Abstract
To determine the extent and structure of genetic variation in dengue viruses (DENV) on a restricted spatial and temporal scale, we sequenced the E (envelope) genes of DENV-1, -2, and -3 isolates collected in 2001 from children enrolled in a prospective school-based study in Kamphaeng Phet, Thailand, and diagnosed with dengue disease. Our analysis revealed substantial viral genetic variation in both time and space, with multiple viral lineages circulating within individual schools, suggesting the frequent gene flow of DENV into this microenvironment. More-detailed analyses of DENV-2 samples revealed strong clustering of viral isolates within individual schools and evidence of more-frequent viral gene flow among schools closely related in space. Conversely, we observed little evolutionary change in those viral isolates sampled over multiple time points within individual schools, indicating a low rate of mutation fixation. These results suggest that frequent viral migration into Kamphaeng Phet, coupled with population (school) subdivision, shapes the genetic diversity of DENV on a local scale, more so than in situ evolution within school catchment areas.
Dengue is the most common mosquito-borne viral disease in tropical and subtropical regions of the world, and hence, dengue virus (DENV) is an emerging human pathogen of major importance (10). The numbers of cases of dengue fever (DF) and the more severe conditions dengue hemorrhagic fever (DHF) and dengue shock syndrome, as well as the number of countries affected by dengue, have increased dramatically. Current estimates place over 2 billion people in areas of dengue endemicity, with more than 50 million DENV infections and over 20,000 deaths each year (8, 9). The majority of dengue cases are characterized by a self-limited febrile illness (DF) associated with viremia, transient mild laboratory abnormalities, and sometimes mild bleeding. In a small percentage of infections, overt plasma leakage occurs, with hemoconcentration, hypoalbuminemia, and extravasation of fluid (DHF). Dengue shock syndrome can result when plasma leakage is severe and is responsible for most of the severe morbidity and mortality associated with DENV. The causative RNA virus (family Flaviviridae, genus Flavivirus) has a single-strand, positive-sense genome approximately 11 kb in length and consists of four antigenically distinct serotypes (DENV-1 to DENV-4), which now cocirculate in many populations.
Over the last 20 years, many studies have documented the extent and structure of genetic variation in all four DENV serotypes (reviewed in reference 11). In general, these studies have revealed that each of the four serotypes contains a number of phylogenetically distinct “subtypes” (or genotypes), the genetic structures of which clearly reflect a complex pattern of viral gene flow (migration among locations) coupled with population subdivision. Most notably, some DENV genotypes appear to be restricted to specific localities, commonly South East Asia, while others have more-cosmopolitan distributions across the tropical and subtropical world. Whether these differences in geographic distribution reflect underlying differences in viral fitness, manifest as differences in transmissibility and/or virulence, is still a subject of debate.
Despite the growing database of partial and complete DENV genome sequences (17) and the increasing number of studies addressing various aspects of the molecular epidemiology of DENV infection, none to date have considered the nature of viral genetic diversity on a scale of populations separated by only a few kilometers and sampled over a time period of months. However, such studies of DENV “microevolution” are essential to obtain a full understanding of the mechanisms shaping DENV evolution. Further, given forthcoming phase III clinical trials of DENV vaccines, which may be imperfect in their protection against all four serotypes and hence shape viral evolution (7), it is critical to document the extent and structure of DENV genetic variation at vaccine trial sites and the pace of viral evolution within a limited time frame. To this end, we performed a detailed genetic analysis of the envelope (E) genes of DENV serotype 1, 2, and 3 isolates collected in Kamphaeng Phet, Thailand, during 2001 as part of a school absence-based prospective study.
MATERIALS AND METHODS
Prospective study with primary school children in Kamphaeng Phet.
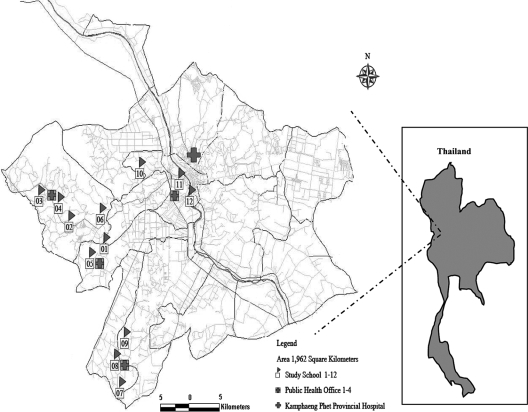
Twelve primary schools from the Kamphaeng Phet Province in northern Thailand were selected to participate in a prospective dengue study (Fig. 1). Details of this study have been previously published (5). Children were recruited during January 1998 from school grades 1 through 5 and were eligible to remain in the study until graduation from sixth grade. New first-grade students were enrolled in the cohort each January. The enrollment criteria were attendance at a study school, enrollment in a grade between first and sixth, and parental informed consent. Blood samples for dengue serology were obtained from the entire population four times each year (January or February, 1 June, 15 August, and 15 November). Active acute-illness-case surveillance occurred during the peak DENV transmission season, from 1 June to 15 November. Absent students were visited by village health workers and evaluated with a symptom questionnaire, and oral temperatures were obtained. Samples collected outside this surveillance period were based on subject visits to the hospital during which a study nurse identified the subjects as enrolled in this study. Acute-phase and 14-day convalescent-phase blood samples were obtained from absent students with histories of fever within 7 days of school absence or oral temperatures of ≥38°C. Laboratory assays, including an immunoglobulin M (IgM)/IgG enzyme-linked immunosorbent assay (ELISA) and PCR, were used to diagnose dengue as previously described (5, 6).
FIG. 1.
Map of study area in subdistrict of Muang, Kamphaeng Phet, Thailand, showing the locations of participating schools.
Classification of dengue disease.
The criteria for DF were school absence associated with a febrile illness and laboratory confirmation of acute DENV infection without evidence of DHF by World Health Organization (WHO) criteria (2). DF was further classified as symptomatic nonhospitalized or symptomatic hospitalized DF. The criteria for DHF were school absence associated with a febrile illness and laboratory confirmation of acute DENV infection with evidence of DHF by WHO criteria (2).
Viral isolation and sequencing.
For all suspected acute dengue cases, DENV serotype-specific PCR was performed on the first serum sample obtained for acute-illness evaluation. Reverse transcription-PCR (RT-PCR) was performed according to the protocol of Lanciotti et al. (14), with the following modifications. Avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI) was used in the first-round RT-PCR. The concentrations of the primers used in the RT-PCR and nested reactions were reduced from 50 pmol to 12.5 pmol per reaction, and the number of nested PCR amplification cycles was increased to 25.
For DENV PCR-positive samples, virus isolation was performed with C6/36 cells and/or Toxorhynchites splendens mosquitoes as previously described (13, 16). DENV serotype identification was performed using a dengue serotype-specific enzyme immunoassay as previously described (18, 20). Viral RNA for sequencing was extracted from patient samples first amplified in T. splendens mosquitoes, followed by one passage in C6/36 cells by use of a QIAamp viral RNA mini kit (Qiagen, Germany) according to the manufacturer's instructions. RT was performed using random hexamer oligonucleotides with the SuperScript first-strand synthesis system (Invitrogen) according to the manufacturer's instructions. The DNA fragments of the envelope gene regions of 9 DENV-1, 40 DENV-2, and 18 DENV-3 isolates were amplified by PCR using 5 μl of cDNA in a 50-μl reaction mixture containing 0.3 mM deoxynucleoside triphosphates, 2.5 U AmpliTaq DNA polymerase (Applied Biosystems), 1× PCR buffer, 1.5 mM MgCl2, and 15 pmol of each forward and reverse primer. The PCR mixtures of DENV-1 and -3 were subjected to 1 cycle of 95°C for 5 min; 35 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 2 min; and 1 cycle of 72°C for 15 min. The PCR mixtures of DENV-2 were subjected to the same thermal conditions as those of DENV-1 and -3, except for the annealing temperature, which was changed to 55°C. The PCR-amplified DNA fragments were purified using QIAquick PCR purification kits (Qiagen) according to the manufacturer's instructions. Purified DNA fragments were used for sequencing.
Sequencing reactions were performed by using a DYEnamic ET dye terminator sequencing kit (GE Healthcare Bio-Sciences) according to the manufacturer's instructions. The sequencing primers are available upon request. The sequencing products were cleaned by standard precipitation before sequencing with a MegaBACE 500 automated DNA sequencer (GE Healthcare Bio-Sciences). Overlapping nucleic acid sequences were combined for analysis and edited by using SEQUENCHER software (Gene Code Corporation).
Unfortunately, the number of viruses available for sequencing was a relatively small percentage of the viruses circulating in our cohort during the 2001 dengue season. In all, 244 infections, including inapparent infections, were detected in our cohort. Of the overt infections, only 86 had virus detectable by PCR, while the remaining 38 were detected based on a serological response consistent with acute dengue. Hence, we were able to sequence 78% of the PCR-positive samples but only 67 of 244 sequences (27%).
Phylogenetic analysis.
The following sets of DENV E-gene sequences from Kamphaeng Phet were compiled for evolutionary analysis: for DENV-1, 9 sequences (genotype I) from three schools; for DENV-2, 40 sequences (genotype Asian I) from nine schools; and for DENV-3, 18 sequences (genotype II) from three schools. To place these isolates within the wider context of DENV evolution, particularly that from South East Asia, we also compiled, from GenBank, phylogenetically representative “background” data sets of sequences from each of the respective genotypes, many of which also came from Thailand. This resulted in final data sets of the following sizes in the phylogenetic analyses: for DENV-1, 116 sequences (1,485 nucleotides [nt]); for DENV-2, 168 sequences (1,485 nt); and for DENV-3, 124 sequences (1,479 nt).
Phylogenetic trees for all three data sets were inferred using the maximum likelihood (ML) method available in PAUP* (19), with the best-fit model of nucleotide substitution (usually GTR+I+Γ4) determined using the MODELTEST program (15) and employing tree bisection-reconnection branch swapping. Bootstrap resampling (1,000-replicate neighbor-joining trees under the ML substitution model) was used to determine the phylogenetic support for individual nodes. All parameter values are available from the authors on request.
Analysis of spatial and temporal structure.
In the case of DENV-2, the larger number of E-gene sequences allowed us to determine whether there were distinct spatial and temporal components to the evolutionary patterns observed. This was achieved by using a parsimony approach which had previously proven highly informative in a study of DENV in the Caribbean (4). In the case of spatial structure, we tested whether there was significant clustering according to the school from which the virus was isolated. In the case of temporal structure, we tested whether viruses were clustered according to their months of isolation. In each of these analyses, DENV-2 sequences were assigned a character state dependent on the school (spatial) or month (temporal) of origin. Given the ML phylogeny for these sequences (determined as described above) and the above-defined isolate states, the minimum number of changes in character state needed to give rise to the observed distribution of states was then estimated using parsimony (with all ambiguous changes excluded). To determine the expected number of changes under the null hypothesis of complete mixing among states by space or time, the states of all isolates were randomized 500 times, and for each randomization, the number of changes in state was calculated in the manner described above. The difference between the mean numbers of observed and expected changes for each pair of states indicates the level of geographic or temporal isolation, with statistical significance calculated by comparing the total number of observed state changes to the number expected under random mixing. All these analyses were performed using PAUP*.
Analysis of selection pressures.
Overall and site-specific selection pressures in the E genes of all three serotypes circulating in Kamphaeng Phet (excluding other Asian sequences) were measured as ratios of nonsynonymous (dN) to synonymous (dS) substitutions per site (dN/dS), estimated using the single-likelihood-ancestor-counting and random-effect-likelihood methods. Both methods utilized the GTR substitution model with input phylogenetic trees inferred using the neighbor-joining method available at the Datamonkey Web facility (12).
Nucleotide sequence accession numbers.
All DENV gene sequences have been submitted to GenBank and assigned accession numbers EU117304 to EU117312, EU117313 to EU117352, and EU117353 to EU117370 for DENV-1, -2, and -3, respectively.
RESULTS AND DISCUSSION
DENV prevalence and serotypes in Kamphaeng Phet.
From 1998 to 2002, a prospective study of DENV infection was conducted with ∼2,000 primary school age children in Kamphaeng Phet, Thailand. Overt illness was detected using school absences to identify sick children, and DENV infections were confirmed using IgM/IgG ELISA, hemagglutination inhibition, and RT-PCR (5). Inapparent infections were identified based on a fourfold rise in hemagglutination inhibition between serial serum samples. Over the 5-year study period, 312 inapparent and 304 overt infections were identified. The overt infections were detected in children attending every school in the study, and all four serotypes were identified by RT-PCR (1, 5, 6).
The highest incidence of disease occurred in 2001, with 120 inapparent infections and 124 cases of overt disease (Table 1). Because of the high dengue incidence during 2001, samples were collected throughout the year, with the majority identified during the peak dengue season of June to November as part of the school absence study and the remainder identified from hospitalization of study participants (Table 2). Through ELISA typing of clinical isolates or RT-PCR of serum samples, we were able to identify the infecting DENV serotype in 86 (69%) symptomatic cases; the remaining 38 (31%) were identified as acute DENV infection by serology only. DENV-2 was the predominant serotype, identified in 52 of the 86 (60%) cases, followed by DENV-3 (25 cases; 29%) and DENV-1 (9 cases; 10%), collected in 10 of the 12 schools.
TABLE 1.
Numbers of DENV infections in school absence-based cohort during 2001
| School | No. of subjects
|
|||
|---|---|---|---|---|
| With inapparent infections | Not hospitalized | Hospitalized for:
|
||
| DF | DHF | |||
| 1 | 1 | 2 | 2 | 0 |
| 2 | 5 | 1 | 1 | 3 |
| 3 | 3 | 2 | 0 | 0 |
| 4 | 36 | 15 | 3 | 1 |
| 5 | 2 | 0 | 6 | 10 |
| 6 | 0 | 0 | 0 | 0 |
| 7 | 5 | 0 | 0 | 0 |
| 8 | 10 | 11 | 0 | 2 |
| 9 | 16 | 9 | 0 | 0 |
| 10 | 4 | 8 | 0 | 4 |
| 11 | 33 | 28 | 1 | 10 |
| 12 | 5 | 4 | 0 | 1 |
| Total | 120 | 80 | 13 | 31 |
TABLE 2.
Numbers of viral isolates sequenced according to school and month of sampling
| Serotype | School (no. of isolates) | Mo of sampling |
|---|---|---|
| DENV-1 | 8 (4) | July, August, November |
| 11 (3) | July, September | |
| 12 (2) | July | |
| DENV-2 | 1 (3) | June |
| 2 (3) | June, July | |
| 4 (6) | June, July, August, November | |
| 5 (5) | February, March | |
| 8 (5) | June, July, August, September | |
| 9 (3) | June, August, September | |
| 10 (5) | May, June, July, August, November | |
| 11 (7) | April, May, June, July, August | |
| 12 (3) | July, September | |
| DENV-3 | 4 (3) | June, July |
| 10 (1) | July | |
| 11 (14) | June, July, August, September, November |
Microevolution of DENV in Kamphaeng Phet.
To determine the extent and structure of genetic diversity during the 2001 dengue season, we sequenced the E genes of all available viruses collected during this year (Table 2). In total, all nine DENV-1 virus samples, collected from students in schools 8, 11, and 12 from July to November 2001, were available. All were assigned to genotype I, which is common in South East Asia, especially in Thailand. Similarly, 40/52 DENV-2 viruses, from 9 of the 12 schools, were available for sequencing, all of which represented the Asian I genotype, which is common in Thailand. The period of collection of these viruses was diverse, ranging from February to November and hence covering periods of both low and high DENV transmission. A total of 18/25 DENV-3 viruses, obtained from three schools in 2001 and largely from school 11 between June and November, were available for sequencing. All DENV-3 viruses were assigned to genotype II, which is again common in South East Asia. Viruses that were unavailable for sequencing were unavailable because of a lack of clinical material, a failure to grow a viral stock during the study, or insufficient genetic material for sequencing.
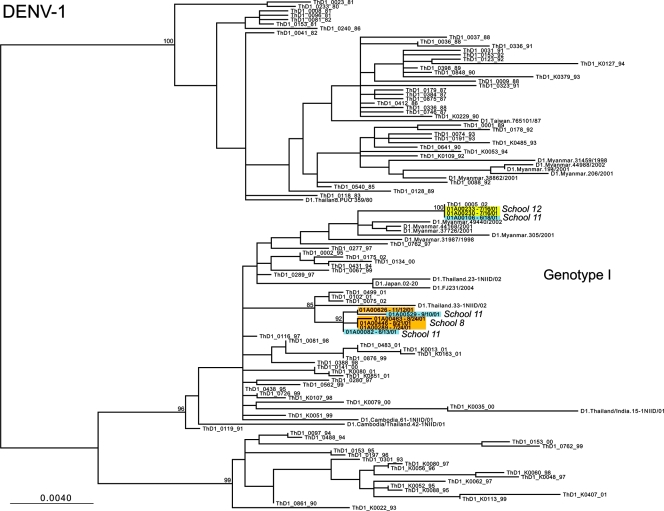
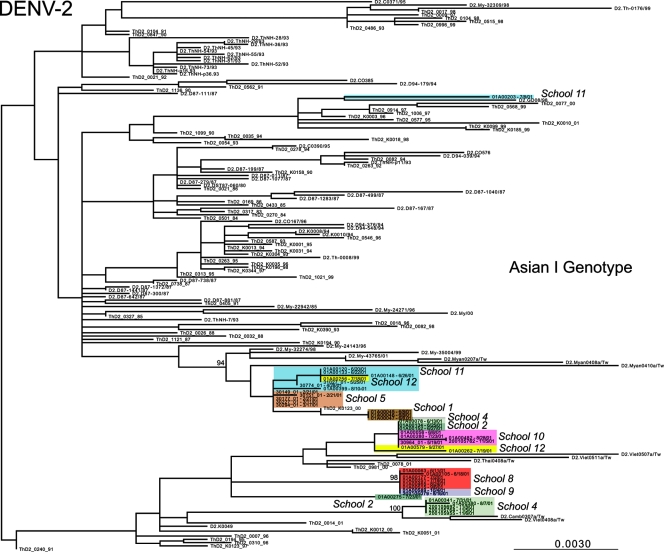
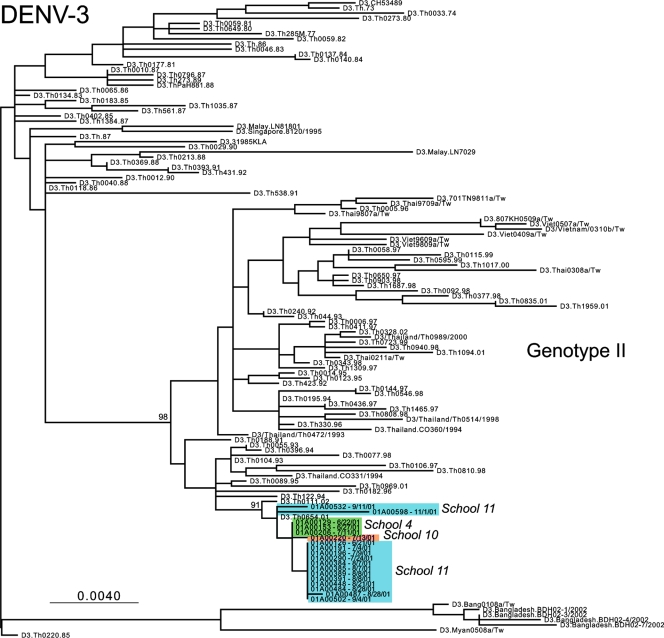
ML phylogenetic trees for the DENV-1, DENV-2, and DENV-3 E-gene sequences from Kamphaeng Phet, combined with the background isolates, are presented in Fig. 2, 3, and 4, respectively. In the case of DENV-1, viral isolates fall into two distinct clades, separated by other Asian viruses. That the DENV-1 isolates from school 11 fall in both clades provides compelling evidence for the independent entry of genetically distinct viruses into this spatially restricted region. A similar pattern was observed in DENV-2. Here, those viruses sampled from schools 1, 5, 8, 9, and 10 formed school-specific clusters (although only those from school 1 were clearly phylogenetically distinct from those from the other schools), while multiple genetic variants were seen in schools 2, 4, 11, and 12 (with one highly divergent variant observed in school 11). Finally, in the case of DENV-3, single clades were observed in schools 4 and 10, while two clades were again observed in school 11, although with weak bootstrap support. Notably, children from school 11 come from a densely populated urban environment at the center of the study area, close to the major transportation throughway and a public health clinic, which may have contributed to the appearance of multiple viral variants in this school.
FIG. 2.
ML tree of selected isolates of genotype I of DENV-1. Different schools from within Kamphaeng Phet are designated by different colors. All horizontal branch lengths are drawn to scale according to the numbers of nucleotide substitutions per site, with bootstrap support values shown next to relevant nodes. The tree is midpoint rooted for purposes of clarity only.
FIG. 3.
ML tree of selected isolates of the Asian I genotype of DENV-2. Different schools from within Kamphaeng Phet are designated by different colors. All horizontal branch lengths are drawn to scale according to the numbers of nucleotide substitutions per site, with bootstrap support values shown next to relevant nodes. The tree is midpoint rooted for purposes of clarity only.
FIG. 4.
ML tree of selected isolates of genotype II of DENV-3. Different schools from within Kamphaeng Phet are designated by different colors. All horizontal branch lengths are drawn to scale according to the numbers of nucleotide substitutions per site, with bootstrap support values shown next to relevant nodes. The tree is midpoint rooted for purposes of clarity only.
Overall, these phylogenetic results reveal a marked clustering by school (reflected in the grouping of colors in Fig. 2 to 4), indicating that multiple genetic variants of DENV circulate within a spatially restricted area during a single dengue season but that individual schools represent distinct evolutionary entities that have experienced clear population subdivision. In only a relatively small number of cases do multiple variants of individual serotypes cocirculate, most notably in some cases involving school 11. Further, the observation that these distinct clusters are often separated by viruses isolated from outside Kamphaeng Phet (and in the cases of DENV-1 and DENV-2 outside Thailand) indicates that there were multiple introductions of DENV into this region during 2001. We believe that this is the first report of evolution on a localized scale for DENV, and this report indicates that even viruses sampled from spatially restricted regions may harbor extensive genetic diversity.
To determine the extent of spatial clustering more rigorously, we examined DENV-2 in more detail as this represented our best-sampled serotype. A parsimony-based analysis confirmed that there is a highly significant clustering of viral isolates by school (P < 0.005). However, despite this strong genetic segregation, there is also clear evidence (P < 0.005) of viral migration between schools 8 and 9. Notably, these two schools are geographically the closest in the study population, separated by only 5 km, and the furthest from the city, at distances of 36 and 33 km from the field site, respectively (Fig. 1). Further, the villages that feed these schools have low population densities, are highly rural, and possess poor road infrastructure, making access to this area more difficult than access to other regions of Kamphaeng Phet. As such, our data suggest that patterns of viral gene flow are determined by local geographic and economic variables. The villages that make up the Kamphaeng Phet region are diverse, with the most developed and densely populated villages lying close to the city center, where frequent travel into and out of the city is common. In contrast, villages become less developed and densely populated further from the city center, in turn restricting travel and resulting in the genetic isolation of viruses from schools 8 and 9.
In contrast, these DENV-2 sequence data exhibited no significant clustering by month of sampling, indicating that there was little replacement of viral lineages at this level of spatial and temporal resolution. Similarly, at the level of the E-gene sequences analyzed, there was little evidence for in situ evolution, such that viruses sampled from the same school over time periods ranging from days to months exhibited few mutational differences (in some cases only one mutation over a 6-month sampling period). Such relative intraschool stability sits in marked contrast to the often strong genetic differentiation among schools. This observation, coupled with the strong population subdivision exhibited by each school, indicates that the importation of DENV into the study population, rather than in situ evolution within school catchment areas, was the most important factor shaping viral genetic diversity on this spatial scale.
To determine the selection pressures acting on the DENV E-gene sequences sampled from Kamphaeng Phet, we compared the relative numbers of nonsynonymous (dN) and synonymous (dS) substitutions per site. This analysis provided no evidence for positive selection acting on any nucleotide site, although all dN/dS methods are inherently conservative. Indeed, the overall picture obtained was that of strong purifying selection, with mean dN/dS values of 0.084, 0.065, and 0.008 for DENV-1, DENV-2 and DENV-3, respectively. Such strong purifying selection has previously been observed for these serotypes in Thailand (21, 22), highlighting the predominance of genetic drift over natural selection in shaping DENV substitution dynamics in the short term. However, the reasons for the greater selective constraints in DENV-3, which is characterized by very low dN/dS values, are unclear. Further, as random genetic drift is the most common process determining substitution dynamics, allele frequencies are not expected to change greatly over the period of sampling, even given the high mutation rate of DENV (3).
Overall, these results have mixed implications for how DENV populations might respond to imperfect vaccination in the near future. DENV sampled on a short spatial and temporal scale evidently exhibits remarkably high levels of genetic diversity, thereby providing the raw material for adaptive evolution. However, this variation is more often generated by migration than by mutation accumulation, with purifying selection the dominant evolutionary force.
Acknowledgments
We acknowledge the invaluable contributions of the clinical, laboratory, and entomological personnel of AFRIMS and Kamphaeng Phet AFRIMS Research Unit (KAVRU). We are indebted to the staff of the Kamphaeng Phet AFRIMS Research Unit (KPP) Governor and Chief, the KPP Provincial Medical Office and Chief, and the KPP Provincial School Office as well as participating village leaders. Finally, we are grateful to the children of the KPP and their parents for their lasting enthusiasm and cooperation.
Funding was partially provided by the U.S. Military Infectious Disease Research Program, Ft. Detrick, MD, and the National Institutes of Health (NIH-P01-AI34533).
The opinions and assertions contained herein are the personal views of the authors and are not to be construed as official or reflecting the views of the Armed Forces Research Institute of Medical Sciences, the U.S. Department of the Army, the U.S. Department of Defense, or the National Institutes of Health.
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Anderson, K. B., S. Chunsuttiwat, A. Nisalak, M. P. Mammen, D. H. Libraty, A. L. Rothman, S. Green, D. W. Vaughn, F. A. Ennis, and T. P. Endy. 2007. Burden of symptomatic dengue infection in children at primary school in Thailand: a prospective study. Lancet 3691452-1459. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control, 2nd ed. World Health Organization, Geneva, Switzerland.
- 3.Bennett, S. N., E. C. Holmes, M. Chirivella, D. M. Rodriguez, M. Beltran, V. Vorndam, D. J. Gubler, and W. O. McMillan. 2003. Selection-driven evolution of emergent dengue virus. Mol. Biol. Evol. 201650-1658. [DOI] [PubMed] [Google Scholar]
- 4.Carrington, C. V., J. E. Foster, O. G. Pybus, S. N. Bennett, and E. C. Holmes. 2005. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J. Virol. 7914680-14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endy, T. P., S. Chunsuttiwat, A. Nisalak, D. H. Libraty, S. Green, A. L. Rothman, D. W. Vaughn, and F. A. Ennis. 2002. Epidemiology of inapparent and symptomatic acute dengue virus infection: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am. J. Epidemiol. 15640-51. [DOI] [PubMed] [Google Scholar]
- 6.Endy, T. P., A. Nisalak, S. Chunsuttiwat, D. H. Libraty, S. Green, A. L. Rothman, D. W. Vaughn, and F. A. Ennis. 2002. Spatial and temporal circulation of dengue virus serotypes: a prospective study of primary school children in Kamphaeng Phet, Thailand. Am. J. Epidemiol. 15652-59. [DOI] [PubMed] [Google Scholar]
- 7.Gandon, S., M. J. Mackinnon, S. Nee, and A. F. Read. 2001. Imperfect vaccines and the evolution of pathogen virulence. Nature 414751-756. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons, R. V., and D. W. Vaughn. 2002. Dengue: an escalating problem. BMJ 3241563-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubler, D. J. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10100-103. [DOI] [PubMed] [Google Scholar]
- 11.Holmes, E. C., and S. S. Twiddy. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 319-28. [DOI] [PubMed] [Google Scholar]
- 12.Kosakovsky Pond, S. L., and S. D. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 221208-1222. [DOI] [PubMed] [Google Scholar]
- 13.Kuno, G., D. J. Gubler, and N. S. Santiago de Weil. 1985. Antigen capture ELISA for the identification of dengue viruses. J. Virol. Methods 1293-103. [DOI] [PubMed] [Google Scholar]
- 14.Lanciotti, R. S., C. H. Calisher, D. J. Gubler, G. J. Chang, and A. V. Vorndam. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 16.Rosen, L., and D. J. Gubler. 1974. The use of mosquitoes to detect and propagate dengue viruses. Am. J. Trop. Med. Hyg. 231153-1160. [DOI] [PubMed] [Google Scholar]
- 17.Schreiber, M. J., S. H. Ong, R. C. Holland, M. L. Hibberd, S. G. Vasudevan, W. P. Mitchell, and E. C. Holmes. 2007. DengueInfo: a web portal to dengue information resources. Infect. Genet. Evol. 7540-541. [DOI] [PubMed] [Google Scholar]
- 18.Singh, K. R., and S. D. Paul. 1969. Isolation of dengue viruses in Aedes albopictus cell cultures. Bull. W. H. O. 40982-983. [PMC free article] [PubMed] [Google Scholar]
- 19.Swofford, D. 2003. Phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 20.Tesh, R. B. 1979. A method for the isolation and identification of dengue viruses, using mosquito cell cultures. Am. J. Trop. Med. Hyg. 281053-1059. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, C., M. P. Mammen, Jr., P. Chinnawirotpisan, C. Klungthong, P. Rodpradit, P. Monkongdee, S. Nimmannitya, S. Kalayanarooj, and E. C. Holmes. 2005. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J. Virol. 7915123-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, C., M. P. Mammen, Jr., P. Chinnawirotpisan, C. Klungthong, P. Rodpradit, A. Nisalak, D. W. Vaughn, S. Nimmannitya, S. Kalayanarooj, and E. C. Holmes. 2006. Structure and age of genetic diversity of dengue virus type 2 in Thailand. J. Gen. Virol. 87873-883. [DOI] [PubMed] [Google Scholar]