Abstract
Papillomaviruses are a family of nonenveloped DNA tumor viruses. Some sexually transmitted human papillomavirus (HPV) types, including HPV type 16 (HPV16), cause cancer of the uterine cervix. Papillomaviruses encode two capsid proteins, L1 and L2. The major capsid protein, L1, can assemble spontaneously into a 72-pentamer icosahedral structure that closely resembles native virions. Although the minor capsid protein, L2, is not required for capsid formation, it is thought to participate in encapsidation of the viral genome and plays a number of essential roles in the viral infectious entry pathway. The abundance of L2 and its arrangement within the virion remain unclear. To address these questions, we developed methods for serial propagation of infectious HPV16 capsids (pseudoviruses) in cultured human cell lines. Biochemical analysis of capsid preparations produced using various methods showed that up to 72 molecules of L2 can be incorporated per capsid. Cryoelectron microscopy and image reconstruction analysis of purified capsids revealed an icosahedrally ordered L2-specific density beneath the axial lumen of each L1 capsomer. The relatively close proximity of these L2 density buttons to one another raised the possibility of homotypic L2 interactions within assembled virions. The concept that the N and C termini of neighboring L2 molecules can be closely apposed within the capsid was supported using bimolecular fluorescence complementation or “split GFP” technology. This structural information should facilitate investigation of L2 function during the assembly and entry phases of the papillomavirus life cycle.
Papillomaviruses are a family of structurally similar nonenveloped DNA viruses that infect stratified squamous epithelial tissues, such as the skin or mucous membranes, of humans and various animal species (reviewed in reference (17). A group of roughly 30 sexually transmitted human papillomavirus (HPV) types tend to infect the skin or the mucosal surfaces of the genitals. Although most genital HPV infections are clinically inapparent and self-limiting, persistent infection with oncogenic HPV types, such as HPV16, can lead to the development of cervical cancer or other anogenital cancers (reviewed in reference 47).
The papillomavirus major capsid protein L1 can spontaneously self-assemble into 72-pentamer virus-like particles (VLPs) that closely resemble the native T=7 icosahedral structure of papillomavirus virions (29). The structure of a truncated T=1 12-pentamer L1 VLP has been solved at 3.5-Å resolution (9, 35). Naturally produced authentic papillomavirus virions also contain a minor capsid protein, L2 (20). Although L2 is dispensable for capsid formation, it has been implicated in encapsidation of the 8-kb circular double-stranded DNA viral genome (25, 54). L2 has also been shown to participate in multiple steps during papillomavirus entry into host cells. Proposed roles for L2 include the induction of conformational changes in cell-bound virions, disruption of endosomal membranes, and the subcellular trafficking of the incoming viral genome (3, 15, 22, 28, 41, 53).
The amino-terminal third of L2 contains sequences that are highly conserved between distantly related human and animal papillomavirus types. In contrast to the HPV type-restricted character of anti-L1 neutralizing antibodies, some antibodies targeting amino-terminal portions of L2 can cross-neutralize a broad range of human and animal papillomavirus types (19, 23, 39, 40, 45). Thus, L2 is a candidate target antigen for the development of broadly protective prophylactic HPV vaccines.
Because the papillomavirus life cycle is closely linked to the differentiation of cells within stratified squamous epithelia, it has not been possible to produce large amounts of authentic papillomavirus virions in cell culture. This is particularly true of cancer-associated genital mucosotropic HPVs (such as HPV16), which tend to produce relatively few virions (19, 34). It has therefore been difficult to systematically determine the number and arrangement of L2 in papillomavirus virions, especially those of genital HPVs.
Most current literature reflects the widespread view that typical HPV virions probably contain about 12 molecules of L2. Support for this belief comes primarily from studies of recombinant L1+L2 VLPs (44, 49). Despite the general consensus, a careful review of the literature reveals several seemingly forgotten early studies of naturally produced HPV virions in which L2 occupancy varied substantially between different virion preparations but in some instances averaged 30 or perhaps more molecules of L2 per virion (37, 38, 42). Furthermore, one recent study has reported baculovirus-based production of VLPs carrying an average of 80 molecules of L2 per VLP (50). The concept that virions might accommodate more than 12 molecules of L2 is also consistent with the observation that recombinant L1 capsomers can associate with L2 at a 5:1 stoichiometry, implying the potential existence of up to 72 L2 binding sites in an assembled virion (21).
Recently our group has developed high-yield methods for producing infectious L1+L2 capsids (pseudoviruses) in cultured mammalian cells (4, 7). In this report, we have developed a system for serial propagation of HPV16 pseudoviruses, allowing production of concentrated capsid preparations. We used this novel system to demonstrate that intracellularly assembled pseudovirions can accommodate up to 72 molecules of L2. Using cryoelectron microscopy (cryo-EM) and difference imaging, we have visualized an icosahedrally ordered portion of L2 within intact L1+L2 capsids. This information should be useful for developing a more-detailed understanding of the biology of L2.
MATERIALS AND METHODS
Cell culture.
293TT cells (4) were maintained in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% 56°C heat-inactivated fetal calf serum (HyClone), nonessential amino acid mixture, Glutamax-I (Invitrogen), and 400 μg/ml hygromycin (Roche) (DMEM-10). Cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions. TT-16L2 cells, which carry a stably integrated HPV16 L2 expression plasmid (p16L2P), were maintained in DMEM-10 supplemented with 0.6 μg/ml puromycin (Clontech).
Capsid production.
Detailed technical protocols and nucleotide maps of plasmids used in this work are available at our laboratory website (http://home.ccr.cancer.gov/Lco/) and in references 5 and 6.
For experiments investigating the association of L2 with L1, capsids were produced by transiently transfecting 75-cm2 flasks of 293TT cells with the plasmids shown at the top of Fig. 1. Lysates were clarified and purified using a previously reported 27-33-39% iodixanol (Optiprep) step gradient method (5). Trypsin (Invitrogen) digestion was performed by spiking capsid samples with trypsin at a final concentration of 125 μg/ml, followed by a 10-min incubation at 37°C.
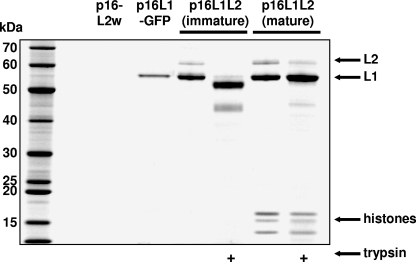
FIG. 1.
L2 is integrally associated with L1. Cells were transfected with the plasmid shown at the top of the figure. After 48 h, the cells were lysed and capsids were allowed to mature in crude lysate (except in lanes marked “immature”). Capsids were then purified using iodixanol gradients. For immature capsids, this procedure results in a loss of encapsidated DNA and associated histone proteins. In some instances, purified capsid preparations were treated briefly with trypsin. The capsids were separated by SDS-PAGE and visualized with Sypro Ruby fluorescent protein staining.
Quantitation of the L2 content of capsid stocks was performed by comparing the intensity of L1 and L2 bands in stained NuPage (Invitrogen) gels using a ChemiImager 4400 (Alpha Innotech) imaging device. Gels were stained with the Microwave Blue colloidal Coomassie stain (Protiga) or Sypro Ruby (Molecular Probes) according to the manufacturer's instructions.
L1-only capsids used for cryo-EM analysis were generated by transfecting four 150-cm2 flasks of 293TT cells with p16L1-GFP (7) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were harvested 48 h after transfection.
Viral seed stocks for production of L1+L2 capsids were generated by cotransfecting 293TT cells with an equal mixture of p16L1L2 and p16L2w. Forty-eight hours after transfection, the cells were suspended at 108 cells/ml in Dulbecco's phosphate-buffered saline (Invitrogen) supplemented with 0.4% Brij58 (Sigma) and a nuclease cocktail (4). The resulting cell lysates were incubated at 37°C overnight to allow capsid maturation (7). After overnight incubation, the lysates were adjusted to 850 mM NaCl and clarified by centrifugation for 10 min at 5,000 × g. Thirty microliters of the resulting seed stock was used to infect a 150-cm2 flask of TT-16L2 cells preplated overnight at a density of 1.5 × 107 cells per flask. Infected cells were harvested 52 h after infection.
Transfected or infected capsid-producing cells were harvested as described for production of seed stocks (above). Capsids were purified out of clarified cell lysates over Optiprep gradients. Gradient fractions were screened for capsid protein and histone content using NuPage 4 to 12% gradient gels (Invitrogen) stained with Microwave Blue. For the L1-only capsids, pooled step-gradient fractions were further concentrated by equilibrium ultracentrifugation in 30% Optiprep for 4 h at 340,000 × g in an NVT65 near-vertical rotor. A single peak capsid-containing gradient fraction was exchanged into Dulbecco's phosphate-buffered saline containing a total of 0.5 M NaCl by gel filtration chromatography (6). The samples were stored briefly on ice and diluted with water for cryo-EM analysis.
Cryo-EM analysis.
Samples (3 μl each) were applied dropwise to grids coated with holey carbon films and then incubated for 1 min, blotted, rapidly frozen in liquid ethane, and imaged in a CM200-FEG electron microscope (FEI) at 120 keV with a 626 cryo-holder (Gatan). Grids were surveyed at low magnification using a charge-coupled-device camera, and micrographs were recorded on film, using low-dose procedures and at magnification ×38,000 (11).
Negatives were scanned with an SCAI scanner (Z/I Imaging, Huntsville, AL) and binned twofold to give 3.68 Å/pixel. Each micrograph typically contained 50 to 200 usable particle images. Particles were extracted using the X3DPreprocess software program (13). Defocus values ranged from 0.7 to 1.2 mm, placing the first contrast transfer function zeros between 16Å−1 and 20Å−1. Orientations were determined and refined by the PFT software program, and three-dimensional (3D) reconstructions were calculated by the EM3DR software program (1). 3D density maps were rendered at 100% of expected mass after removal of blobs of mass not connected to the capsid shell with the UNBLOB software program (13). Scaling of the L1-only maps to the L1+L2 maps was fine-tuned via their one-dimensional radial averages.
A 3D reconstruction (3DR) of L1-only capsids was computed from 530 images out of a total of 1,950 images extracted from 18 micrographs, with a resolution of 25.1 Å (Fourier shell correlation = 0.3). Analysis of L1+L2 capsids was based on 722 images out of a total of 4,858 images extracted from 24 micrographs, with a resolution of 22.7 Å (Fourier shell correlation = 0.3).
BiFC.
Enhanced yellow fluorescent protein (YFP) (BD-Clontech) was mutated (F64→L; numbering is with respect to wild-type green fluorescent protein) and dissected at E172/D173 into N- and C-terminal fragments according to a strategy presented in a patent application (51). The YFP fragments were fused to the C terminus of L2 (constructs pL2-NYFP and pL2-CYFP) with an LGGTGSG linker peptide. In separate constructs, YFP fragments were fused to the N terminus of L2 (constructs pNYFP-L2 and CYFP-L2) with a GSGGTRAT linker peptide. Capsids were produced by cotransfecting pairs of L2 bimolecular fluorescence complementation (BiFC) plasmids together with an L1 expression plasmid (pOhuL) into 293TT cells. Capsids were harvested and purified as described above. The presence of the YFP fusion partners did not grossly affect the L1 capsid yield or L2 incorporation levels relative to those of wild-type L2 (data not shown).
Fluorescence of purified BiFC capsid preparations or transfected HeLa cells was measured in black 96-well plates (Nunc) using a Spectramax Gemini XPS (Molecular Devices) plate fluorometer with excitation and emission set at 488 and 527 nm, respectively, and a cutoff of 515 nm.
RESULTS
Analysis of L2 incorporation into pseudovirions.
Methods for production of HPV pseudovirions using human embryonic kidney 293TT cells are now well established (6). In the course of purifying pseudovirions for other studies, we noticed that the ratio of L1 to L2 often appeared to be substantially lower than 30:1. In other words, a variety of different pseudovirion preparations appeared to have more than the widely presumed average of 12 molecules of L2 per capsid.
One possible explanation for the appearance of additional L2 might be copurification of free L2 along with L1 capsids. We utilized two approaches to verify that the L2 in purified capsid preparations is integrally associated with L1. First, when 293TT cells were transfected with an L2 expression plasmid (p16L2w) alone and lysates were subjected to ultracentrifugation over Optiprep step gradients, L2 did not migrate to gradient fractions containing L1 only or L1+L2 capsids (Fig. 1). Second, we determined that a substantial fraction of L2 in purified mature capsids was resistant to proteolytic digestion with trypsin while L2 (and L1) in purified immature capsids was highly susceptible to trypsin, consistent with the disrupted configuration of immature capsids after ultracentrifugation (Fig. 1) (5). These results show that the presence of L2 in purified capsid stocks is due to its integral association with L1 capsids.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified capsids produced by transfecting 293TT cells with p16L1L2 revealed an L1:L2 molar ratio of about 10:1 (unpublished result available on our laboratory website [http://home.ccr.cancer.gov/Lco/L2CryoSupp.htm]). In four independent p16L1L2-based capsid preparations, L1:L2 ratio estimates ranged from about 9:1 to 11:1. The results imply an average of about 36 molecules of L2 per capsid.
Improvement of capsid yield.
We have previously speculated that single L2 molecules might be arranged in association with each of the 12 pentavalent (vertex) L1 capsomers (48). The apparent presence of an average of 36 molecules of L2 per capsid was inconsistent with this prediction and suggested the possibility that L2 might occupy other areas within the capsid. To address this question, we set out to perform cryo-EM difference imaging to localize L2 within the virion. Localization of L2 using cryo-EM difference imaging requires computational subtraction of 3D reconstructions of L1-only capsids from 3D reconstructions of L1+L2 capsids. To begin these analyses, we produced L1-only capsids by transient transfection of 293TT cells. The capsids were allowed to mature, purified using iodixanol (Optiprep) gradients, and then subjected to cryo-EM analysis. Although the resulting L1-only capsids were sufficiently uniform to perform 3D reconstructions (see below), the low concentration of capsids in the samples made the analysis arduous.
To increase the yield of L1+L2 capsids, we developed a system for viral propagation of the p16L1L2 plasmid. Since p16L1L2 carries the simian virus 40 (SV40) origin of replication, the plasmid replicates in 293TT cells, which express high levels of the SV40 T antigen. Because p16L1L2 is under 8 kb in size, it can be efficiently packaged into infectious L1+L2 pseudovirions. This allows production of an initial viral “seed” stock by self-packaging of p16L1L2 in transiently transfected 293TT cells. When seed stocks were used to infect fresh 293TT cells, high yields of L1+L2 particles were recovered 72 h postinfection. Typically, 107 293TT cells infected with 15 μl of crude seed stock (containing several μg of capsids) ultimately yielded several milligrams of purified capsids.
Production of capsids with increased L2 content.
Given the possible existence of 72 L2 binding sites within the capsid, we reasoned that capsid preparations containing an average of 36 copies of L2 per capsid might be submaximally occupied with L2. In p16L1L2, the L1 expression cassette is transcribed under the control of a recombinant human elongation factor 1 alpha (EF1α) promoter while L2 is under the control of the SV40 early promoter. In general, expression of proteins from the SV40 promoter is less efficient than expression from the EF1α promoter in 293TT cells (unpublished observation), perhaps due in part to the fact that the SV40 T antigen can exert negative regulatory effects on transcription from the SV40 early promoter (12). To test the hypothesis that the incorporation of L2 into p16L1L2-produced capsids was limited by L2 expression levels, we performed transient-cotransfection experiments in which p16L1L2 was transiently cotransfected with a 6.1-kb plasmid expressing HPV16 L2 under the control of the EF1α promoter (p16L2w). Cotransfection of the two plasmids resulted in increased L2 content in purified capsids (data not shown). Capsids generated by copropagation of a p16L1L2 and p16L2w viral “swarm” also displayed greater levels of L2 incorporation. This increase is presumably attributable to both plasmids being individually encapsidated to produce infectious pseudovirions that then coinfect the 293TT cells in the second-round production.
A second successful strategy for increasing the L2 content of capsids was to harvest infected 293TT cells 48 to 56 h after infection with the L1+L2 seed stock. A third successful strategy was to perform the capsid production in 293TT cells engineered to stably express L2. When all three strategies were employed simultaneously in pilot experiments, it was possible to generate capsid preparations with an average of 72 molecules of L2 per capsid (Fig. 2).
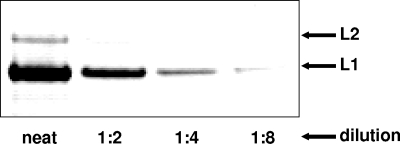
FIG. 2.
Capsids with an average of 72 molecules of L2 per capsid. A small pilot batch of capsids with greater L2 occupancy was generated as described in the text. The capsids were separated by SDS-PAGE and stained with Sypro Ruby. Visual inspection and digital analysis of L1 and L2 band intensities suggested an L1:L2 ratio of 5:1.
Cryo-EM difference imaging of L1-only and L1+L2 capsids.
A scaled-up preparation of capsids with an average of 60 molecules of L2 per capsid (Fig. 3) was subjected to cryo-EM difference comparison to L1-only capsids. The cryo-EM analysis was complicated by the presence of subsets of larger, less-ordered capsids within both the L1+L2 and L1-only capsid preparations. Although the capsid preparations were allowed to mature overnight, we speculated that the larger capsids might represent intermediate degrees of capsid maturity. An analysis of capsid maturation is the subject of other work (B. Trus, C. Buck, N. Cheng, C. Thompson, D. Lowy, J. Schiller, and A. Steven, unpublished data).
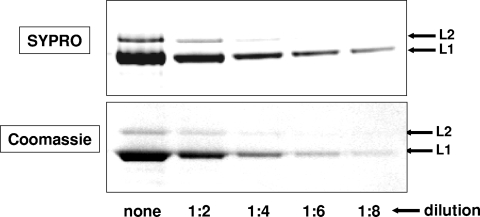
FIG. 3.
L2 content of capsids used for cryo-EM analysis. A scaled-up capsid preparation was diluted and separated by SDS-PAGE. Replicate gels were stained with Sypro-Ruby (top panel) or colloidal Coomassie (bottom panel). Visual inspection and digital analysis of L1 and L2 band intensities suggested an L1:L2 ratio of 6:1.
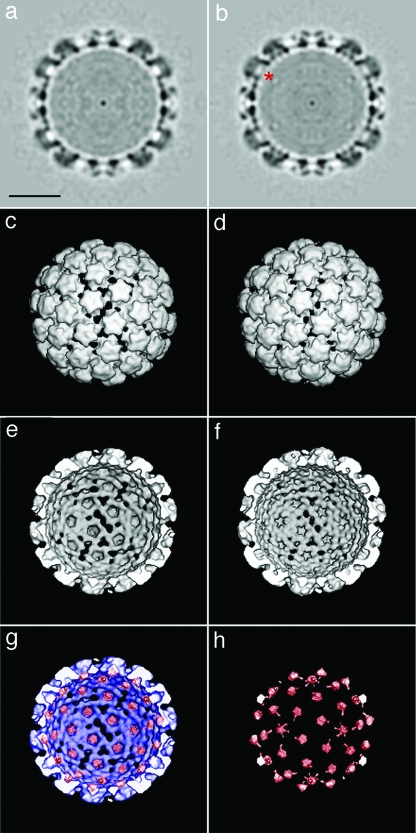
Visual comparison of raw cryo-EM micrographs of L1-only capsids to those of L1+L2 capsids showed no obvious difference due to the presence of L2 (unpublished result shown on our laboratory website [http://home.ccr.cancer.gov/Lco/L2CryoSupp.htm]). Comparison of planar sections of L1-only capsids (Fig. 4a) versus those of L1+L2 capsids (Fig. 4b) shows additional density at the base of each capsomer lumen in the L1+L2 capsids (red asterisk in panel b). Although there are no obvious differences in the exterior views of the L1-only and L1+L2 reconstructions (Fig. 4c and d, respectively), an interior view also shows additional, L2-specific density around the base of each capsomer lumen (compare Fig. 4c and d). The L2-specific difference density (in red) is shown superimposed on the L1-only reconstruction (blue) in Fig. 4g and is also shown alone in panel h.
FIG. 4.
3DR of more capsids with an average of 60 molecules of L2 per capsid. Computerized reconstructions of L1-only capsids (a, c, and e) and L1+L2 capsids (b, d, and f) are shown. The top images (a and b) are of the central section through vertex (pentavalent) capsomers. An area of additional density present in L1+L2 capsids (b) is marked with a red asterisk. Panels c and d show exterior views of each type of capsid. Panels e and f show interior/cutaway views of an L1-only or L1+L2 capsid, respectively. DNA and histone density have been computationally removed from the interior views. The bottom images show L2-specific density in red either alone (h) or superimposed on an interior view of the L1-only 3DR in blue (g). Bar = 200 Å.
The estimated mass of each L2-specific density button is roughly 12 kDa, suggesting that it represents only about a quarter of the volume predicted to be occupied by the entire 50-kDa L2 protein. Furthermore, estimates based on comparison of the average protein density inside the button to the protein density in the capsid shell revealed that the occupancy of the L2 button was about 65%. A cryo-EM difference comparison of L1-only capsids to capsids containing an average of 36 copies of L2 per capsid revealed an identical pattern of L2-specific density buttons in which each button was about 50% occupied (unpublished result displayed on our laboratory website [http://home.ccr.cancer.gov/Lco/L2CryoSupp.htm]). The results support the concept that up to 72 L2 molecules can be randomly permuted among 72 binding sites within the capsid lumen.
Bimolecular fluorescence analysis of L2 arrangement.
The observation that L2 can occupy binding sites in adjacent capsomers raises the possibility that neighboring L2 molecules might contact one another. Low-threshold difference comparisons of the more fully occupied L1+L2 capsids to L1-only capsids showed filaments of L2-specific density extending beneath the floor of the capsid (Fig. 4; also data not shown). The filaments were poorly occupied and became less prominent as the models were refined and could have been artifactual. However, the initial observation of L2-specific filaments in early difference images served to reinforce the hypothesis that neighboring L2 molecules might contact one another.
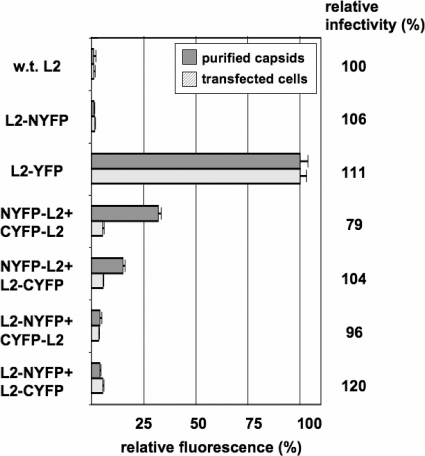
To further investigate the hypothesis that neighboring L2 molecules might contact one another within the capsid lumen, we made use of the fact that L2 can accommodate genetic fusion to other proteins, such as green fluorescent protein, without compromising infectivity (8, 15). This permitted the use of a technique known as BiFC, in which a fluorescent protein, such as enhanced YFP, is genetically split into two halves, which, when closely apposed, can associate to allow formation of the YFP fluorochrome (26, 51). N-terminal and complementary C-terminal fragments of YFP were genetically fused to the N or C terminus of L2 in four separate constructs. When pairs of L2 fusion constructs were cotransfected into 293TT cells with an HPV16 L1 expression plasmid, the resulting capsids exhibited various degrees of fluorescence, indicating BiFC of neighboring L2-YFP fusion proteins (Fig. 5). The results indicate that the N and C termini of neighboring L2 molecules can be closely apposed within the capsid.
FIG. 5.
Bimolecular fluorescence complementation. The graph depicts the fluorescence of purified capsids (gray bars) produced in 293TT cells by cotransfecting an L1 expression plasmid together with the L2 constructs shown to the left of the figure. Light gray bars show the fluorescence of live HeLa cells transfected with the L2 constructs shown (without any L1 plasmid). The terms “NYFP” and “CYFP” refer to N-terminal and C-terminal fragments of YFP, respectively. Fluorescence intensity was standardized to the fluorescence of L2 fused to full-length YFP (L2-YFP). The relative infectious titer of each capsid stock is given to the right of the graph. w.t., wild type.
BiFC was much less evident when various L2-YFP fusion constructs were transfected into cells in the absence of an L1 construct. Thus, the close apposition of the termini of neighboring L2 molecules observed in purified capsids appears to be an arrangement that is forced upon L2 during capsid assembly.
DISCUSSION
While much the fine structure of L1 is well characterized (9, 35), analysis of the arrangement of L2 within the capsid has been complicated by a variety of factors. A major complication has been the significant uncertainty about how many molecules of L2 can be incorporated into the capsid. VLP studies have the inherent ambiguity that such particles are not infectious, while pseudoviruses and authentic virions both circumvent this problem. Our data show that capsids in highly infectious pseudovirus preparations can accommodate up to 72 molecules of L2. Given the appearance of an L2-specific density button beneath each of the 72 L1 capsomers, it is likely that this represents a maximum L2 occupancy. In capsids with lower L2 occupancy, our data indicate that L2 molecules are, on average, randomly distributed among 72 binding sites.
L2 is required for production of fully infectious HPV virions and pseudovirions, and the infectivity of both types of virion can be neutralized by a similar spectrum of L2-specific antibodies (7, 19, 25, 40). Furthermore, the L2 occupancy levels we have observed in pseudoviruses are comparable to L2 occupancy levels observed in various preparations of authentic papillomavirus virions (37, 38, 42). Thus, it seems likely that naturally produced papillomavirus virions and the pseudoviruses we have studied in this report carry L2 in a similar configuration.
The implications of these findings for viral infectivity are not yet clear. We have observed that in general, pseudovirus stocks with low average numbers of L2 molecules are less infectious than those with higher average levels of L2 (unpublished result). However, the precise relationship between the number of L2 molecules in a given virion and the efficiency of its infectivity remains to be determined.
Our results suggest the possibility that L2 may be radially permuted relative to the L1 capsomer axis. This model for the organization of L2 is strikingly similar to the arrangement of the polyomavirus minor capsid proteins, VP2 and VP3, relative to the major capsid protein, VP1 (10, 32). Although the similar external appearances of the capsids of polyomaviruses (including SV40) and papillomaviruses initially led to their phylogenetic classification within a single family, Papovaviridae, the uncovering of major differences in the genetic organization of the two virus groups eventually led the International Committee on the Taxonomy of Viruses to conclude that the two families never shared a common viral ancestor (discussed in reference 16). However, this judgment has recently been called into question by the discovery of a chimeric polyomavirus/papillomavirus occurring naturally in an Australian marsupial (52). Furthermore, the view that polyomaviruses and papillomaviruses never shared a common viral ancestor doesn't preclude the possibility of shared genetic components in the early histories of the two virus families. For example, it seems likely that both virus families originated when an episomal genetic element fortuitously captured a cellular gene that facilitated intercellular transmission of the episome. One class of proteins that might theoretically serve as a protocapsid is embodied by the cellular chromatin chaperone nucleoplasmin, a pentameric protein whose core fold closely resembles that of the polyomavirus and papillomavirus major capsid proteins (9, 18, 32). In this scenario, the cellular precursor of the minor capsid proteins would presumably have been captured in a second recombination event. Thus, even if the polyomaviridae and papillomaviridae never shared a common viral ancestor, it seems possible that the similar structural interrelationship of their minor and major capsid proteins reflects a similar series of gene-capture events occurring during the early histories of the two virus families.
L2 is thought to have at least two L1 interaction domains, including a well-characterized hydrophobic interaction domain toward the carboxy end of L2 and a less well understood L1 interaction domain in the amino half of L2 (21, 36, 46). Although it is not clear which of these interaction domains (if either) might be represented in the icosahedrally ordered portion of L2 detected in our cryo-EM studies, it is interesting to note that the polyomavirus minor capsid proteins interact with the axial lumen of VP1 capsomers through a strong hydrophobic interaction (2, 10).
It is thought that a small portion of L2 is exposed on the surface of mature capsids (24, 31, 33). However, surface-exposed L2 density was not detected in our cryo-EM difference images. It is possible that surface-exposed portions of L2 might be relatively disordered, might occupy a volume too small to be detected at the resolutions we have achieved, or might have too low an occupancy to be detectable in an icosahedrally averaged structure.
It has previously been shown that L2 can facilitate the assembly of purified L1 capsomers into particles (21, 27, 30). Our BiFC studies suggest that both the N and C termini of L2 can be closely apposed within the capsid, consistent with the concept that L2 might form a subnetwork of contacts within the capsid. The relative lack of BiFC effects in transfected cells suggests a model in which the termini of L2 molecules are actively drawn together during capsid morphogenesis.
It has recently been shown that during infectious entry, a motif near the N terminus of L2 must be cleaved by the cellular protease furin (41). However, the furin cleavage motif is inaccessible in mature capsid preparations. Likewise, epitope tags fused to the N or C termini of L2 remain inaccessible to antibody binding until many hours after initial capsid interaction with cells (15). A membrane-destabilizing motif near the C terminus of L2 is also thought to become exposed only after internalization of the virion into an endosomal compartment (28). Taken together, the data strongly suggest that during cell entry, conformational changes in the capsid allow extrusion of L2 terminal domains. Given the possibility of inter-L2 contacts within the capsid, it seems possible that these extrusion events are coordinated between neighboring L2 molecules.
Analysis of the relative abundance and arrangement of L2 was facilitated by our development of replication-competent synthetic papillomaviruses. Since propagation of synthetic papillomaviruses allows for inexpensive, high-yield production of capsids, this method may be useful for future structural studies, such as X-ray analysis of crystallized papillomavirus capsids. Since reporter plasmids can be copropagated alongside the L1/L2 expression plasmid, the method may also be useful for reducing the potential expense of large-scale production of HPV pseudoviruses. With additional safety modifications, such as copropagation of separate L1 and L2 plasmids and construction of cell lines carrying less-oncogenic T-antigen mutants (14), such propagated pseudoviruses might be suitable for use as gene transfer vectors in vivo (6, 43).
Acknowledgments
This research was funded by the Intramural Research Program of the NIH, with support from the Center for Cancer Research (National Cancer Institute), the Center for Information Technology, and the National Institute of Arthritis, Musculoskeletal and Skin Diseases.
We thank Richard Roden, Patricia Day, and Ted Pierson for helpful advice. We also thank Tim Baker, David Belnap, James Conway, and J. Bernard Heymann for sharing their reconstruction software.
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116120-130. [DOI] [PubMed] [Google Scholar]
- 2.Barouch, D. H., and S. C. Harrison. 1994. Interactions among the major and minor coat proteins of polyomavirus. J. Virol. 683982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossis, I., R. B. Roden, R. Gambhira, R. Yang, M. Tagaya, P. M. Howley, and P. I. Meneses. 2005. Interaction of tSNARE syntaxin 18 with the papillomavirus minor capsid protein mediates infection. J. Virol. 796723-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buck, C. B., D. V. Pastrana, D. R. Lowy, and J. T. Schiller. 2004. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck, C. B., D. V. Pastrana, D. R. Lowy, and J. T. Schiller. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 119445-462. [DOI] [PubMed] [Google Scholar]
- 6.Buck, C. B., and C. D. Thompson. 2007. Production of papillomavirus-based gene transfer vectors. Current protocols in cell biology, unit 26.1. John Wiley & Sons, Inc., Hoboken, NJ. [DOI] [PubMed]
- 7.Buck, C. B., C. D. Thompson, Y. Y. Pang, D. R. Lowy, and J. T. Schiller. 2005. Maturation of papillomavirus capsids. J. Virol. 792839-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, C. B., C. D. Thompson, J. N. Roberts, M. Muller, D. R. Lowy, and J. T. Schiller. 2006. Carrageenan is a potent inhibitor of papillomavirus infection. PLoS Pathog. 2e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5557-567. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X. S., T. Stehle, and S. C. Harrison. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 173233-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, N., B. L. Trus, D. M. Belnap, W. W. Newcomb, J. C. Brown, and A. C. Steven. 2002. Handedness of the herpes simplex virus capsid and procapsid. J. Virol. 767855-7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, C. N., and S. D. Conzen. 2001. Polyomaviridae: the viruses and their replication, p. 2141-2174. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Conway, J. F., B. L. Trus, F. P. Booy, W. W. Newcomb, J. C. Brown, and A. C. Steven. 1993. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J. Struct. Biol. 111222-233. [DOI] [PubMed] [Google Scholar]
- 14.Cooper, M. J., M. Lippa, J. M. Payne, G. Hatzivassiliou, E. Reifenberg, B. Fayazi, J. C. Perales, L. J. Morrison, D. Templeton, R. L. Piekarz, and J. Tan. 1997. Safety-modified episomal vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 946450-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day, P. M., C. C. Baker, D. R. Lowy, and J. T. Schiller. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. USA 10114252-14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. Zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 17.Doorbar, J. 2005. The papillomavirus life cycle. J. Clin. Virol. 32(Suppl 1)S7-S15. [DOI] [PubMed] [Google Scholar]
- 18.Dutta, S., I. V. Akey, C. Dingwall, K. L. Hartman, T. Laue, R. T. Nolte, J. F. Head, and C. W. Akey. 2001. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8841-853. [DOI] [PubMed] [Google Scholar]
- 19.Embers, M. E., L. R. Budgeon, T. D. Culp, C. A. Reed, M. D. Pickel, and N. D. Christensen. 2004. Differential antibody responses to a distinct region of human papillomavirus minor capsid proteins. Vaccine 22670-680. [DOI] [PubMed] [Google Scholar]
- 20.Favre, M., F. Breitburd, O. Croissant, and G. Orth. 1975. Structural polypeptides of rabbit, bovine, and human papillomaviruses. J. Virol. 151239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnen, R. L., K. D. Erickson, X. S. Chen, and R. L. Garcea. 2003. Interactions between papillomavirus L1 and L2 capsid proteins. J. Virol. 774818-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florin, L., K. A. Becker, C. Lambert, T. Nowak, C. Sapp, D. Strand, R. E. Streeck, and M. Sapp. 2006. Identification of a dynein interacting domain in the papillomavirus minor capsid protein l2. J. Virol. 806691-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gambhira, R., P. E. Gravitt, I. Bossis, P. L. Stern, R. P. Viscidi, and R. B. Roden. 2006. Vaccination of healthy volunteers with human papillomavirus type 16 L2E7E6 fusion protein induces serum antibody that neutralizes across papillomavirus species. Cancer Res. 6611120-11124. [DOI] [PubMed] [Google Scholar]
- 24.Hagensee, M. E., N. Yaegashi, and D. A. Galloway. 1993. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J. Virol. 67315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmgren, S. C., N. A. Patterson, M. A. Ozbun, and P. F. Lambert. 2005. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J. Virol. 793938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9789-798. [DOI] [PubMed] [Google Scholar]
- 27.Ishii, Y., S. Ozaki, K. Tanaka, and T. Kanda. 2005. Human papillomavirus 16 minor capsid protein L2 helps capsomeres assemble independently of intercapsomeric disulfide bonding. Virus Genes 31321-328. [DOI] [PubMed] [Google Scholar]
- 28.Kämper, N., P. M. Day, T. Nowak, H. C. Selinka, L. Florin, J. Bolscher, L. Hilbig, J. T. Schiller, and M. Sapp. 2006. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J. Virol. 80759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 8912180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirnbauer, R., J. Taub, H. Greenstone, R. Roden, M. Durst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 676929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo, K., Y. Ishii, H. Ochi, T. Matsumoto, H. Yoshikawa, and T. Kanda. 2007. Neutralization of HPV16, 18, 31, and 58 pseudovirions with antisera induced by immunizing rabbits with synthetic peptides representing segments of the HPV16 minor capsid protein L2 surface region. Virology 358266-272. [DOI] [PubMed] [Google Scholar]
- 32.Liddington, R. C., Y. Yan, J. Moulai, R. Sahli, T. L. Benjamin, and S. C. Harrison. 1991. Structure of simian virus 40 at 3.8-Å resolution. Nature 354278-284. [DOI] [PubMed] [Google Scholar]
- 33.Liu, W. J., L. Gissmann, X. Y. Sun, A. Kanjanahaluethai, M. Muller, J. Doorbar, and J. Zhou. 1997. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology 227474-483. [DOI] [PubMed] [Google Scholar]
- 34.McLaughlin-Drubin, M. E., N. D. Christensen, and C. Meyers. 2004. Propagation, infection, and neutralization of authentic HPV16 virus. Virology 322213-219. [DOI] [PubMed] [Google Scholar]
- 35.Modis, Y., B. L. Trus, and S. C. Harrison. 2002. Atomic model of the papillomavirus capsid. EMBO J. 214754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okun, M. M., P. M. Day, H. L. Greenstone, F. P. Booy, D. R. Lowy, J. T. Schiller, and R. B. Roden. 2001. L1 interaction domains of papillomavirus l2 necessary for viral genome encapsidation. J. Virol. 754332-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orth, G., and M. Favre. 1985. Human papillomaviruses. Biochemical and biologic properties. Clin. Dermatol. 327-42. [DOI] [PubMed] [Google Scholar]
- 38.Orth, G., M. Favre, and O. Croissant. 1977. Characterization of a new type of human papillomavirus that causes skin warts. J. Virol. 24108-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer, K. E., A. Benko, S. A. Doucette, T. I. Cameron, T. Foster, K. M. Hanley, A. A. McCormick, M. McCulloch, G. P. Pogue, M. L. Smith, and N. D. Christensen. 2006. Protection of rabbits against cutaneous papillomavirus infection using recombinant tobacco mosaic virus containing L2 capsid epitopes. Vaccine 245516-5525. [DOI] [PubMed] [Google Scholar]
- 40.Pastrana, D. V., R. Gambhira, C. B. Buck, Y. Y. Pang, C. D. Thompson, T. D. Culp, N. D. Christensen, D. R. Lowy, J. T. Schiller, and R. B. Roden. 2005. Cross-neutralization of cutaneous and mucosal Papillomavirus types with anti-sera to the amino terminus of L2. Virology 337365-372. [DOI] [PubMed] [Google Scholar]
- 41.Richards, R. M., D. R. Lowy, J. T. Schiller, and P. M. Day. 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc. Natl. Acad. Sci. USA 1031522-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rippe, R. A., and W. J. Meinke. 1989. Identification and characterization of the BPV-2 L2 protein. Virology 171298-301. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, J. N., C. B. Buck, C. D. Thompson, R. Kines, M. Bernardo, P. L. Choyke, D. R. Lowy, and J. T. Schiller. 2007. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat. Med. 13857-861. [DOI] [PubMed] [Google Scholar]
- 44.Roden, R. B., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 705875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roden, R. B., W. H. T. Yutzy, R. Fallon, S. Inglis, D. R. Lowy, and J. T. Schiller. 2000. Minor capsid protein of human genital papillomaviruses contains subdominant, cross-neutralizing epitopes. Virology 270254-257. [DOI] [PubMed] [Google Scholar]
- 46.Sapp, M., C. Volpers, M. Muller, and R. E. Streeck. 1995. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J. Gen. Virol. 762407-2412. [DOI] [PubMed] [Google Scholar]
- 47.Schiffman, M., and S. K. Kjaer. 2003. Chapter 2: natural history of anogenital human papillomavirus infection and neoplasia. J. Natl. Cancer Inst. Monogr. 200314-19. [DOI] [PubMed] [Google Scholar]
- 48.Trus, B. L., R. B. Roden, H. L. Greenstone, M. Vrhel, J. T. Schiller, and F. P. Booy. 1997. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 Å resolution. Nat. Struct. Biol. 4413-420. [DOI] [PubMed] [Google Scholar]
- 49.Volpers, C., P. Schirmacher, R. E. Streeck, and M. Sapp. 1994. Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology 200504-512. [DOI] [PubMed] [Google Scholar]
- 50.Wang, M., L. L. Wang, L. F. Chen, Y. H. Han, Y. H. Zou, J. Y. Si, and G. X. Song. 2003. Expression of human papillomavirus type 6 L1 and L2 isolated in China and self assembly of virus-like particles by the products. Acta Biochim. Biophys. Sinica 3527-34. [PubMed] [Google Scholar]
- 51.Weis, D. S., and T. B. Robert. October 2003. Two green fluorescent protein fragments and their use in a method for detecting protein-protein interactions. International patent WO03089464.
- 52.Woolford, L., A. Rector, M. Van Ranst, A. Ducki, M. D. Bennett, P. K. Nicholls, K. S. Warren, R. A. Swan, G. E. Wilcox, and A. J. O'Hara. 2007. A novel virus detected in papillomas and carcinomas of the endangered western barred bandicoot (Perameles bougainville) exhibits genomic features of both the Papillomaviridae and Polyomaviridae. J. Virol. 8113280-13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, R., W. H. T. Yutzy, R. P. Viscidi, and R. B. Roden. 2003. Interaction of L2 with beta-actin directs intracellular transport of papillomavirus and infection. J. Biol. Chem. 27812546-12553. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, J., D. J. Stenzel, X. Y. Sun, and I. H. Frazer. 1993. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J. Gen. Virol. 74763-768. [DOI] [PubMed] [Google Scholar]