis of MRN.
Proteasome-dependent MRN degradation has been observed for cells infected by other DNA viruses (24, 54, 55, 69, 80), but the discovery that this occurs in SV40-infected cells is novel and its role in SV40 infection is unknown.
MRN colocalization with Tag on chromatin in viral replication centers (Fig. 3) is consistent with the functional interactions between MRN and ATM in DNA damage signaling (50, 90), the association of MRN with host replication forks (51, 60), and the physical interaction of MRN with Tag (19, 49, 92). The proposed role of MRN in host replication fork surveillance might suggest that MRN acts in viral replication centers to maintain ATM activation, consistent with the loss of viral replication centers in the presence of ATMi (Fig. 6B). Activation of ATM by MRN in the centers may also be needed to maintain the infected cell in S/G2 phase for viral replication (15), but additional work will be required to address these possibilities.
Our data argue strongly that CUL7 interaction with Tag plays a crucial role in proteolytic targeting of MRN subunits. However, it is not clear whether CUL7, a primarily cytoplasmic protein (3), enters the nucleus together with Tag, gains access to the nucleus in some other way, or perhaps interacts with Tag in the cytoplasm. Similarly, it is not known whether MRN is recognized as a target for CUL7 through its association with Tag or viral replication centers or through CUL7-associated substrate recognition factors, e.g., Fbxw8 (17). Lastly, it remains to be shown directly that MRN subunits undergo ubiquitin modification and that CUL7 catalytic activity is required.
Our data suggest indirectly that MRN degradation benefits SV40 propagation in infected monkey cells, since Tag variants that bind poorly to CUL7 and fail to direct MRN degradation reduce viral yield (Table 1) or abolish it (59). However, it is not clear how MRN depletion facilitates virus production. Nbs1 has been suggested to suppress Tag-induced amplification of SV40 origin DNA present in an integrated retroviral construct in human cells (92). Thus, proteolytic reduction of MRN levels in infected cells may facilitate viral DNA replication. On the other hand, as noted above, MRN depletion in viral replication centers might be predicted to decrease ATM activity required to maintain the centers. Moreover, in the absence of MRN depletion, replication of unit-length F98A mutant viral DNA is clearly detectable in monkey cells (Fig. 4D) (X. Zhao and J. M. Pipas, unpublished data). Thus, MRN proteolysis might promote a later step in infection, e.g., encapsidation of viral chromatin. Furthermore, it should be noted that our data do not rule out the possibility that Tag-CUL7 directs proteolysis of additional unknown proteins in infected cells, which could also affect viral yield.
Tag-CUL7 interaction is clearly required for SV40-induced cell transformation (2, 42, 46), but the mechanism by which this interaction promotes transformation is not known. One possibility, analogous to Tag inactivation of p53 and pRB tumor suppressor activities, is that Tag binding to CUL7 blocks a potential tumor suppressor activity of CUL7. On the other hand, mutations that inactivate CUL7 in human patients give rise to growth retardation (37, 57) rather than the cancer susceptibility associated with p53 and pRB mutations. The key role of MRN in maintaining host genomic stability (50, 90) suggests an alternative possibility: MRN depletion directed by Tag-CUL7 complexes could contribute to SV40 transformation activity. Consistent with this possibility, wt Tag expression in human fibroblasts has been reported to result in karyotypic abnormalities, whereas mutational inactivation of multiple Tag functions, including but not limited to p53 and pRB binding, reduced the frequency of chromosome damage (70, 71). Evaluation of this possibility, as well as others (21, 44, 45), will be needed to elucidate the role of CUL7-Tag interaction in cell transformation.
SV40 DNA replication centers: repair centers for damaged DNA?
A significant body of evidence suggests that SV40 chromatin replicates in discrete subnuclear factories that reside near PML bodies (Fig. 2; also see references 38, 40, and 82). The biochemical mechanisms that position SV40 minichromosomes near PML bodies are not known, but this spatial relationship bears some resemblance to the juxtaposition of damaged chromosomal DNA and PML bodies, which is thought to facilitate SUMO-regulated recruitment of Bloom's helicase and other repair proteins from PML bodies to damaged chromatin (7, 8, 16, 23). In addition, PML bodies contain proteasomes and are the primary centers for nuclear proteolysis (91). A growing body of evidence demonstrates that DNA damage checkpoints, in particular recovery from checkpoints after repair, are regulated through the ubiquitin-proteasome system (12, 41, 47, 61). Perhaps Tag-bound SV40 minichromosomes are recognized as damaged chromatin to be repaired, consistent with the requirement for ATM signaling to assemble viral replication centers in primate cells (Fig. 6; also data not shown). Consistent with this speculation, the localization of γ-H2AX in SV40 replication centers suggests that it is incorporated in viral chromatin and may help to recruit the host repair proteins that carry out viral DNA replication (29). From this viewpoint, Tag-CUL7-directed proteolysis of MRN could represent a step in recovery from checkpoint signaling after “repair” in viral replication centers. However, the potential role of PML bodies in SV40 DNA replication remains uncertain, since small interfering RNA-directed silencing of PML expression did not significantly reduce viral plasmid DNA replication (40). Moreover, which host proteins sense the incoming viral minichromosomes as damage, which features of the viral chromatin are recognized, and how the juxtaposition with PML bodies is determined are some of the many unanswered questions raised by this speculation.
Acknowledgments
We thank AstraZeneca for providing KU-55933; Timothy R. Peters, Michael Botchan, Sean Schaffer, Kevin Harvey (EMD Bioscience), Paul Cantalupo, David Spector, Jennifer Pietenpol, David Cortez, Nathan Ellis, and David Bazett-Jones for generously sharing reagents and advice; Deniz Yavas for sharing preliminary data; Kun Zhao for help with data management; and members of the Fanning lab for discussion.
We gratefully acknowledge financial support from the VICC (NCI 5P30 CA068485-11), NIH (GM52948 to E.F. and CA40586 to J.M.P.), HHMI Professors Program (52003905), Vanderbilt Discovery program, and Vanderbilt University.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Ahuja, D., M. T. Saenz-Robles, and J. M. Pipas. 2005. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 247729-7745. [DOI] [PubMed] [Google Scholar]
- 2.Ali, S. H., J. S. Kasper, T. Arai, and J. A. DeCaprio. 2004. Cul7/p185/p193 binding to simian virus 40 large T antigen has a role in cellular transformation. J. Virol. 782749-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, P., Y. J. He, and Y. Xiong. 2006. Cytoplasmic localized ubiquitin ligase cullin 7 binds to p53 and promotes cell growth by antagonizing p53 function. Oncogene 254534-4548. [DOI] [PubMed] [Google Scholar]
- 4.Bartek, J., J. Bartkova, and J. Lukas. 2007. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 267773-7779. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71333-374. [DOI] [PubMed] [Google Scholar]
- 6.Bensch, K. G., S. Tanaka, S. Z. Hu, T. S. Wang, and D. Korn. 1982. Intracellular localization of human DNA polymerase alpha with monoclonal antibodies. J. Biol. Chem. 2578391-8396. [PubMed] [Google Scholar]
- 7.Bischof, O., S. H. Kim, J. Irving, S. Beresten, N. A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boe, S. O., M. Haave, A. Jul-Larsen, A. Grudic, R. Bjerkvig, and P. E. Lonning. 2006. Promyelocytic leukemia nuclear bodies are predetermined processing sites for damaged DNA. J. Cell Sci. 1193284-3295. [DOI] [PubMed] [Google Scholar]
- 9.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32503-568. [DOI] [PubMed] [Google Scholar]
- 10.Bush, K. T., A. L. Goldberg, and S. K. Nigam. 1997. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 2729086-9092. [DOI] [PubMed] [Google Scholar]
- 11.Chattopadhyay, S., and A. K. Bielinsky. 2007. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol. Biol. Cell 184085-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury, D., M. C. Keogh, H. Ishii, C. L. Peterson, S. Buratowski, and J. Lieberman. 2005. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20801-809. [DOI] [PubMed] [Google Scholar]
- 13.Cortez, D. 2005. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 191007-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotsiki, M., R. L. Lock, Y. Cheng, G. L. Williams, J. Zhao, D. Perera, R. Freire, A. Entwistle, E. A. Golemis, T. M. Roberts, P. S. Jat, and O. V. Gjoerup. 2004. Simian virus 40 large T antigen targets the spindle assembly checkpoint protein Bub1. Proc. Natl. Acad. Sci. USA 101947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl, J., J. You, and T. L. Benjamin. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J. Virol. 7913007-13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dellaire, G., and D. P. Bazett-Jones. 2007. Beyond repair foci: subnuclear domains and the cellular response to DNA damage. Cell Cycle 61864-1872. [DOI] [PubMed] [Google Scholar]
- 17.Dias, D. C., G. Dolios, R. Wang, and Z. Q. Pan. 2002. CUL7: a DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. USA 9916601-16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickmanns, A., A. Zeitvogel, F. Simmersbach, R. Weber, A. K. Arthur, S. Dehde, A. G. Wildeman, and E. Fanning. 1994. The kinetics of simian virus 40-induced progression of quiescent cells into S phase depend on four independent functions of large T antigen. J. Virol. 685496-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Digweed, M., I. Demuth, S. Rothe, R. Scholz, A. Jordan, C. Grotzinger, D. Schindler, M. Grompe, and K. Sperling. 2002. SV40 large T-antigen disturbs the formation of nuclear DNA-repair foci containing MRE11. Oncogene 214873-4878. [DOI] [PubMed] [Google Scholar]
- 20.Dobbelstein, M., A. K. Arthur, S. Dehde, K. van Zee, A. Dickmanns, and E. Fanning. 1992. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene 7837-847. [PubMed] [Google Scholar]
- 21.Dowell, J. D., S. C. Tsai, D. C. Dias-Santagata, H. Nakajima, Z. Wang, W. Zhu, and L. J. Field. 2007. Expression of a mutant p193/CUL7 molecule confers resistance to MG132- and etoposide-induced apoptosis independent of p53 or Parc binding. Biochim. Biophys. Acta 1773358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn, S. D. 1986. Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal. Biochem. 157144-153. [DOI] [PubMed] [Google Scholar]
- 23.Eladad, S., T. Z. Ye, P. Hu, M. Leversha, S. Beresten, M. J. Matunis, and N. A. Ellis. 2005. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 141351-1365. [DOI] [PubMed] [Google Scholar]
- 24.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 796207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett, R. D. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8365-374. [DOI] [PubMed] [Google Scholar]
- 26.Fanning, E., V. Klimovich, and A. R. Nager. 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 344126-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fanning, E., and J. M. Pipas. 2006. DNA replication and human disease. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Fanning, E., K. H. Westphal, D. Brauer, and D. Corlin. 1982. Subclasses of simian virus 40 large T antigen: differential binding of two subclasses of T antigen from productively infected cells to viral and cellular DNA. EMBO J. 11023-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fillingham, J., M. C. Keogh, and N. J. Krogan. 2006. GammaH2AX and its role in DNA double-strand break repair. Biochem. Cell Biol. 84568-577. [DOI] [PubMed] [Google Scholar]
- 30.Gai, D., R. Zhao, D. Li, C. V. Finkielstein, and X. S. Chen. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 11947-60. [DOI] [PubMed] [Google Scholar]
- 31.Gjoerup, O. V., J. Wu, D. Chandler-Militello, G. L. Williams, J. Zhao, B. Schaffhausen, P. S. Jat, and T. M. Roberts. 2007. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc. Natl. Acad. Sci. USA 1048334-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray, S. J., J. Gerhardt, W. Doerfler, L. E. Small, and E. Fanning. 2007. An origin of DNA replication in the promoter region of the human fragile X mental retardation (FMR1) gene. Mol. Cell. Biol. 27426-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurney, E. G., R. O. Harrison, and J. Fenno. 1980. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct subclasses of large T antigen and for similarities among nonviral T antigens. J. Virol. 34752-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hennessy, F., W. S. Nicoll, R. Zimmermann, M. E. Cheetham, and G. L. Blatch. 2005. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 141697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermeking, H., D. A. Wolf, F. Kohlhuber, A. Dickmanns, M. Billaud, E. Fanning, and D. Eick. 1994. Role of c-myc in simian virus 40 large tumor antigen-induced DNA synthesis in quiescent 3T3-L1 mouse fibroblasts. Proc. Natl. Acad. Sci. USA 9110412-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hickson, I., Y. Zhao, C. J. Richardson, S. J. Green, N. M. Martin, A. I. Orr, P. M. Reaper, S. P. Jackson, N. J. Curtin, and G. C. Smith. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 649152-9159. [DOI] [PubMed] [Google Scholar]
- 37.Huber, C., D. Dias-Santagata, A. Glaser, J. O'Sullivan, R. Brauner, K. Wu, X. Xu, K. Pearce, R. Wang, M. L. Uzielli, N. Dagoneau, W. Chemaitilly, A. Superti-Furga, H. Dos Santos, A. Megarbane, G. Morin, G. Gillessen-Kaesbach, R. Hennekam, I. Van der Burgt, G. C. Black, P. E. Clayton, A. Read, M. Le Merrer, P. J. Scambler, A. Munnich, Z. Q. Pan, R. Winter, and V. Cormier-Daire. 2005. Identification of mutations in CUL7 in 3-M syndrome. Nat. Genet. 371119-1124. [DOI] [PubMed] [Google Scholar]
- 38.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith, J. Lukas, and S. P. Jackson. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 837-45. [DOI] [PubMed] [Google Scholar]
- 40.Jul-Larsen, A., T. Visted, B. O. Karlsen, C. H. Rinaldo, R. Bjerkvig, P. E. Lonning, and S. O. Boe. 2004. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp. Cell Res. 29858-73. [DOI] [PubMed] [Google Scholar]
- 41.Kapetanaki, M. G., J. Guerrero-Santoro, D. C. Bisi, C. L. Hsieh, V. Rapic-Otrin, and A. S. Levine. 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 1032588-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasper, J. S., H. Kuwabara, T. Arai, S. H. Ali, and J. A. DeCaprio. 2005. Simian virus 40 large T antigen's association with the CUL7 SCF complex contributes to cellular transformation. J. Virol. 7911685-11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, H.-Y., B.-Y. Ahn, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, S. S., M. Shago, L. Kaustov, P. C. Boutros, J. W. Clendening, Y. Sheng, G. A. Trentin, D. Barsyte-Lovejoy, D. Y. Mao, R. Kay, I. Jurisica, C. H. Arrowsmith, and L. Z. Penn. 2007. CUL7 is a novel antiapoptotic oncogene. Cancer Res. 679616-9622. [DOI] [PubMed] [Google Scholar]
- 45.Koch, H. B., R. Zhang, B. Verdoodt, A. Bailey, C. D. Zhang, J. R. Yates III, A. Menssen, and H. Hermeking. 2007. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 6205-217. [DOI] [PubMed] [Google Scholar]
- 46.Kohrman, D. C., and M. J. Imperiale. 1992. Simian virus 40 large T antigen stably complexes with a 185-kilodalton host protein. J. Virol. 661752-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krogan, N. J., M. H. Lam, J. Fillingham, M. C. Keogh, M. Gebbia, J. Li, N. Datta, G. Cagney, S. Buratowski, A. Emili, and J. F. Greenblatt. 2004. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell 161027-1034. [DOI] [PubMed] [Google Scholar]
- 48.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 49.Lanson, N. A., Jr., D. B. Egeland, B. A. Royals, and W. C. Claycomb. 2000. The MRE11-NBS1-RAD50 pathway is perturbed in SV40 large T antigen-immortalized AT-1, AT-2 and HL-1 cardiomyocytes. Nucleic Acids Res. 282882-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavin, M. F., and S. Kozlov. 2007. ATM activation and DNA damage response. Cell Cycle 6931-942. [DOI] [PubMed] [Google Scholar]
- 51.Lee, A. Y., E. Liu, and X. Wu. 2007. The Mre11/Rad50/Nbs1 complex plays an important role in the prevention of DNA rereplication in mammalian cells. J. Biol. Chem. 28232243-32255. [DOI] [PubMed] [Google Scholar]
- 52.Lee, J. H., and T. T. Paull. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 267741-7748. [DOI] [PubMed] [Google Scholar]
- 53.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423512-518. [DOI] [PubMed] [Google Scholar]
- 54.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15119-126. [DOI] [PubMed] [Google Scholar]
- 55.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 7914004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat. Struct. Biol. 31034-1039. [DOI] [PubMed] [Google Scholar]
- 57.Maksimova, N., K. Hara, A. Miyashita, I. Nikolaeva, A. Shiga, A. Nogovicina, A. Sukhomyasova, V. Argunov, A. Shvedova, T. Ikeuchi, M. Nishizawa, R. Kuwano, and O. Onodera. 2007. Clinical, molecular and histopathological features of short stature syndrome with novel CUL7 mutation in Yakuts: new population isolate in Asia. J. Med. Genet. 44772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129278-287. [DOI] [PubMed] [Google Scholar]
- 59.Maulbecker, C., I. Mohr, Y. Gluzman, J. Bartholomew, and M. Botchan. 1992. A deletion in the simian virus 40 large T antigen impairs lytic replication in monkey cells in vivo but enhances DNA replication in vitro: new complementation function of T antigen. J. Virol. 662195-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirzoeva, O. K., and J. H. Petrini. 2003. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1207-218. [PubMed] [Google Scholar]
- 61.Mu, J. J., Y. Wang, H. Luo, M. Leng, J. Zhang, T. Yang, D. Besusso, S. Y. Jung, and J. Qin. 2007. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J. Biol. Chem. 28217330-17334. [DOI] [PubMed] [Google Scholar]
- 62.Okubo, E., J. M. Lehman, and T. D. Friedrich. 2003. Negative regulation of mitotic promoting factor by the checkpoint kinase chk1 in simian virus 40 lytic infection. J. Virol. 771257-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pacek, M., A. V. Tutter, Y. Kubota, H. Takisawa, and J. C. Walter. 2006. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21581-587. [DOI] [PubMed] [Google Scholar]
- 64.Paulsen, R. D., and K. A. Cimprich. 2007. The ATR pathway: fine-tuning the fork. DNA Repair (Amsterdam) 6953-966. [DOI] [PubMed] [Google Scholar]
- 65.Perry, M. B., and J. M. Lehman. 1998. Activities of SV40 T antigen necessary for the induction of tetraploid DNA content in permissive CV-1 cells. Cytometry 31251-259. [PubMed] [Google Scholar]
- 66.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 69-20. [DOI] [PubMed] [Google Scholar]
- 67.Pipas, J. M. 1985. Mutations near the carboxyl terminus of the simian virus 40 large tumor antigen alter viral host range. J. Virol. 54569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poulin, D. L., and J. A. DeCaprio. 2006. The carboxyl-terminal domain of large T antigen rescues SV40 host range activity in trans independent of acetylation. Virology 349212-221. [DOI] [PubMed] [Google Scholar]
- 69.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 153104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ray, F. A., D. S. Peabody, J. L. Cooper, L. S. Cram, and P. M. Kraemer. 1990. SV40 T antigen alone drives karyotype instability that precedes neoplastic transformation of human diploid fibroblasts. J. Cell. Biochem. 4213-31. [DOI] [PubMed] [Google Scholar]
- 71.Ray, F. A., M. J. Waltman, J. M. Lehman, J. B. Little, J. A. Nickoloff, and P. M. Kraemer. 1998. Identification of SV40 T-antigen mutants that alter T-antigen-induced chromosome damage in human fibroblasts. Cytometry 31242-250. [PubMed] [Google Scholar]
- 72.Reddel, R. R. 2007. A SUMO ligase for ALT. Nat. Struct. Mol. Biol. 14570-571. [DOI] [PubMed] [Google Scholar]
- 73.Rock, K. L., C. Gramm, L. Rothstein, K. Clark, R. Stein, L. Dick, D. Hwang, and A. L. Goldberg. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78761-771. [DOI] [PubMed] [Google Scholar]
- 74.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 7339-85. [DOI] [PubMed] [Google Scholar]
- 75.Sartori, A. A., C. Lukas, J. Coates, M. Mistrik, S. Fu, J. Bartek, R. Baer, J. Lukas, and S. P. Jackson. 2007. Human CtIP promotes DNA end resection. Nature 450509-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider, J., and E. Fanning. 1988. Mutations in the phosphorylation sites of simian virus 40 (SV40) T antigen alter its origin DNA-binding specificity for sites I or II and affect SV40 DNA replication activity. J. Virol. 621598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi, Y., G. E. Dodson, S. Shaikh, K. Rundell, and R. S. Tibbetts. 2005. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J. Biol. Chem. 28040195-40200. [DOI] [PubMed] [Google Scholar]
- 78.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 5575-134. [DOI] [PubMed] [Google Scholar]
- 79.Stenlund, A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 4777-785. [DOI] [PubMed] [Google Scholar]
- 80.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 81.Sullivan, C. S., and J. M. Pipas. 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 749694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tornow, J., M. Polvino-Bodnar, G. Santangelo, and C. N. Cole. 1985. Two separable functional domains of simian virus 40 large T antigen: carboxyl-terminal region of simian virus 40 large T antigen is required for efficient capsid protein synthesis. J. Virol. 53415-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tremblay, J. D., K. F. Sachsenmeier, and J. M. Pipas. 2001. Propagation of wild-type and mutant SV40. Methods Mol. Biol. 1651-7. [DOI] [PubMed] [Google Scholar]
- 85.Tsai, S. C., K. B. Pasumarthi, L. Pajak, M. Franklin, B. Patton, H. Wang, W. J. Henzel, J. T. Stults, and L. J. Field. 2000. Simian virus 40 large T antigen binds a novel Bcl-2 homology domain 3-containing proapoptosis protein in the cytoplasm. J. Biol. Chem. 2753239-3246. [DOI] [PubMed] [Google Scholar]
- 86.Valle, M., X. S. Chen, L. E. Donate, E. Fanning, and J. M. Carazo. 2006. Structural basis for the cooperative assembly of large T antigen on the origin of replication. J. Mol. Biol. 3571295-1305. [DOI] [PubMed] [Google Scholar]
- 87.Waga, S., and B. Stillman. 1994. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369207-212. [DOI] [PubMed] [Google Scholar]
- 88.Weisshart, K., S. Friedl, P. Taneja, H.-P. Nasheuer, B. Schlott, F. Grosse, and E. Fanning. 2004. Partial proteolysis of simian virus 40 T antigen reveals intramolecular contacts between domains and conformation changes upon hexamer assembly. J. Biol. Chem. 27938943-38951. [DOI] [PubMed] [Google Scholar]
- 89.Williams, G. L., T. M. Roberts, and O. V. Gjoerup. 2007. Bub1: escapades in a cellular world. Cell Cycle 61699-1704. [DOI] [PubMed] [Google Scholar]
- 90.Williams, R. S., J. S. Williams, and J. A. Tainer. 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85509-520. [DOI] [PubMed] [Google Scholar]
- 91.Wojcik, C., and G. N. DeMartino. 2003. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35579-589. [DOI] [PubMed] [Google Scholar]
- 92.Wu, X., D. Avni, T. Chiba, F. Yan, Q. Zhao, Y. Lin, H. Heng, and D. Livingston. 2004. SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev. 181305-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ziegler, K., T. Bui, R. J. Frisque, A. Grandinetti, and V. R. Nerurkar. 2004. A rapid in vitro polyomavirus DNA replication assay. J. Virol. Methods 122123-127. [DOI] [PubMed] [Google Scholar]