Abstract
In plants, small RNA-guided processes referred to as RNA silencing control gene expression and serve as an efficient antiviral mechanism. Plant viruses are inducers and targets of RNA silencing as infection involves the production of functional virus-derived small interfering RNAs (siRNAs). Here we investigate the structural and genetic components influencing the formation of Tobacco rattle virus (TRV)-derived siRNAs. TRV siRNAs are mostly 21 nucleotides in length and derive from positive and negative viral RNA strands, although TRV siRNAs of positive polarity are significantly more abundant. This asymmetry appears not to correlate with the presence of highly structured regions of single-stranded viral RNA. The Dicer-like enzyme DCL4, DCL3, or DCL2 targets, alone or in combination, viral templates to promote synthesis of siRNAs of both polarities from all regions of the viral genome. The heterogeneous distribution profile of TRV siRNAs reveals differential contributions throughout the TRV genome to siRNA formation. Indirect evidence suggests that DCL2 is responsible for production of a subset of siRNAs derived from the 3′ end region of TRV. TRV siRNA biogenesis and antiviral silencing are strongly dependent on the combined activity of the host-encoded RNA-dependent RNA polymerases RDR1, RDR2, and RDR6, thus providing evidence that perfectly complementary double-stranded RNA serves as a substrate for siRNA production. We conclude that the overall composition of viral siRNAs in TRV-infected plants reflects the combined action of several interconnected pathways involving different DCL and RDR activities.
RNA silencing includes a set of small RNA-guided pathways that regulate gene expression in eukaryotes and control processes such as development, genome stability, stress-induced responses, and defense against molecular parasites (4, 6, 10). An essential component of RNA silencing is RNase III Dicer, an enzyme that cleaves RNA with double-stranded (ds) features into 21- to 24-nucleotide (nt) duplex small interfering RNA (siRNA) or microRNA (miRNA). The Arabidopsis thaliana genome encodes four Dicer-like (DCL) enzymes and six RNA-dependent RNA polymerases (RDR), other components of the silencing machinery in plants (26, 39, 46, 55, 58). DCL1 produces 21-nt miRNAs derived from RNA polymerase II transcripts with imperfect self-complementary fold-back structures (22). Endogenous siRNAs are likely generated from dsRNA substrates because of the requirement for a specific RDR for the production of each siRNA class (32, 33). The 24-nt cis-acting siRNA class derives mostly from retroelements, transposons, and DNA repeats and depends on DCL3 and RDR2 that use RNA polymerase IV transcripts as a template for dsRNA production (33). Synthesis of 21-nt trans-acting siRNAs is DCL4 dependent and occurs through an initial DCL1-dependent, miRNA-directed cleavage of trans-acting siRNA precursor transcripts, which are subsequently processed into long, perfect dsRNA by RDR6 (5, 32).
In plants, RNA interference is guided by two distinct classes of siRNAs, named primary siRNAs, which result from DCL-mediated cleavage of the initial trigger, and secondary siRNAs, whose biogenesis requires an RDR enzyme (19, 55). DCL4, DCL2, and DCL3 process exogenous hairpin RNAs into siRNAs of 21, 22, and 24 nt, respectively, although cleavage of target transcripts is mainly caused by DCL4 siRNAs (19, 23). In Arabidopsis, RDR6 and RDR2 are required for RNA silencing of transgenes, the spread of long-range cell-to-cell silencing, and the reception of the long-distance mRNA silencing signal (11, 12, 29, 42).
Among their functions, RDR and DCL enzymes participate in viral-induced silencing and antiviral defense (17, 20). RNA and DNA viruses produce virus-associated siRNAs upon infection in plants (17, 54) though the structural and genetic requirements for their biogenesis are only partially understood. Infection with several RNA viruses results in the accumulation of both sense and antisense viral siRNAs, consistent with DCL-mediated processing of perfectly complementary dsRNA (7, 9, 15, 25, 31, 52). The predominant plus-strand polarity and genomic distribution of siRNAs produced by Cymbidium ringspot virus and Cucumber mosaic virus (CMV) satellite RNAs suggest that viral siRNA may also be produced by direct DCL cleavage of imperfect duplexes originating from highly base-paired structures from the positive-strand viral RNA (18, 41, 51). A recent study showed that the structured, 35S RNA leader region of Cauliflower mosaic virus contributes significantly to virus-derived siRNA production (40).
Analysis of Arabidopsis loss-of-function dcl mutants provides genetic evidence that RNA viruses are mainly targeted by DCL4, DCL2, and DCL3 to generate virus-derived siRNAs of 21, 22, and 24 nt, respectively (7, 9, 13, 18, 58). DCL4- and DCL2-dependent siRNAs are capable of recruiting an antiviral effector complex to promote degradation of viral genomes (9, 13), whereas the antiviral role of DCL3 might be related to the perception of the non-cell-autonomous silencing signal (11, 15). Infection with several single-stranded DNA geminiviruses and dsDNA caulimoviruses also results in accumulation of different siRNA size classes, which reflects the activity of distinct DCL proteins (2, 7).
In plants, RDRs are postulated to strengthen primary silencing-based antiviral responses, probably by using viral templates to produce dsRNA substrates for siRNA synthesis (42, 60). Nicotiana benthamiana and Arabidopsis plants with compromised RDR6 activity display enhanced susceptibility to infection by several RNA viruses (42, 47, 49). It was proposed that RDR6 responds to incoming long-distance silencing signals to trigger a rapid response to viral infection that involves production of secondary siRNAs (29, 49, 52). Likewise, inhibition of RDR1 function correlates with increased susceptibility to several RNA viruses (57, 59, 60). Although RDR1 and RDR6 seem to be dispensable for the production of virus-derived siRNAs from several RNA viruses (7, 13, 60), Ding and colleagues recently demonstrated that without interference of the virus-encoded 2b silencing suppressor protein, accumulation of virus-specific siRNAs produced in response to infection with CMV is RDR1 dependent (15). For most RNA viruses, however, the role of RDR enzymes in the biogenesis of viral siRNAs remains unclear.
Tobacco rattle virus (TRV) is a bipartite, single-stranded, positive-sense RNA plant virus and the type member of the genus Tobravirus. The genomic RNA1 encodes four open reading frames (38). The 134- and 194-kDa replicase proteins are expressed directly from the genomic RNA, whereas the 29-kDa cell-to-cell movement protein and the 16-kDa silencing suppressor protein are translated from subgenomic RNAs. The genomic RNA2 usually contains the coat protein, a nematode-transmission factor 2b, and factor 2c, a protein of unknown function. A characteristic of TRV is that RNA1 is capable of replicating and spreading in host plants in the absence of RNA2 (27). RNA1 and RNA2 are herein referred to as TRV1 and TRV2, respectively.
In this paper, we carried out a molecular characterization of TRV-derived siRNAs produced during viral infection in Arabidopsis and N. benthamiana to understand the structural and genetic factors necessary for viral siRNA formation. This study reveals that several pathways for siRNA biogenesis function coordinately to produce viral siRNAs. We analyzed target specificities throughout the virus genome and the contribution of each DCL to the synthesis of siRNA of sense and antisense polarities and present genetic evidence that the biogenesis of TRV siRNAs was largely affected by redundant activities of host-encoded RDR enzymes.
MATERIALS AND METHODS
Plant materials and TRV inoculation.
Arabidopsis mutants lines for dcl2-1, dcl3-1, dcl4-2, dcl2 dcl3, dcl2 dcl4, dcl3 dcl4, dcl2 dcl3 dcl4, rdr1-1, rdr2-1, rdr6-15 were described previously (3, 13, 44, 45, 48, 53, 56, 58). Mutants rdr1 rdr6, rdr2 rdr6, and rdr1 rdr2 rdr6 were kindly provided by Jim Carrington (Center for Gene Research and Biocomputing, Oregon State University). All mutants were in the ecotype Columbia background.
The infectious clone of TRV-phytoene desaturase (PDS) was described before (34). N. benthamiana plants were inoculated by infiltration of Agrobacterium cultures containing pTRV1 and pTRV2-PDS mixed in a 1:1 ratio as described previously (16). Arabidopsis plants were sap inoculated on rosette leaves using inoculum prepared from N. benthamiana systemically infected leaves. In Arabidopsis, the genomic and subgenomic TRV RNAs as well as TRV siRNAs accumulated consistently in upper, noninoculated inflorescence tissue, as opposed to rosette leaves, where they showed a rather erratic accumulation pattern (data not shown).
RNA isolation and blot analysis.
Total RNA was extracted with Trizol reagent (Invitrogen) from pools of 10 to 20 plants using upper, noninoculated TRV-infected inflorescence tissue of Arabidopsis or systemically infected N. benthamiana leaves. Low- and high-molecular-weight RNAs were further isolated by anion-exchange chromatography using an RNA/DNA Midi Kit (Qiagen) according to the manufacturer's instructions.
Blot hybridization was performed as described previously (36) with some modifications. For analysis of viral RNAs, high-molecular-weight RNA (20 μg) was separated on 1.5% formaldehyde-HEPES agarose gels and blotted onto positively charged, nylon membranes (Roche). For analysis of virus-derived siRNAs, low-molecular-weight RNA (30 μg) was separated in denaturing polyacrylamide gels and transferred to nylon membranes using a Trans-Blot semidry transfer cell (Bio-Rad). Blots were prehybridized and hybridized at 42°C using Perfect-Hyb buffer (Sigma). Radiolabeled DNA probes from cloned sequences were made by random priming reactions in the presence of [α-32P]dCTP. Oligonucleotides complementary to positive or negative viral RNA strands were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (PNK; New England Biolabs). Unincorporated nucleotides were removed using Micro Bio-Spin Chromatography columns (Bio-Rad). Ethidium bromide staining of gels before blot transfer was used to visualize rRNA and ensure even loading of RNA samples.
For purification and labeling of small RNAs, ∼250 μg of total RNA was subjected to denaturing electrophoresis as described above. A gel slice containing RNAs of ∼15 to 40 nt (based on RNA oligonucleotide size standards) was excised and eluted overnight in 0.3 M NaCl at 4°C. The small RNAs were dephosphorylated using calf intestinal phosphatase (New England Biolabs) and subsequently labeled with [γ-32P]ATP by using PNK. The labeled small RNAs were used for hybridization of DNA blots containing PCR-amplified fragments of ∼500 bp corresponding to the TRV1 genomic sequence, as described above. DNA properties of the PCR fragments and primer sequences used for amplification are available in Table S2 in the supplemental material. Relative siRNA accumulation was measured by densitometry of RNA blots exposed to autoradiographic film.
Cloning and sequencing of TRV siRNAs.
Small RNA was gel purified from total RNA preparations and cloned as described previously (35) with minor modifications. Briefly, purified small RNA was dephosphorylated by calf intestinal phosphatase and ligated to a 5′ phosphorylated 3′ RNA oligonucleotide adaptor using T4 RNA ligase (New England Biolabs). The ligation product was gel purified, 5′ phosphorylated using PNK, and ligated to a 5′ RNA oligonucleotide adaptor containing 5′ and 3′ hydroxyl groups. The new ligation product was reverse transcribed and PCR amplified. The PCR product was then digested with BanI and concatemerized using T4 DNA ligase (Promega). Concatemers larger than ∼200 bp were eluted from low-melting-point agarose gels, and the ends were filled by incubation with Taq DNA polymerase (Perkin Elmer). The DNA product was ligated into a pGEM-T Easy vector (Promega) using T4 DNA ligase. Individual clones were randomly selected and sequenced. TRV siRNAs were identified by a BLAST search against the TRV genome (accession numbers NC_003805 and AF406991; isolate PpK20), and only sequences with perfect matches were considered.
Statistical procedures.
Correlation between variables was tested using Pearson coefficients. Differences in the accumulation of TRV siRNAs between DNA fragments and between genetic backgrounds were analyzed using one-way analysis of variance (ANOVA). Significant differences between classes within each factor were analyzed using a least significant difference (LSD) test. The proportion of total variance explained by each factor was calculated as percentage by variance component (VC) analyses in the corresponding model. Broad-sense heritability (h2b) was estimated as the percentage of the total variance accounted by genetic (background) differences (h2b = σ2G/σ2P, where σ2G is the genetic VC of σ2P total phenotypic variance). σ2P and σ2G were derived by VC analysis using separated univariate analyses for each fragment. All statistical analyses were performed using the statistical software Statgraphics Plus, version 5.1 (Statistical Graphics Corp.).
RESULTS
The formation of virus-derived siRNAs in plants likely involves the production of virus-derived dsRNA. Non-mutually excluding pathways can be envisioned whereby dsRNA can be formed upon infection, including replication of RNA viruses, which is hypothesized to produce dsRNA of positive and negative strands of genomic RNA; partial annealing of complementary regions between viral RNAs of positive and negative polarities; RDR-dependent synthesis of dsRNA from virus templates; and local base pairing of imperfectly complementary regions in the positive-strand viral RNA. We used a recombinant TRV carrying a fragment of the N. benthamiana PDS gene to gain insight into the biosynthetic pathways of virus-derived siRNAs in N. benthamiana and Arabidopsis plants and to assess the relative contribution of each of the above mechanisms to TRV siRNA formation (Fig. 1A). TRV infection was associated with accumulation of functional virus-derived siRNAs in both plant species, which targeted cRNA sequences for degradation, as evidenced by decreased viral titer over time and, in the case of N. benthamiana, extensive photobleaching due to PDS silencing (Fig. 1B and C) (14).
FIG. 1.
Production and molecular characterization of virus-derived siRNAs in TRV-infected plants. (A) Schematic representation of the TRV-PDS recombinant system. (B) Bleached phenotype caused by PDS silencing in TRV-infected N. benthamiana plants. (C) RNA gel blot analysis showing time course accumulation of genomic (g) and subgenomic (sg) TRV RNAs (top) and TRV siRNAs (bottom) after infection with TRV in N. benthamiana. Blots were probed with a radiolabeled DNA fragment corresponding to the 3′ UTR of TRV (top) and the 134-kDa protein-coding region of TRV1 (bottom). The positions of RNA size markers are shown on the left side of each panel. Ethidium bromide-stained RNA (prior to transfer) is shown as a loading control. dpi, days postinoculation. (D) Histogram of sizes of cloned TRV siRNAs from infected N. benthamiana and Arabidopsis. (E) Distribution of cloned sense (S) and antisense (A) TRV siRNA sequences along the TRV1 and TRV2 genomes from infected N. benthamiana and Arabidopsis plants.
Composition of the TRV siRNA population in TRV-infected plants.
To investigate the size and origin of TRV siRNAs, we analyzed a pool of siRNAs with perfect homology to the TRV genome cloned from N. benthamiana and Arabidopsis plants systemically infected with TRV (see Table S1 in the supplemental material). The siRNAs ranged in length between 17 and 24 nt, with the most common lengths being 21 and 22 nt (Fig. 1D). Interestingly, TRV siRNAs of 21 nt (42.5%) were cloned to the same extent as 22 nt (44.7%) in TRV-infected N. benthamiana leaves, whereas 21-nt siRNA species were overrepresented (65.2%) in TRV-infected Arabidopsis (Fig. 1D). Viral siRNAs mapped throughout the full-length TRV genome in both sense and antisense orientations (Fig. 1E). In N. benthamiana, 45.6% of the total TRV1 siRNAs cloned were of positive polarity, while 54.4% were of negative polarity, suggesting that viral RNAs of both polarities contributed to a similar degree to siRNA formation (Fig. 1E). In contrast, 73% of the cloned siRNAs derived from TRV2 were of positive polarity, which indicated an asymmetrical distribution. Several clusters of partially overlapping sense siRNA sequences were particularly evident throughout TRV2 (Fig. 1E; see also Table S1 in the supplemental material). Among the TRV2 siRNA sequences, three sense/antisense pairs were fully complementary in base pairing and contained either blunt ends or 3′ overhangs ranging between 1 and 3 nt in length (see Table S1 in the supplemental material). This suggests that these siRNA sequences might arise as complementary strands from a single DCL processing event. Asymmetry in strand polarity was also found in Arabidopsis, where 64% of the cloned TRV1 siRNAs were derived from the positive strand, and 36% were derived from the negative viral strand. Only nine TRV2 siRNA sequences of positive (44.4%) and negative (55.6%) polarities were identified in the library from TRV-infected Arabidopsis.
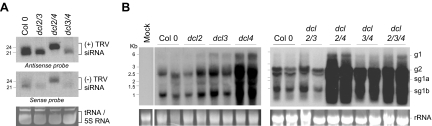
To corroborate these preliminary data and avoid misinterpretation due to a potential bias in the cloning method (43, 50), low-molecular-weight RNA from TRV-infected Arabidopsis plants was sequentially hybridized with a mixture of TRV-specific, sense or antisense ∼21-nt DNA probes encompassing the entire TRV1 genome (see Table S2 in the supplemental material). RNA blots showed that viral siRNAs of both polarities migrated as a heterogeneous population with two predominant bands of 21 and 24 nt (Fig. 2A). Densitometric analysis of the hybridization signals revealed that TRV infection in Arabidopsis triggered nearly 50% more 21-nt siRNAs of sense and antisense polarities relative to 24-nt siRNAs (Fig. 2A). Identical results were obtained from several independent experiments using TRV1- and TRV2-specific probes (data not shown). Similarly, there were more siRNAs of 21 to 22 nt than siRNAs of 24 nt in TRV-infected N. benthamiana (Fig. 1C). Using oligonucleotide probes normalized for total radioactivity, RNA blot assays revealed that plus-strand siRNAs that hybridized to antisense probes represented nearly 80% of the total TRV siRNA population in TRV-infected Arabidopsis tissue (Fig. 2A). Furthermore, 32P-labeled small RNAs from TRV-infected Arabidopsis plants hybridized more strongly (three- to fourfold) to negative-strand TRV1 and TRV2 transcripts than to the corresponding positive-strand transcripts (data not shown). Therefore, in general, the hybridization-based analyses of sense and antisense siRNAs showed good consistency with the results obtained from the sequence data and demonstrated that TRV siRNAs accumulating in the infected tissue were mostly 21 nt in length and derived predominantly from viral positive-strand RNA.
FIG. 2.
Accumulation of TRV siRNAs and viral TRV RNAs in virus-infected plants. (A) RNA gel blot analysis of sense (+) and antisense (−) TRV siRNAs in wild-type Arabidopsis and dcl double mutants. The RNA blot was sequentially hybridized with a mixture of TRV-specific sense or antisense 32P-labeled oligonucleotide probes as indicated. (B) Accumulation of genomic (g) and subgenomic (sg) TRV RNAs from inflorescence tissue of wild-type and single, double, and triple dcl Arabidopsis mutants infected with TRV. Duplicate samples were analyzed at 16 days postinoculation. Radiolabeled probes correspond to the 3′ UTR of TRV. The positions of RNA size markers are shown on the left side of each panel. Ethidium bromide-stained RNA (prior to transfer) is shown as a loading control. dcl2/3, dcl2 dcl3; dcl2/4, dcl2 dcl4; dcl3/4, dcl3 dcl4; dcl2/3/4, dcl2 dcl3 dcl4.
This asymmetry in strand polarity apparently fits well with the hypothesis that a large fraction of TRV siRNAs might originate from regions within the plus-strand viral RNA with a high probability of forming secondary structures, as was previously shown for other RNA viruses (18, 31, 41). We used Mfold, version 3.2 (61), to generate secondary structure maps from diverse regions of the TRV genome containing siRNA sequences. The Mfold prediction indicated that siRNA clusters in TRV1 and TRV2 corresponded to regions with relatively high local base pairing as well as regions predicted to lack extensive base pairing (see Fig. S1 in the supplemental material). Therefore, there was no obvious correlation between proximity to base-paired sequences, including stem-loops or T-shaped hairpins, and siRNA production. The above observation was well illustrated by the 3′ untranslated region (UTR) of TRV2, which contains three clusters of two to seven cloned, overlapping siRNA sequences with a predominant plus polarity (see Table S1 in the supplemental material). Prediction of secondary structures within the 3′ UTR of TRV2 revealed the occurrence of three stable stem structures, named stem-loops A (ΔG of −24.50 Kcal/mol), B (ΔG of −25.20 Kcal/mol), and C (ΔG of −30.20 Kcal/mol) (see Fig. S2 in the supplemental material). The majority of the cloned siRNAs mapping to the 3′ UTR clustered in a region that apparently lacks local secondary structure, whereas the other two siRNA clusters mapped to stem-loops C and A, implying that the presence of siRNA did not necessarily correlate with local secondary structures (see Fig. S2 in the supplemental material).
DCL requirements for the production of plus- and minus-strand TRV siRNAs in Arabidopsis.
Recent studies using TRV and other plant RNA viruses demonstrated that DCL4 is the main producer of 21-nt siRNAs of potent antiviral activity and that DCL2 and DCL3 generate siRNAs of 22 and 24 nt, respectively, whereas DCL1 is not directly required for viral siRNA biogenesis (7, 9, 13). To gain additional insight into the production of viral siRNAs, we investigated the contribution of each DCL enzyme to the synthesis of sense and antisense TRV siRNAs. First, we studied the effects of the various DCLs on the outcome of viral infection using combinations of dcl mutants. Northern blot analysis using a probe specific for both TRV1 and TRV2 showed that accumulation of full-length genomic TRV1 and TRV2 and the two subgenomic RNAs of TRV1 did not differ substantially between wild-type plants and single dcl2 and dcl3 and double dcl2 dcl3 mutants (Fig. 2B). However, single, double, and triple mutants involving dysfunctional DCL4 activity (dcl2 dcl4, dcl3 dcl4, and dcl2 dcl3 dcl4) exhibited higher TRV RNA titers than wild-type plants (Fig. 2B).
We next carried out northern hybridizations of low-molecular-weight RNA extracted from TRV-infected Arabidopsis carrying dcl knockout double mutations. A mixture of sense or antisense TRV-specific DNA oligonucleotides normalized for total radioactivity was used as a probe. RNA blots showed that in dcl2 dcl3 mutants infected with TRV, DCL4 produced TRV siRNA of 21 nt of sense polarity that accumulated to the same high levels as in wild-type plants (Fig. 2A). A band of 21-nt minus-strand siRNA that hybridized with sense probes was also detected in dcl2 dcl3 although this was decreased by nearly 70% in the mutant compared to wild-type Arabidopsis in two independent experiments (Fig. 2A). A possible explanation for this observation is that production of antisense, but not sense, DCL4-dependent 21-nt siRNAs might be assisted, although indirectly, by DCL2 or DCL3. In dcl2 dcl4 mutants, increased levels of TRV genomic and subgenomic RNAs and increased developmental defects (data not shown) correlated with the absence of DCL4-dependent, 21-nt TRV siRNAs (Fig. 2A and B), confirming that this siRNA species targets viral RNA for silencing-mediated cleavage (9, 13). DCL3 generated increasing amounts of 24-nt species of both sense and antisense polarity in dcl2 dcl4 compared to wild-type plants infected with TRV, probably due to the high levels of virus genomic and subgenomic RNA in the mutant tissue (Fig. 2A and B). DCL4 was the major producer of siRNA of both positive and negative polarities in wild-type plants, as indicated by the finding that DCL4-dependent, 21-nt siRNAs were more abundant than DCL3-dependent, 24-nt siRNAs (Fig. 2A). DCL2-dependent siRNAs of both polarities accumulated to equivalent levels in dcl3 dcl4 and wild-type plants (Fig. 2A). Despite the increasing levels of viral RNAs in the dcl3 dcl4 mutant, these data indicated that the global contribution of DCL2 to siRNA formation and antiviral defense is lower than that of DCL4 and DCL3. These results confirmed DCL specificities to produce different sizes of viral siRNAs and showed that DCL4, DCL3, or DCL2 was each alone capable of targeting viral templates to promote synthesis of siRNAs of both polarities.
In agreement with our previous observations in wild-type plants, sense and antisense siRNA species produced by each DCL were not equally represented in the pool of TRV siRNAs in the infected tissue. Data from two independent experiments revealed that there was an approximately fivefold increase in the amount of positive-strand TRV siRNAs (antisense probes) with respect to siRNAs derived from negative strands (sense probes), irrespective of the size class and the DCL involved. These results were confirmed by northern hybridization of 5′ 32P-labeled siRNAs isolated from TRV-infected dcl tissue to viral transcripts of both positive and negative polarities. Labeled TRV siRNAs hybridized consistently to negative transcripts derived from TRV1 and TRV2 with higher intensities (data not shown).
Spatial distribution profile of TRV siRNAs.
To gather further knowledge on the distribution profile of TRV siRNAs in infected Arabidopsis plants, a reverse RNA blot assay was used. The TRV1 genome was amplified by PCR into 14 DNA fragments, ∼500 bp each, using sequence-specific primers (Fig. 3; see also Table S2 in the supplemental material). Normalized amounts of each DNA fragment were separated by electrophoresis, immobilized onto a nylon membrane, and hybridized to purified 32P-labeled small RNAs. The results in Fig. 3 show the spatial distribution profile of TRV siRNAs in several independent experiments. siRNAs isolated from TRV-infected Arabidopsis hybridized to each of the DNA fragments although to different intensities, suggesting a heterogeneous contribution of the TRV genome to siRNA formation (Fig. 3). One-way ANOVA showed significant differences in the production of siRNAs between fragments (F13,55 = 3.29; P = 0.0017) in Arabidopsis. An LSD test revealed that genomic regions corresponding to fragments 1 to 12 produced comparable levels of TRV siRNAs, whereas fragments 13 and 14 accumulated significantly higher levels of viral siRNAs. In this analysis, fragments 7, 10, and 11 exhibited a tendency to accumulate low levels of siRNA, and fragment 12 had a tendency to overaccumulate siRNAs. This observation suggests that some special features might be associated with these genomic areas that ultimately prevent or stimulate formation of siRNAs, perhaps through differential substrate affinities for specific DCL/RDR activities. No hybridization signals were detected using labeled small RNA from mock-inoculated plants (Fig. 3).
FIG. 3.
Spatial distribution profile of TRV1-derived siRNAs. A schematic of the TRV1 genome and the location of PCR-amplified DNA fragments are shown. DNA blots of PCR fragments were probed with 5′ radiolabeled siRNAs purified from TRV-infected Arabidopsis inflorescence tissue. A DNA fragment corresponding to the β-glucoronidase gene (GUS) was used as a negative hybridization control. The ethidium bromide-stained agarose gel is shown as a loading control. Blots were exposed to autoradiographic film, and the hybridization signals were measured by densitometry using Quantity One-4.2.3 (Bio-Rad). Histograms below the DNA gel blot represent the percentages of the intensities of the hybridization signals corresponding to each PCR fragment relative to the total intensity of all the fragments. Intensity values are the averages of four independent experiments. Error bars represent standard deviations. Numbers below the histograms indicate the PCR-amplified DNA fragments.
To determine whether the higher/lower abundance of siRNA was associated with genomic regions with extensive secondary structure, we used Mfold to calculate the minimum folding free energy corresponding to each of the above 14 TRV1 fragments as well as that of 14 RNA sequences of similar size derived from randomly selected Arabidopsis mRNAs (see Table S3 in the supplemental material). We found no statistically significant difference in the minimum free energy of folding between TRV-derived sequences and the Arabidopsis transcriptome (t = −1.38; P = 0.18). These data suggest that none of the TRV fragments contained structural features involving extensive base pairing that differed substantially from those found in host mRNAs. Indeed, the Mfold calculations revealed the existence of equivalent base-paired structures in the form of local fold-back or T-shaped structures in both TRV and Arabidopsis transcripts (data not shown). Moreover, there was no significant correlation between the relative abundance of siRNAs from each TRV fragment and their corresponding minimum folding energy (r = 0.39; P = 0.16).
DCL target specificities throughout the TRV genome.
We next investigated whether the various DCL pathways in Arabidopsis exhibited their processing activities along the entire viral genome or, by contrast, whether each DCL was associated with specific genomic regions. Single, double, and triple dcl mutants were infected with TRV, and the corresponding small RNA population was isolated from inflorescence tissue and 32P labeled. Northern blot analysis showed that TRV siRNAs from single as well as double dcl mutants hybridized with each of the ∼500-bp DNA fragments (Fig. 4; see also Fig. S3 in the supplemental material). Therefore, the sole or combined activity of DCL2, DCL3, and DCL4 was sufficient to produce siRNAs corresponding to all regions of the TRV1 genome, thus reinforcing the silencing-mediated antiviral effect exerted against the viral RNA. No detectable levels of siRNAs were found to hybridize with any of the TRV fragments in the dcl2 dcl3 dcl4 mutant, confirming that DCL1 is not directly involved in the synthesis of significant amounts of virus-derived siRNAs (see Fig. S3 in the supplemental material).
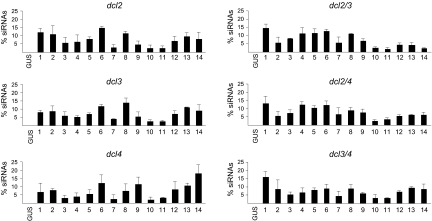
FIG. 4.
Spatial distribution profile of TRV1-derived siRNAs in Arabidopsis dcl mutants. Histograms represent the percentage of siRNA accumulation in each PCR fragment relative to the total accumulation in all the fragments, as determined by DNA blotting of PCR-amplified DNA fragments hybridized with 5′ radiolabeled siRNAs purified from single, double, and triple dcl Arabidopsis mutants infected with TRV. Hybridization signals were measured by densitometry from three independent experiments. Mean values and standard deviations are shown. Figure content is as described in the legend of Fig. 3. dcl2/3, dcl2 dcl3; dcl2/4, dcl2 dcl4; dcl3/4, dcl3 dcl4; GUS, β-glucoronidase gene.
We then examined the spatial distribution pattern of TRV siRNAs associated with each DCL enzyme. Using three independent biological replicates per mutant, we found that most of the mutants tested displayed viral siRNA distribution profiles that essentially resembled those produced in wild-type plants infected with TRV (Fig. 4). One-way ANOVA indicated that in all single and double dcl mutants tested, there were statistically significant differences in the accumulation of TRV siRNAs between fragments (F13,41 > 2.87; P < 0.0094). An LSD test showed that fragments 7, 10, and 11 consistently formed a homogeneous group that differed significantly from other fragments by producing small amounts of TRV siRNAs. Besides, there was a manifest tendency, which was highly representative in certain mutants, for siRNAs to overaccumulate in fragments 1, 4, 6, and 8. In single dcl2, dcl3, and dcl4 mutants and double dcl3 dcl4 mutants, siRNA levels from fragments 12 to 14 were significantly different from those from fragments 10 and 11 and were classified into different homogeneous groups. In contrast, in double dcl2 dcl3 and dcl2 dcl4 mutants, fragments 12 to 14 produced amounts of siRNAs that were as small as the amounts from fragments 10 and 11. In addition, differences between fragments explained a significant fraction of the variance in siRNA production (VC of 0.36 to 0.79, depending on genetic background).
We next used the same statistical procedures to analyze our data using the genetic background as a factor to determine if the siRNA accumulation profile differed between wild-type and the various dcl knockout mutants tested. Consistent with our previous analysis, one-way ANOVA showed that in fragments 1 to 12, no significant differences were found in all mutants tested (F6,21 < 2.78; P > 0.0509). However, there was a significant difference in the amount of viral siRNA that accumulated in fragments 13 and 14 between genetic backgrounds (F6,21 > 3.73; P < 0.017). An LSD test showed that wild-type Arabidopsis accumulated high levels of siRNAs in fragments 13 and 14 and formed a homogeneous group with single dcl and double dcl3 dcl4 mutants, whereas double dcl2 dcl3 and dcl2 dcl4 mutants formed a distinct homogeneous group that accumulated lower levels of siRNAs in these fragments. In these fragments, siRNA accumulation showed moderate broad-sense heritability (h2b of 0.47 and h2b of 0.52 for fragments 13 and 14, respectively). Thus, genetic background explained a significant fraction of the variance in siRNA accumulation in our experimental system. Although our analysis did not identify significant differences in the accumulation of siRNAs from fragment 12 between genetic backgrounds, the lowest mean values corresponded to dcl2 dcl3 and dcl2 dcl4 mutants, which reflected a tendency to accumulate less siRNAs in this region.
Collectively, these findings suggested that the three DCL enzymes would be capable of processing, albeit with different affinities, dsRNA structures generated along the entire viral RNA to yield similar patterns of siRNA distribution. However, a substantial amount of TRV siRNAs originating at the 3′ end of the TRV1 genome specifically required DCL2-mediated processing of viral templates.
RDR requirements for the biogenesis of TRV siRNAs in Arabidopsis.
In plants, host-encoded RDRs may use viral templates to produce dsRNAs that serve as substrates for the formation of secondary siRNAs, hence amplifying initial silencing responses. We studied the involvement of host RDR enzymes in TRV siRNA biogenesis by infecting loss-of-function single, double, and triple mutants of rdr1, rdr2, and rdr6 in Arabidopsis. Northern blot analysis showed that accumulation of the two major 21- and 24-nt TRV siRNA classes in upper, noninoculated inflorescence tissue was not affected by single loss-of-function rdr mutations (Fig. 5A). Accordingly, accumulation of TRV genomic RNA in single rdr mutants was not substantially different from that in wild-type plants (Fig. 5B). However, while TRV siRNAs were only slightly less abundant in rdr1 rdr6 and rdr2 rdr6 mutants, they were strongly reduced in the rdr1 rdr2 rdr6 triple mutant compared to wild-type plants at 10 and 16 days postinfection (Fig. 5A). This result was reproducible using a probe corresponding to TRV2 (data not shown). The low accumulation of siRNAs was not due to lack of TRV replication or movement in the tissue analyzed, as double and triple rdr mutants accumulated significantly higher levels of full-length viral genomic and subgenomic RNAs over time than did the wild-type plants (Fig. 5B). This result reflects a considerable difference in the ratio of viral genomic and subgenomic RNA relative to TRV-specific siRNAs between the wild-type and each of the double and triple rdr mutants. We suspect that in the absence of functional RDRs, only a limited amount of viral-derived dsRNA was formed, and consequently less substrate was available for DCL-mediated cleavage and siRNA generation despite increasing amounts of viral RNAs. TRV siRNA detected in the rdr1 rdr2 rdr6 triple mutant probably represented a pool of primary and secondary siRNAs derived from cleavage of the silencing inducer viral RNA and redundant activities of other endogenous RDR enzymes. From these data, we conclude that biogenesis of a large fraction of the TRV siRNA population is largely influenced by the activity of at least three RDR homologs in Arabidopsis, which presumably exhibit partially complementary functions.
FIG. 5.
Accumulation of TRV siRNAs and viral TRV RNAs in Arabidopsis rdr mutants. RNA gel blot analysis of TRV siRNAs (A) and genomic (g) and subgenomic (sg) TRV RNAs (B) from inflorescence tissue of wild-type and single, double, and triple rdr Arabidopsis mutants infected with TRV. Duplicate samples were analyzed using a radiolabeled probe specific for the 3′ UTR of TRV. The relative accumulations (RA) of TRV siRNAs were calculated from band intensities and are indicated below the siRNA bands. Band intensities in tissue from wild-type-inoculated plants with the highest hybridization signal were normalized to 1.0. Composite images from single blots are shown. Figure content is as described in the legend of Fig. 2. (C) Spatial distribution profile of TRV1-derived siRNAs in Arabidospsis rdr mutants. Histograms represent the percentage of siRNA accumulation in each PCR fragment relative to the total accumulation in all the fragments, as determined for Fig. 3 and 4, using double and triple rdr mutants infected with TRV. Mean values and standard deviations from three independent experiments are shown. Figure content is as described in the legend of Fig. 3. dpi, days postinoculation; GUS, β-glucoronidase; rdr1/6, rdr1 rdr6; rdr2/6, rdr2 rdr6, rdr1/2/6, rd1 rdr2 rdr6.
We further examined whether the loss of specific RDRs altered substantially the spatial distribution of TRV siRNAs. Northern analysis revealed that 32P-labeled TRV siRNAs from rdr mutants hybridized to each of the ∼500-bp TRV-derived DNA fragments according to the same pattern observed in wild-type plants (Fig. 5C; see also Fig. S4 in the supplemental material). One-way ANOVA, using the genetic background as a factor, indicated that there were not statistically significant differences in the siRNA accumulation profile between wild-type and the rdr mutant series tested (F6,21 < 2.45; P > 0.0745). Nevertheless, these results are difficult to interpret since other RDR enzymes among the six RDR genes in Arabidopsis may compensate, at least partially, for RDR deficiencies during the production of viral siRNAs, complicating the identification of target regions for specific RDR activities. Further studies using mutant combinations involving rdr3, rdr4, and rdr5 should provide a more comprehensive scenario on the implication of the different plant RDRs on siRNA biogenesis.
DISCUSSION
In this study, we investigated the origin and the genetic components implicated in the biogenesis of viral siRNAs using TRV. Our data revealed a global population of TRV siRNAs in systemically infected plants that was influenced by distinct interconnected RNA silencing pathways involving coordinated DCL and RDR activities.
We used two experimental approaches to analyze the population of viral siRNAs in two host species systemically infected with TRV. In Arabidopsis, cloning of small RNAs and RNA hybridization-based analyses revealed a set of TRV siRNAs derived from positive and negative viral RNA strands, although TRV siRNAs of positive polarity were significantly more abundant than those derived from the complementary strand of negative polarity. Formation of both sense and antisense, 21- to 24-nt siRNAs was associated with the processing activity of DCL4, DCL3, and DCL2, which all functioned throughout the viral genome to produce multiple siRNA sequences. DCL4-dependent, siRNA of 21 nt was the dominant species, while the DCL3-dependent, 24-nt class accumulated to lower levels in the infected tissue. In general, data from RNA hybridizations and sequence analysis were consistent and indicated that the profile of TRV siRNAs was fairly representative of the entire siRNA population in TRV-infected plants. Interestingly, in N. benthamiana, asymmetry in strand polarity was evident only for TRV2, whereas TRV1 generated equivalent numbers of sense and antisense siRNA sequences in the cloned set. Another particularity of the cloned library in N. benthamiana is that siRNAs of 22 nt accumulated to the same high levels as 21-nt siRNAs in the infected tissue. It will be interesting to investigate whether this symmetrical distribution in strand polarity and size class distribution accurately reflects the siRNA population and to determine if the biosynthetic pathways of siRNAs in N. benthamiana differ from those in Arabidopsis.
The preferential enrichment of viral positive-strand siRNAs in response to virus infection observed for many plant viruses, including the CMV satellite RNA, supports a model whereby secondary structures within viral single-stranded RNA strands serve as a substrate for DCL-mediated cleavage (17, 18, 31, 40, 41, 51). In the case of TRV, we wonder whether the dominance of sense siRNAs in the infected tissue reflects a biogenesis mechanism that preferentially targets single-stranded RNAs or, by contrast, is the result of maturation events downstream of cleavage of perfectly complementary dsRNA that ultimately favors the accumulation of sense siRNAs to the detriment of antisense siRNAs. In this regard, it is plausible that structural and/or biochemical modifications particularly associated to siRNAs derived from positive viral strands or preferential incorporation of sense siRNAs into effector complexes might account for the higher incidence of these siRNA species. Using Mfold to predict RNA secondary structures, we found that there was little, if any, correlation between regions of predicted local base pairing within the sense TRV and the sequences of siRNAs. Therefore, the direct contribution of internal folded regions to the final population of TRV siRNAs is not clear. Considering that analysis and designation of structural RNA features by means of computational algorithms must be interpreted with caution, it will be necessary to demonstrate experimentally that these proposed base-paired structures exist in vivo and can be processed by DCL into viral siRNAs. On the other hand, there is no reason to presume that the presence of siRNA clusters, which is also associated with processing of perfectly complementary dsRNA in plants and animals (21, 32, 35, 43), is a reliable indicator of cleavage of folded RNA.
In addition to sense siRNAs, accumulation of antisense viral siRNAs in the infected tissue has been reported for most, if not all, plant viruses tested, including TRV (this study), demonstrating that synthesis of negative-stranded RNA is also a common step in the formation of virus-derived siRNAs (7, 15, 25, 31, 52). This leads us to the question of whether DCL enzymes use different types of substrates to generate siRNAs of different polarities. Derivation of antisense siRNA from single negative-strand viral RNA may involve processing of internal folded regions, as proposed for certain siRNAs derived from CMV satellite RNAs (18). If this is case, the relatively low abundance of antisense siRNA would correlate well with the assumption that negative-strand viral RNA is much less abundant than positive-strand viral RNA. Another possibility is that siRNAs of both polarities derive from dsRNA substrates that result from intermolecular base pairing of positive viral RNAs and the complementary negative strands. It is assumed that dsRNA intermediates can be formed during the infectious cycle in the host cell and can potentially be processed into viral siRNAs (1, 17). The existence of long dsRNA replicative forms and their accessibility for DCL processing have not yet been proven experimentally, and therefore their contribution to siRNA formation is debatable. However, we consider it likely that other shorter viral RNA species of positive and negative polarities (subgenomic RNA or partly degraded or partly synthesized pieces of genomic RNA) anneal to create dsRNA molecules with the potential to initiate a silencing response.
A complementary mechanism for dsRNA production involves host-encoded RDRs using positive-strand viral RNA as a template for synthesis of negative-strand RNA (54). Host-encoded RDRs play a crucial role in the maintenance stage of virus-induced RNA silencing in plants, which is linked to the formation of secondary siRNAs (12, 29, 42, 49, 52, 60). The results presented in this study indicated that a large portion of TRV siRNAs was RDR dependent and identified RDR1, RDR2, and RDR6 as host factors required for biogenesis of TRV siRNAs and antiviral silencing. But what is the overall contribution of the RDR-mediated pathways to viral siRNA formation? The reduction in viral siRNA accumulation in contrast to the increased accumulation of viral genomic and subgenomic RNAs (which should potentially serve as additional substrates for direct DCL-mediated cleavage) in the rdr1 rdr2 rdr6 triple mutant argues against secondary structures as the main source of siRNAs and emphasizes the relevance of RDR-dependent silencing pathways in the formation of secondary siRNAs. An important issue to clarify is the specific role of each of the remaining Arabidopsis RDR genes since they might function redundantly in siRNA biogenesis. Previous studies showed that silencing of RDR6 alone in N. benthamiana enhances susceptibility to Potato virus X, Potato virus Y, and CMV in combination with its Y satellite but not to TRV and Tobacco mosaic virus (49). Similarly, the rdr6 Arabidopsis mutant is hypersusceptible to CMV but not to Tobacco mosaic virus, TRV, Turnip mosaic virus, or the Turnip vein clearing virus (12, 13, 42). The fact that inactivation of a single RDR does not substantially alter susceptibility to all RNA viruses belonging to different groups may be due to functional redundancy in mediating antiviral silencing among RDR enzymes. Consistent with this idea, we found that diminished siRNA accumulation and hypersusceptibility to TRV infection were evident in double and triple rdr mutants but not in single rdr Arabidopsis mutants. This finding firmly suggests that more than one RDR-dependent pathway functions in the biogenesis of TRV siRNAs and subsequent antiviral defense. Alternatively, virus-encoded proteins with silencing suppressor properties may inhibit one or more RDR pathways of siRNA biogenesis, as reported for the CMV-encoded 2b protein, which specifically interferes with RDR1-dependent synthesis of secondary viral siRNAs (15).
Our study reveals a heterogeneous distribution profile of viral siRNAs along the TRV genome, presumably due to the existence of genomic areas with some special features that may influence accessibility, affinity, or enzymatic activity of one or more components of the RNA silencing machinery required for siRNA biogenesis. For instance, we show in this study that TRV siRNAs were particularly abundant at the 3′ end of TRV1 (fragments 12 to 14). This region encompasses the 29-kDa- and 16-kDa-coding genes which are translated from two subgenomic RNAs (1a and 1b) (Fig. 1A). Therefore, high accumulation of siRNAs at the 3′ end of TRV1 was relatively easy to envision since both subgenomic RNAs most probably provided additional templates for the formation of viral dsRNA substrates. The accumulation of siRNAs at this region was significantly reduced in double dcl mutants lacking DCL2 activity. A possible scenario for this target specificity is that certain dsRNA substrates derived from this genomic region possess structural features that favor preferential DCL2 targeting. For instance, complementary regions from overlapping subgenomic RNAs of positive and negative polarity might anneal either together or with complementary sequences derived from longer viral RNAs to form dsRNA substrates. This hypothetical dsRNA would resemble that produced by annealing of complementary regions in converging transcripts from antisense genes, which is the endogenous target for the DCL2 pathway (8). This substrate specificity is also presumably conditioned by the association of each DCL enzyme with any of the dsRNA-binding proteins that function as essential cofactors in RNA silencing (30). Nevertheless, since we did not observe any statistically noticeable effect on the accumulation of siRNA at the 3′ end of TRV1 in single dcl2 mutants, it is reasonable to think that the three DCL enzymes contributed significantly to siRNA formation along this region, perhaps by targeting different types of dsRNA generated through distinct mechanisms of dsRNA synthesis.
Closely related to possible preferential cleavage zones in siRNA precursors is the issue of whether viral sequences containing dsRNA structure extend to all regions of the virus genome. Besides the putative sites of RNA folding within viral sequences, the involvement of host RDR activities and the hypothetical presence of optimal and suboptimal sites for RDR activity to initiate synthesis of minus RNA strands appear to be critical in determining the extent of dsRNA formation along the virus template. Several reports proposed that viral RNAs that lack features of host transcripts such as a 5′ cap or poly(A) tail may enter the RNA silencing pathway because they are converted to dsRNA by host RDR enzymes (5, 24, 28, 37, 60). These aberrant viral RNAs might be formed during virus replication or as a result of initial cleavage events guided by primary viral siRNAs originated at the earliest steps of the infectious cycle. Alternatively, we cannot totally rule out the possibility that RDRs may also function to generate secondary siRNAs of antisense polarity directly from non-RNA-induced silencing complex-cleaved RNA templates, as described in Caenorhabditis elegans (43, 50).
The results presented here may well support a model in which primary siRNAs result from DCL-mediated cleavage of the original viral RNA trigger in the earliest stages of virus infection. The trigger might appear in the form of secondary structures along the viral RNA and/or dsRNA formed through base pairing of complementary positive and negative viral strands. Secondary siRNAs would represent a large fraction of the population of TRV siRNAs which would derive either from processing of viral dsRNAs generated by the action of endogenous RDRs or as direct RDR products. The challenge ahead is to determine how broadly applicable our findings with TRV may be to different viruses and host species in order to build a general model for virus-derived siRNA biogenesis in plants.
Supplementary Material
Acknowledgments
We thank Michael A. Phillips, Francisco Tenllado, Fernando García-Arenal, and Tomás Canto for valuable comments and critical readings of the manuscript; Jim Carrington and Olivier Voinnet for providing Arabidopsis mutant seeds; and S. P. Dinesh-Kumar for providing the TRV-PDS system.
C.L. dedicates this work to the memory of Juan Carlos Lorente.
This work was supported by grants GEN2003-20222-C02-01 and BIO2006-13107 from the M.E.C. (Spain) and GR/SAL/0821/2004 from the C.A.M. (Spain). L.D. was the recipient of a predoctoral fellowship from the C.A.M.
Footnotes
Published ahead of print on 19 March 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 2961270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Akbergenov, R., A. Si-Ammour, T. Blevins, I. Amin, C. Kutter, H. Vanderschuren, P. Zhang, W. Gruissem, F. Meins, Jr., T. Hohn, and M. M. Pooggin. 2006. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 34462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, E., Z. Xie, A. M. Gustafson, and J. C. Carrington. 2005. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121207-221. [DOI] [PubMed] [Google Scholar]
- 4.Ambros, V., and X. Chen. 2007. The regulation of genes and genomes by small RNAs. Development 1341635-1641. [DOI] [PubMed] [Google Scholar]
- 5.Axtell, M. J., C. Jan, R. Rajagopalan, and D. P. Bartel. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127565-577. [DOI] [PubMed] [Google Scholar]
- 6.Baulcombe, D. 2005. RNA silencing. Trends Biochem. Sci. 30290-293. [DOI] [PubMed] [Google Scholar]
- 7.Blevins, T., R. Rajeswaran, P. V. Shivaprasad, D. Beknazariants, A. Si-Ammour, H. S. Park, F. Vazquez, D. Robertson, F. Meins, Jr., T. Hohn, and M. M. Pooggin. 2006. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 346233-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borsani, O., J. Zhu, P. E. Verslues, R. Sunkar, and J. K. Zhu. 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 1231279-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouché, N., D. Lauressergues, V. Gasciolli, and H. Vaucheret. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 253347-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodersen, P., and O. Voinnet. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22268-280. [DOI] [PubMed] [Google Scholar]
- 11.Brosnan, C. A., N. Mitter, M. Christie, N. A. Smith, P. M. Waterhouse, and B. J. Carroll. 2007. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 10414741-14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalmay, T., A. Hamilton, S. Rudd, S. Angell, and D. C. Baulcombe. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101543-553. [DOI] [PubMed] [Google Scholar]
- 13.Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 31368-71. [DOI] [PubMed] [Google Scholar]
- 14.Demmig-Adams, B., and W. W. Adams. 1992. Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43599-626. [Google Scholar]
- 15.Diaz-Pendon, J. A., F. Li, W. X. Li, and S. W. Ding. 2007. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 192053-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinesh-Kumar, S. P., R. Anandalakshmi, R. Marathe, M. Schiff, and Y. Liu. 2003. Virus-induced gene silencing. Methods Mol. Biol. 236287-294. [DOI] [PubMed] [Google Scholar]
- 17.Ding, S. W., and O. Voinnet. 2007. Antiviral immunity directed by small RNAs. Cell 130413-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du, Q. S., C. G. Duan, Z. H. Zhang, Y. Y. Fang, R. X. Fang, Q. Xie, and H. S. Guo. 2007. DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J. Virol. 819142-9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunoyer, P., C. Himber, and O. Voinnet. 2005. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 371356-1360. [DOI] [PubMed] [Google Scholar]
- 20.Dunoyer, P., C. Himber, and O. Voinnet. 2006. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 38258-263. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahlgren, N., M. D. Howell, K. D. Kasschau, E. J. Chapman, C. M. Sullivan, J. S. Cumbie, S. A. Givan, T. F. Law, S. R. Grant, J. L. Dangl, and J. C. Carrington. 2007. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fusaro, A. F., L. Matthew, N. A. Smith, S. J. Curtin, J. Dedic-Hagan, G. A. Ellacott, J. M. Watson, M. B. Wang, C. Brosnan, B. J. Carroll, and P. M. Waterhouse. 2006. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 71168-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazzani, S., T. Lawrenson, C. Woodward, D. Headon, and R. Sablowski. 2004. A link between mRNA turnover and RNA interference in Arabidopsis. Science 3061046-1048. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286950-952. [DOI] [PubMed] [Google Scholar]
- 26.Henderson, I. R., X. Zhang, C. Lu, L. Johnson, B. C. Meyers, P. J. Green, and S. E. Jacobsen. 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38721-725. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez, C., J. E. Carette, D. J. Brown, and J. F. Bol. 1996. Serial passage of tobacco rattle virus under different selection conditions results in deletion of structural and nonstructural genes in RNA 2. J. Virol. 704933-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herr, A. J., A. Molnar, A. Jones, and D. C. Baulcombe. 2006. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA 10314994-15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Himber, C., P. Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 224523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiraguri, A., R. Itoh, N. Kondo, Y. Nomura, D. Aizawa, Y. Murai, H. Koiwa, M. Seki, K. Shinozaki, and T. Fukuhara. 2005. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57173-188. [DOI] [PubMed] [Google Scholar]
- 31.Ho, T., D. Pallett, R. Rusholme, T. Dalmay, and H. Wang. 2006. A simplified method for cloning of short interfering RNAs from Brassica juncea infected with turnip mosaic potyvirus and turnip crinkle carmovirus. J. Virol. Methods 136217-223. [DOI] [PubMed] [Google Scholar]
- 32.Howell, M. D., N. Fahlgren, E. J. Chapman, J. S. Cumbie, C. M. Sullivan, S. A. Givan, K. D. Kasschau, and J. C. Carrington. 2007. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19926-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasschau, K. D., N. Fahlgren, E. J. Chapman, C. M. Sullivan, J. S. Cumbie, S. A. Givan, and J. C. Carrington. 2007. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y., M. Schiff, R. Marathe, and S. P. Dinesh-Kumar. 2002. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30415-429. [DOI] [PubMed] [Google Scholar]
- 35.Llave, C., K. D. Kasschau, M. A. Rector, and J. C. Carrington. 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 141605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llave, C., Z. Xie, K. D. Kasschau, and J. C. Carrington. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2972053-2056. [DOI] [PubMed] [Google Scholar]
- 37.Luo, Z., and Z. Chen. 2007. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19943-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacFarlane, S. A. 1999. Molecular biology of the tobraviruses. J. Gen. Virol. 802799-2807. [DOI] [PubMed] [Google Scholar]
- 39.Margis, R., A. F. Fusaro, N. A. Smith, S. J. Curtin, J. M. Watson, E. J. Finnegan, and P. M. Waterhouse. 2006. The evolution and diversification of Dicers in plants. FEBS Lett. 5802442-2450. [DOI] [PubMed] [Google Scholar]
- 40.Moissiard, G., and O. Voinnet. 2006. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. USA 10319593-19598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Molnar, A., T. Csorba, L. Lakatos, E. Varallyay, C. Lacomme, and J. Burgyan. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 797812-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mourrain, P., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon, J. B. Morel, D. Jouette, A. M. Lacombe, S. Nikic, N. Picault, K. Remoue, M. Sanial, T. A. Vo, and H. Vaucheret. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101533-542. [DOI] [PubMed] [Google Scholar]
- 43.Pak, J., and A. Fire. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315241-244. [DOI] [PubMed] [Google Scholar]
- 44.Park, W., J. Li, R. Song, J. Messing, and X. Chen. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 121484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peragine, A., M. Yoshikawa, G. Wu, H. L. Albrecht, and R. S. Poethig. 2004. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 182368-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi, Y., A. M. Denli, and G. J. Hannon. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19421-428. [DOI] [PubMed] [Google Scholar]
- 47.Qu, F., X. Ye, G. Hou, S. Sato, T. E. Clemente, and T. J. Morris. 2005. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 7915209-15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinhart, B. J., E. G. Weinstein, M. W. Rhoades, B. Bartel, and D. P. Bartel. 2002. MicroRNAs in plants. Genes Dev. 161616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwach, F., F. E. Vaistij, L. Jones, and D. C. Baulcombe. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 1381842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sijen, T., F. A. Steiner, K. L. Thijssen, and R. H. Plasterk. 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315244-247. [DOI] [PubMed] [Google Scholar]
- 51.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaistij, F. E., L. Jones, and D. C. Baulcombe. 2002. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vazquez, F., V. Gasciolli, P. Crete, and H. Vaucheret. 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14346-351. [DOI] [PubMed] [Google Scholar]
- 54.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6206-220. [DOI] [PubMed] [Google Scholar]
- 55.Wassenegger, M., and G. Krczal. 2006. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 11142-151. [DOI] [PubMed] [Google Scholar]
- 56.Xie, Z., E. Allen, A. Wilken, and J. C. Carrington. 2005. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 10212984-12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie, Z., B. Fan, C. Chen, and Z. Chen. 2001. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA 986516-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie, Z., L. K. Johansen, A. M. Gustafson, K. D. Kasschau, A. D. Lellis, D. Zilberman, S. E. Jacobsen, and J. C. Carrington. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, S. J., S. A. Carter, A. B. Cole, N. H. Cheng, and R. S. Nelson. 2004. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 1016297-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, D., B. Fan, S. A. MacFarlane, and Z. Chen. 2003. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant-Microbe Interact. 16206-216. [DOI] [PubMed] [Google Scholar]
- 61.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.