Abstract
VP1-2 is a large structural protein assembled into the tegument compartment of the virion, conserved across the herpesviridae, and essential for virus replication. In herpes simplex virus (HSV) and pseudorabies virus, VP1-2 is tightly associated with the capsid. Studies of its assembly and function remain incomplete, although recent data indicate that in HSV, VP1-2 is recruited onto capsids in the nucleus, with this being required for subsequent recruitment of additional structural proteins. Here we have developed an antibody to characterize VP1-2 localization, observing the protein in both cytoplasmic and nuclear compartments, frequently in clusters in both locations. Within the nucleus, a subpopulation of VP1-2 colocalized with VP26 and VP5, though VP1-2-positive foci devoid of these components were observed. We note a highly conserved basic motif adjacent to the previously identified N-terminal ubiquitin hydrolase domain (DUB). The DUB domain in isolation exhibited no specific localization, but when extended to include the adjacent motif, it efficiently accumulated in the nucleus. Transfer of the isolated motif to a test protein, β-galactosidase, conferred specific nuclear localization. Substitution of a single amino acid within the motif abolished the nuclear localization function. Deletion of the motif from intact VP1-2 abrogated its nuclear localization. Moreover, in a functional assay examining the ability of VP1-2 to complement growth of a VP1-2-ve mutant, deletion of the nuclear localization signal abolished complementation. The nuclear localization signal may be involved in transport of VP1-2 early in infection or to late assembly sites within the nucleus or, considering the potential existence of VP1-2 cleavage products, in selective localization of subdomains to different compartments.
The tegument compartment of herpesvirus virions is defined as the region lying between the capsid and the envelope and comprises a complex repertoire of proteins recruited in different stoichiometries (22, 29). In addition to being structural components, tegument proteins are proving to play multiple diverse roles in virus replication, including gene regulation, host cell suppression, immune evasion, virion transport, etc. While not all the tegument proteins are essential for replication in tissue culture, one tegument protein which is both conserved across the herpesvirus family and essential in those viruses where it has been examined is the very large tegument protein (VP1-2) encoded by the UL36 gene (4, 6, 14, 16, 19). VP1-2 is synthesized late during infection, dependent on viral DNA replication, and is recruited into virions at a relatively low abundance of between 60 and 120 copies per virus (10, 20, 21).
Early results indicating an essential role for VP1-2 in herpes simplex virus (HSV) came from examination of the phenotype of the temperature-sensitive mutant tsB7 (1, 14). During infection with this mutant virus at the nonpermissive temperature, no virus gene expression was observed and capsids were reported to accumulate at the nuclear pore. A second defect in tsB7 was identified from temperature shift experiments, where initial infection was at the permissive temperature, allowing entry and gene expression, followed by a shift to the nonpermissive, which resulted in a shutoff of late protein synthesis. Both defects were reported by marker rescue experiments to reside in the gene for UL36.
The essential nature of UL36 was further demonstrated by the construction of a deletion mutant (4) lacking the full-length gene (but phenocopied by growth in complementing lines). The mutant virus enters cells, but the lack of de novo-synthesized VP1-2 results in a failure to recruit other tegument proteins to the capsid and to undergo normal assembly in the cytoplasm of infected cells.
Recently, based upon studies with both HSV and pseudorabies virus (PrV), the proposal has been made that the tegument region may be qualitatively subdivided into inner and outer layers, with the inner layer being composed of those tegument proteins which are acquired first in morphogenesis during exit and which remain tightly bound throughout early phases of infection during entry (13, 22, 30, 38). VP1-2 has been proposed to be one of the inner tegument proteins, required for further recruitment of additional components and, at least from the evidence in PrV, tightly bound during entry (7, 8, 13, 18, 25). Consistent with this, visualization of the tegument-capsid structures of herpes simplex virus by cryoelectron microscopy indicated that certain tegument proteins may be selectively recruited onto capsid pentons and that one possible candidate for this interaction was VP1-2 (39), although more recent data indicate that the penton-associated density is due to UL25 (31).
The question of the site and mechanism of tegument protein recruitment is the subject of considerable investigation and some debate (5, 17, 22, 23, 27, 30, 37). With regard to recruitment within the nucleus, a defect in some stage of nuclear capsid assembly or nuclear exit in a mutant lacking a particular tegument protein would likely indicate that the protein in question was assembled onto particles in the nucleus and required for exit. Such evidence has been reported for a minority of tegument proteins, including VP1-2 (19). An alternative explanation could be that if not actually recruited to the particle, the protein was somehow required for nuclear exit, for example, by modifying the nuclear envelope.
In earlier localization studies, VP1-2 has been reported to be present in both the cytoplasm and nucleus of infected cells both by simple fractionation studies and by immunofluorescence (21). A subsequent study indicated that VP1-2 exhibited a strong perinuclear pattern of localization immediately after infection, with de novo-synthesized VP1-2 present in both perinuclear and cytoplasm accumulations, although little protein was detected in the nucleus (26). As indicated above, HSV-1 mutants lacking intact VP1-2 form DNA-filled capsids in the cytoplasm, but such capsids failed to undergo normal maturation and envelope acquisition (4). Given earlier observations on the localization of VP1-2 in the nucleus or nuclear periphery, the authors speculated that VP1-2 may be acquired as capsids exit the nucleus and required for trafficking to a cytoplasmic compartment where subsequent tegument assembly/envelopment takes place (4). For PrV, Luxton et al. (19) reported that deletion of UL36 has a more profound effect on nuclear exit, with capsids being largely restricted to the nucleus and only 8% of infected cells observed to have any cytoplasmic capsids. Fuchs et al., however, in similar studies of a PRV UL36-ve virus, reported the main defect to be in cytoplasmic maturation (6).
Recent work on the acquisition of VP1-2 in HSV has supported earlier proposals for an involvement with normal nuclear assembly of capsids which are competent to progress along the maturation pathway. In examining capsid composition of nuclear type B and type C capsids, Bucks et al. (2) demonstrated that VP1-2 is associated primarily with nuclear C capsids and proposed that this association and that of UL37 may represent initial events in the tegumentation process.
As part of our aim to understand the multiplicity of role(s) of VP1-2 and to dissect functional domains within the protein, we examine VP1-2 localization and demonstrate that VP1-2 contains a nuclear localization signal (NLS) adjacent to the N-terminal ubiquitin hydrolase domain. While not a typical NLS, the motif is highly conserved and functions very efficiently in nuclear import. The motif is also required in a functional assay for VP1-2 in complementation of a VP1-2-ve mutant of HSV-1. The potential of this motif in nuclear localization of VP1-2 in assembly and as part of a functional cleaved domain at the N terminal end of VP1-2 is discussed.
MATERIALS AND METHODS
Cells, virus, infections, and transfection.
Vero, HS30 (5), and COS cells were grown in Dulbecco's modified minimal essential medium (DMEM; Gibco) containing 10% newborn calf serum (NCS) and penicillin-streptomycin. HSV type 1 (HSV-1) strain 17 and the recombinant expressing VP26-green fluorescent protein (GFP) based on strain 17 (a gift from Gillian Elliott) were used in the infections. Infections were routinely performed at 37°C and 5% CO2 at a multiplicity of infection (MOI) of 5. After 1 h, the monolayers were washed with DMEM and incubation continued in DMEM containing 2% NCS. The UL36-ve virus, HSV-1 KΔUL36 (5), was propagated on the complementing line HS30. For expression studies using the cloned gene, cells were routinely transfected (1 μg plasmid DNA) using the calcium phosphate method as previously described (9).
Complementation assay.
Replica plates of COS cells (triplicates for each construct) were seeded into 12-well plates and transfected with 1 μg of pEGFP, as a control, pcDNA3SV5-UL36fl, or pcDNA3SV5-UL36ΔNLS. After 24 h, cells were infected with HSV-1 KΔUL36 at a MOI of 5. Unabsorbed virus was inactivated by an acid wash (40 mM citric acid, 135 mM NaCl, 10 mM KCl, pH 3.0) and the cells incubated in DMEM-10% NCS for 16 h. Cell monolayer media were harvested together. Virus was harvested from each replica plate by freeze/thawing and the viral yields determined by plaque assay on HS30 cells. To confirm equal levels of expression of VP1-2 wild type (w/t) and VP1-2 ΔNLS, one well of each test was harvested directly in sodium dodecyl sulfate (SDS) buffer and the expression levels determined by Western blotting using anti-V5 antibody and goat anti-mouse immunoglobulin G (IgG) Dylight 800 or goat anti-rabbit IgG Dylight 700 (Pierce Biotechnology). Expression was then determined using a Li-Cor Odyssey imaging system.
Construction of VP1-2-expressing plasmids.
For the expression of VP1-2 protein, we first cloned the full-length UL36 gene into pUC19 to create the plasmid pTD1. Oligonucleotides used in this work are listed together in Table 1 rather than in the body of the text. The HSV-1 17 strain purified viral genome was digested with XbaI and SpeI and the fragment containing nucleotides 69247 and 80722 isolated and inserted into pUC19 digested with XbaI to form pTD1. For the construction of an epitope (SV5)-tagged VP1-2 expression vector from pTD1, two cloning steps were necessary. First, a fragment was amplified by PCR using pTD1 as the template with oligo.NT0 (5′ and 3′) and the fragment digested with BamHI. The resulting product was then cloned into BamHI-digested pUC19 to generate the plasmid pUC19-NT0. Secondly, pTD1 was digested with BamHI and HpaI and the corresponding fragment inserted into pcDNASV5, which had been digested with the same enzymes. This intermediate plasmid was then digested with BamHI, and into it was inserted the product of BamHI digestion of pUC19-NT0. The resulting plasmid now expresses a full-length VP1-2 tagged at its N terminus with an epitope tag to facilitate detection by the monoclonal antibody against the paramyxovirus V protein, a gift from R. Randall. This expression vector was named pTD3 (pcDNASV5-UL36).
TABLE 1.
Oligonucleotides used in this work
| Oligonucleotide | Sequence |
|---|---|
| Oligo.NT0 (5′) | TACGGATCCATGGGTGGCGGAAACAACAC |
| Oligo.NT0 (3′) | GGGGGATCCGTGGGTGGGGT |
| Oligo.NT1 (5′) | TACGGATCCATGGGTGGCGGAAACAACAC |
| Oligo.NT1 (3′) | GAAGATCTTTACGCCTCGGCGACCGGCGG |
| Oligo.NT2 (5′) | GAAGATCTATGGGTGGCGGAAACAACAC |
| Oligo.NT2 (3′) | GAAGATCTTTACGCCTCGGCGACCGGCGG |
| Oligo.NT3 (5′) | GCCCTGAAGGGTACCCCACCCACG |
| Oligo.NT3 (3′) | CCGGAATTCTTAGCCGAGGCGTGTATACAG |
| Oligo.NT5 (5′) | CGCGGATCCGCCGGGCCGCCGGTCGCC |
| Oligo.NT5 (3′) | GCCCTGAAGGGTACCCCACCCACG |
| Oligo.NLS.1 | CCCAAGCTTACCATGCGGCACCGGGCACGCTACTCG |
| Oligo.NLS.2wt | CCCAAGCTTACCATGCTTCCCAAGCGCCGCCGACCCACC |
| Oligo.NLS.2mut | CCCAAGCTTACCATGCTTCCAAGCCGCCGCCGACCC |
| Oligo.NLS.3 | CCCAAGCTTACCATGACTTCGGGGGAGAAAACGAAAC |
| Oligo.NLS.4 | GAAGATCTCAGGTCTTCGACGCTGGAAGG |
| Oligo.NLS.5 | GAAGATCTGACGGGGGTTTTGCCTTTGGG |
| Oligo.NLS.6 (5′) | CCCAAGCTTACCATGAAGCGCCGCCGAAGATCTTC |
| Oligo.NLS.6 (3′) | GAAGATCTTCGGCGGCGCTTCATGGTAAGCTTGGG |
| Oligo.NLS.7 (5′) | CCCAAGCTTACCATGCTTCCCAAGCGCCGCCGACCCAGATCTTC |
| Oligo.NLS.7 (3′) | GAAGATCTGGGTCGGCGGCGCTTGGGAAGCATGGTAAGCTTGGG |
| Oligo.NLS.8 (5′) | CGGGCACGCTACTCGGCCGGCACCTGGACTCCGCCTTCCAGC |
| Oligo.NLS.8 (3′) | GCTGGAAGGCGGAGTCCAGGTGCCGGCCGAGTAGCGTGCCCG |
For the cloning of the different subregions of VP1-2, we generated a series of intermediate vectors which lack the epitope tag, followed by additional cloning to create the tagged versions. Appropriate fragments amplified by PCR using pTD1 as the template (see Table 1 for PCR fragments and corresponding oligonucleotides) were then digested with BamHI and BglII (for NT1 and NT2) or BamHI and EcoRI (for NT5). The digested products were cloned into pcDNA3 (Invitrogen) digested with the corresponding enzymes. These steps resulted in the following intermediate vectors: pcDNA3-NT1, pcDNA3-NT2, and pcDNA3-NT5. Additional intermediate vectors were obtained by further subcloning from these starting vectors. pcDNA3-NT3 was constructed by digestion of pcDNA3-NT2 with EcoRI and BamHI and insertion of the appropriate PCR fragment from pTD1. Finally, pcDNA3-NT4 was generated by internal digestion of pcDNA3-NT2 with EcoRI and BamHI and the resulting fragment subcloned into pcDNA3 digested with the corresponding enzymes.
To generate the corresponding tagged versions, we used the pcDNA3-SV5 vector with the SV5 epitope. The pcDNASV5-NT1 vector was obtained by insertion of the BamHI and EcoRI fragment from pcDNA3-NT1 into pcDNASV5 digested with the same enzymes. For the generation of pcDNASV5-NT2, we first created the vector pTrcHisB-NT2. A fragment was amplified by PCR using oligo.NT2 (5′ and 3′) and then digested with BglII and inserted into pTrcHisB (Invitrogen) digested with the same enzyme. The resulting vector was then digested with BglII and the appropriate fragment cloned into BamHI-digested pcDNASV5 to create pcDNASV5-NT2. The plasmid pcDNASV5-NT3 was constructed by digestion of pcDNASV5-NT2 with BamHI and EcoRI and insertion of the appropriate BamHI/EcoRI fragment from pcDNA3-NT3. The plasmid pcDNASV5-NT4 was created by isolation of the appropriate BamHI fragment from pcDNA-NT2, blunt ending with S1 nuclease, and subsequent digestion with EcoRV. This fragment was cloned into EcoRV-digested pcDNASV5. The plasmid pcDNASV5-NT5 was generated by insertion of the appropriate BamHI and EcoRI fragment from pcDNA3-NT5 and insertion into pcDNASV5 digested with the same enzymes. To generate C-terminal expression vectors for VP1-2 subregions, the plasmid pcDNASV5-CT1 was created by digestion of pTD3 with EcoRI and BstBI, subsequent treatment with Klenow to blunt end, and insertion into pcDNASV5 which had been digested with BamHI and treated with S1 nuclease. The plasmid pcDNASV5-CT2 was obtained by digestion of pTD3 digested with ClaI and BstBI, isolation of the appropriate fragment, blunt ending with Klenow, and insertion of the fragment into pcDNASV5 which had been digested with BamHI and treated with Klenow.
For analysis of the effect of deletion of the NLS in the context of full-length VP1-2, the internal deletion of the minimal NLS region (amino acids LPKRRRP) was performed in several linked cloning steps. First, pEGFP-UL36fl was constructed. The BamHI fragment from pTD3 (pcDNA3SV5-UL36 full length) was subcloned into the pEGFP-C1 vector digested with BglII and BamHI. The resulting plasmid was BamHI digested, and the BamHI fragment from pTD1 was subcloned into it, rendering the final pEGFP-UL36fl. The plasmid pEGFP-UL36ΔCT1 was then created by digestion of pEGFP-UL36fl with EcoRI and XbaI, blunt ending with Klenow, and religation. Direct mutagenesis within pEGFP-UL36ΔCT1 was carried out using Oligo.NLS.8 primers, the resulting pEGFP-UL36ΔCT1ΔNLS was sequenced, and the fragment encompassing the N-terminal NLS deletion (RsrII/EcoRI) was subcloned back into pcDNASV5-UL36fl digested with same enzymes to create the final pcDNASV5-UL36ΔNLS.
For the analyses of the transferability of the VP1-2 NLS, we created a set of β-galactosidase (β-Gal)-VP1-2 fusion vectors derived from the β-Gal expression plasmid pJV53, a kind gift of Gill Elliott (32). The plasmid NT4-β-gal was generated by digestion of pcDNASV5-NT4 with BspEI and subsequent Klenow treatment and then digested with HindIII, and the resulting fragment was inserted into pJV53 digested with BglII and subsequent Klenow treatment and then digested with HindIII. Regions containing the potential NLS were amplified by PCR (see Table 1 for oligonucleotides), digested with HindIII and BglII, and then inserted into the pJV53 β-Gal vector digested with the same enzymes. As a result, we created a set of different vectors as described above, named as follows: region 1-β-gal (oligonucleotides Oligo.NLS.1 and Oligo.NLS.5), region 2-β-gal (oligonucleotides Oligo.NLS.2wt and -mut and Oligo.NLS.5), region 3-β-gal (oligonucleotides Oligo.NLS.1 and Oligo.NLS.4), region 4-β-gal (oligonucleotides Oligo.NLS.2wt and -mut and Oligo.NLS.4), and region 5-β-gal (oligonucleotides Oligo.NLS.3 and Oligo.NLS.5). The fusions of regions 6 and 7 were created by insertion of synthetic oligonucleotides, which had been annealed and digested with HindIII/BglII into pJV53 digested with the same enzymes. As a positive NLS control, we used a β-Gal fusion containing the simian virus 40 (SV40) NLS (32).
The vector used for bacterial expression of the glutathione S-transferase (GST)-VP1-2 N-terminal domain fusion protein (pGEX-6P-NT1) was generated by insertion of the appropriate BamHI and EcoRI fragment from pcDNASV5-NT1 into the GST expression vector pGEX-6P-3 (Amersham) digested with the same enzymes.
Production and purification of GST fusion to the N-terminal end of the VP1-2 protein (GST-NT1).
The plasmid encoding the GST-NT1 fusion protein (pGEX-6P-NT1) was transformed into the Escherichia coli bacterial strain CodonPlus (Stratagene). GST fusion protein purification was performed according to the manufacturer's instructions. Briefly, a colony of the transformed pGST-NT1 construct was picked and grown as a starter culture in LB medium in the presence of ampicillin (100 μg/ml) and chloramphenicol (10 μg/ml) overnight at 37°C. A 400-ml culture was inoculated with 2 ml of the overnight culture and grown until an absorbance at 600 nm of 0.6 was reached. Expression was then induced by the addition of isopropyl-β-d-thiogalactoside to a final concentration of 0.1 mM. Incubation was continued for an additional 16 h at 16°C. Cultures then were centrifuged at 3,000 rpm for 20 min at 4°C and the bacterial pellet resuspended in ice-cold PBSN (phosphate-buffered saline [PBS] plus 1% NP-40 and a cocktail of protease inhibitors; Roche). Extracts were then made by ultrasonication at 4°C to ensure complete lysis and clarified at 10,000 rpm for 20 min at 4°C. The supernatants were transferred to fresh tubes and glutathione Sepharose 4B (previously washed with PBSN) added to a 50% slurry. Extracts were incubated with gentle mixing for 1 h at 4°C. The beads were then washed at least three times with PBSN. Binding and purification were monitored analytically by resuspending the beads in SDS loading buffer followed by SDS-polyacrylamide gel electrophoresis (PAGE) gel and total protein staining with Coomassie brilliant blue.
Production of antibody against the NT1 product of the VP1-2 protein.
GST-NT1 purified protein was subjected to further purification in two separate steps. First, the fusion protein was cleaved from the glutathione Sepharose column using PreScission protease (Amersham) and eluted. Briefly, after washing the beads with PBSN buffer (three washes), followed by PBS (three washes), the beads were incubated in protease cleavage buffer (PCB) (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol [DTT]). PreScission protease enzyme in PCB was then added and incubation continued overnight at 4°C. The cleaved protein was then eluted by the addition of 3 volumes of PCB and collection in 0.5-ml fractions. NT1 fractions were pooled and concentrated in Centricon tubes (cutoff, 10 kDa) and the resulting sample purified by fast-protein liquid chromatography anion exchange chromatography on a Mono-Q column equilibrated in a buffer consisting of 1 mM EDTA (pH 7.5), 20 mM Tris-HCl (pH 7.0), 50 mM NaCl, and 1 mM DTT. NT1 was purified with a salt gradient in the same buffer, eluting at approximately 400 mM NaCl. The peak fractions of NT1 (approximately 95% pure) were pooled and used for immunization of rabbits (Sigma-Aldrich) to obtain polyclonal antibody against VP1-2.
Immunofluorescence studies.
Cells were plated in 12-well plates (Costar) containing glass coverslips and infected with HSV-1 or transfected using the calcium phosphate method. At different times, the coverslips were washed with cold PBS and fixed with methanol for 5 min at −20°C. The coverslips were extensively washed with PBS and blocked with PBSB (PBS plus 10% newborn calf serum) and 0.5 mg/ml human IgG (Sigma) for 1 h at room temperature. The coverslips were then incubated with the primary antibody diluted in PBSB (45 min at room temperature), washed with PBS, and incubated with the fluorochrome-conjugated secondary antibody. After 45 min at room temperature, the coverslips were washed with PBS, dried, and mounted with Mowiol containing antifade reagent. The slides were observed using an inverted confocal microscope (LSM410; Carl Zeiss Ltd.). Images were routinely acquired using a Plan-apochromat ×63 oil immersion objective lens, numerical aperture 1.4, and zoom factors ranging from 3 to 8 of the LSM410 acquisition software.
SDS-PAGE and Western blotting.
Mock-infected and infected cells were washed in cold PBS, and total lysates were prepared by adding SDS loading buffer containing 25 mM DTT. Samples were boiled for 5 min and briefly sonicated prior to electrophoresis. Equal amounts of sample were fractionated using 3-to-8%-gradient Tris-acetate SDS-PAGE and transferred onto Immobilon-P (Millipore) membranes. Primary antibodies for immunodetection were diluted in PBST (PBS plus Tween 20 [0.1%]) containing milk (5%). Target proteins were visualized by enhanced chemiluminescence (ECL; Pierce). In addition to the anti-VP1-2 antibody generated in this work, other antibodies included ant-V5 (Invitrogen), anti-AP-1 (gamma adaptin; Sigma), and actin (Sigma).
Cellular fractionation.
Mock-infected and infected cells were washed in PBS, scraped in PBS, and pelleted at 4°C for 5 min (2,300 rpm). The cells were then lysed with hypertonic lysis buffer containing a cocktail of proteases inhibitors (Roche) (HLB) (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 2.5 mM MgCl2) plus 0.5% NP-40 for 5 min on ice. Extracts of the lysed cells were then overlaid on HLB-NP-40-10% sucrose and centrifuged at 4°C for 5 min (500 × g). Cytoplasmic fractions were collected from the top of the sucrose cushion. The nuclear fraction (pellet) was resuspended in HLB-NP-40 and underlaid a second time over HLB-NP-40-10% sucrose to raise a cleaner nuclear pellet that was finally resuspended in HLB-NP-40. Both fractions were then diluted in SDS loading buffer containing DTT for analysis by SDS-PAGE.
RESULTS
VP1-2 localization.
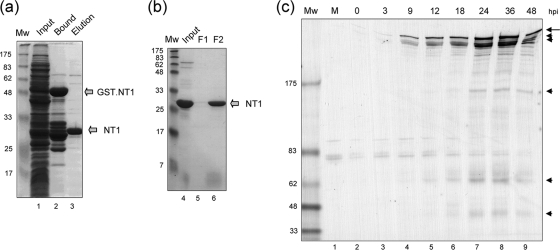
To pursue aspects of VP1-2 biochemical and cellular localization studies, we first generated an anti-VP1-2 rabbit antibody. Based on strong homology within the extreme N-terminal region, we cloned and expressed a region corresponding to the first 287 amino acids of VP1-2 (designated NT1) in frame with the gene for GST. The GST-NT1 fusion protein was purified by standard glutathione chromatography and the isolated NT1 product then eluted after cleavage using Prescission protease (Fig. 1a, lanes 1 to 3). NT1 was further purified by ion exchange chromatography on Mono Q columns to yield highly purified protein (Fig. 1b, lanes 4 to 6). Suitable fractions were then used for immunization and the production of rabbit anti-VP1-2 antiserum. The resulting polyclonal antiserum, αVP1-2NT1r, was then used first to examine the accumulation of VP1-2 by Western blotting after infection in Vero cells with HSV-1 17. The results (Fig. 1c) show that by 9 h postinfection (hpi), several specific high-molecular-weight bands could be observed, increasing in a time-dependent manner. The major high-molecular species were observed as an upper band together with several very closely migrating species, consistent with earlier results (21, 24). Although it has not been reported in previous characterizations, since many figures on VP1-2 expression usually limit presentation to the upper portions of the blots, we observed the appearance of lower-molecular-weight specific bands (the most prominent around ∼160, 62, and ∼45 kDa) which were likely to represent some level of nonspecific VP1-2 cleavage of this large protein within cells. We note also, however, that previous results (12) have indicated that VP1-2 may be specifically processed (see Discussion).
FIG. 1.
VP1-2 NT1 fragment purification and antibody generation. (a) Total protein staining of the soluble fraction of bacterial extracts expressing the GST-NT1 fusion protein (lane 1), the bound, purified GST-NT1 fusion protein (lane 2), or the final NT1 protein after specific protease cleavage and elution (lane 3). This last sample was then loaded onto a Mono Q column and fractionated as described in Materials and Methods. The Mono Q input and sample fractions are shown in panel b. Peak fractions were pooled and used to immunize rabbits. (c) Vero cells were mock infected or infected with the HSV-1 17 strain. At different times postinfection, samples were analyzed by Western blotting with the antibody raised against NT1, αVP1-2NT1r. The long arrow indicates full-length VP1-2, with cleavage products indicated by arrowheads.
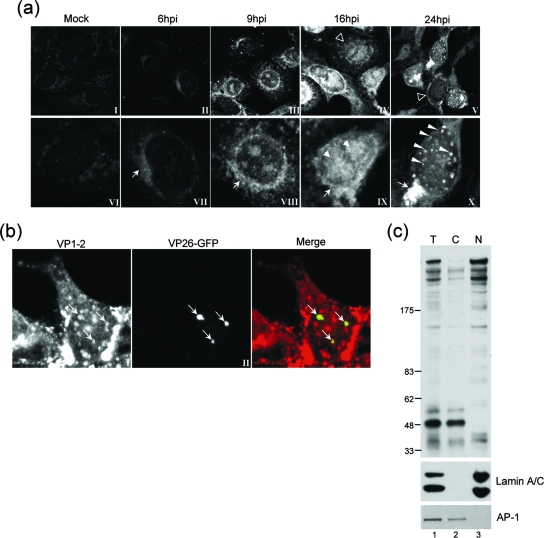
We next used the αVP1-2NT1r antibody to examine localization under similar conditions of infection. Cells were infected, fixed in methanol at different times, and stained with αVP1-2NT1r. Representative images are shown in Fig. 2a (with the lower row showing higher magnifications of the upper images). While we observed a minor background (which was difficult to eliminate by altering conditions of fixation or blocking), we detected a clear increase in signal between 6 and 9 hpi (panels II, III, VII, and VIII). VP1-2 was initially detected in a cytoplasmic perinuclear signal (more clearly seen at 9 hpi, panel VIII), progressing at later times to a combination of cytoplasmic perinuclear together with intranuclear localization (panel IX). The nuclear pattern became more obvious as infection progressed, exhibiting a heterogeneous speckled pattern, sometimes combined with distinct larger punctate foci late in infection, observed in approximately 10% of cells expressing nuclear VP1-2 (panel X). Our results are largely consistent with previous results reporting combined cytoplasmic and nuclear distribution (21).
FIG. 2.
Subcellular localization and fractionation of VP1-2 during virus infection. (a) Monolayers of Vero cells grown on coverslips were mock infected (panels I and VI) or infected with the HSV-1 17 strain (panels II to V and VII to X). At different times postinfection, coverslips were washed, fixed, and probed for VP1-2 using αVP1-2NT1r. Lower panels show higher-magnification images of the top row. Features of VP1-2 localization as discussed in the text are illustrated with different arrows, as follows: cytoplasmic perinuclear signal, arrow, e.g., panels VII and VIII; diffuse intranuclear localization, filled arrowhead, panel IX; nuclear punctate, long arrowheads. Cells exhibiting a diffuse localization are indicated by open arrowheads in the lower-magnification images in panels IV and V. (b) Monolayers of Vero cells were infected with a recombinant virus HSV-1 strain 17 expressing VP26-GFP and cells examined (18 hpi) for VP26 localization by GFP fluorescence and VP1-2 by immunostaining with αVP1-2NT1r. (c) Monolayers of Vero cells were infected with the HSV-1 17 strain harvested at 18 hpi and fractionated as described in Materials and Methods. Total extract (T) and cytoplasmic (C) and nuclear (N) fractions were separated on a 3 to 8% SDS-PAGE gradient gel and subsequently blotted with the specific anti-VP1-2 antibody. Fractionation was assessed using anti-lamin A/C and anti-AP-1 antibodies (lower panel) as markers for nuclear and Golgi compartments, respectively.
Punctate foci on the periphery of replication compartments have been reported in many studies, and these foci have been proposed as “assemblons” or otherwise as potential sites of assembly (11, 15, 16, 36). We therefore wished to examine colocalization with one of the markers of these foci, such as VP26, and to this end we infected cells with a recombinant virus expressing VP26-GFP and analyzed VP1-2 localization in relation to VP26. The results (Fig. 2b) demonstrate that the bright VP26-positive foci contain VP1-2 (panels I, II, and merged). Similar results were obtained by staining with anti-VP5 antibody (data not shown). Analysis of numerous fields indicated that in most cells (>90%) which contained VP1-2 in nuclear foci, a population of the foci colocalized with VP26-GFP. However, a distinct population of VP1-2-positive foci was also observed lacking discernible VP26. The possible relevance of this heterogeneity is discussed below.
To confirm the presence of VP1-2 in the nucleus and to clarify that the signal was due to the presence of the full-length product of the protein, we analyzed VP1-2 distribution by fractionation of infected cells into a detergent-soluble cytoplasmic fraction and a nuclear fraction and examination of partitioning by Western blotting (Fig. 2c). The results indicate full-length VP1-2 was found in the nuclear fraction. Control studies (Fig. 2c) demonstrated little or no segregation of nuclear material (e.g., lamin A/C) in the cytosolic fraction, nor of a typical Golgi marker (e.g., AP-1) with the nuclear fraction. We note that the ratio of the smaller VP1-2 products increased during fractionation, perhaps by nonspecific proteolysis, though this could not be prevented by inclusion of protease inhibitors. Intact VP1-2 may therefore be lost from cytosolic (and-or nuclear) fractions. What was clear, however, was that intact VP1-2 was detected in the nuclear fraction.
A conserved N-terminal NLS.
To pursue determinants which may be involved in VP1-2 localization, we examined the amino acid sequence, noting a single motif close to the N terminus (position 425). This motif, while not conforming to the consensus single or bipartite NLSs, nevertheless contained a run of basic amino acids and was very well conserved across the VP1-2 homologues in all herpesvirus family classes (see Fig. 4a). We constructed a series of vectors to express either the full-length protein or various N-terminal or C-terminal regions (as summarized in Fig. 3a) and examined localization by transfection and immunolocalization (Fig. 3b).
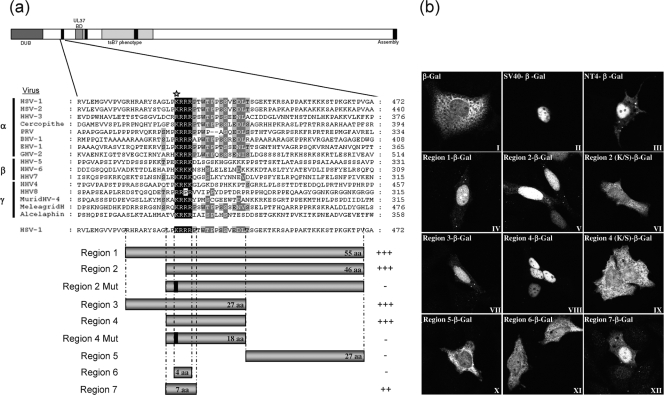
FIG. 4.
A functional NLS in VP1-2 transferable to a heterologous protein. (a) Schematic diagram illustrating the alignment of the N-terminal motif of VP1-2 from the three different herpesvirus families. Regions encompassing the highly conserved basic region of HSV VP1-2 were fused to the β-Gal gene, as illustrated in the bar diagrams below. Region 2 and region 4 were also analyzed, with a single substitution in a conserved lysine (K→S), represented as a solid line within the corresponding schematic and by a star on the sequence line. (b) Monolayers of Vero cells were transfected with the plasmids expressing the different regions of VP1-2 NT4 as β-Gal fusion proteins. Cells were fixed with methanol 24 h later and analyzed for β-Gal localization. Representative images are shown for each fusion region. Summary conclusions on the efficiency of the candidate regions are illustrated next to the schematic in a semiquantitative manner: +++, very efficient nuclear localization (at least as efficient as that of the SV40 NLS-β-gal vector); ++, mainly nuclear localization but also some weaker cytoplasmic distribution of the signal; -, no significant nuclear accumulation.
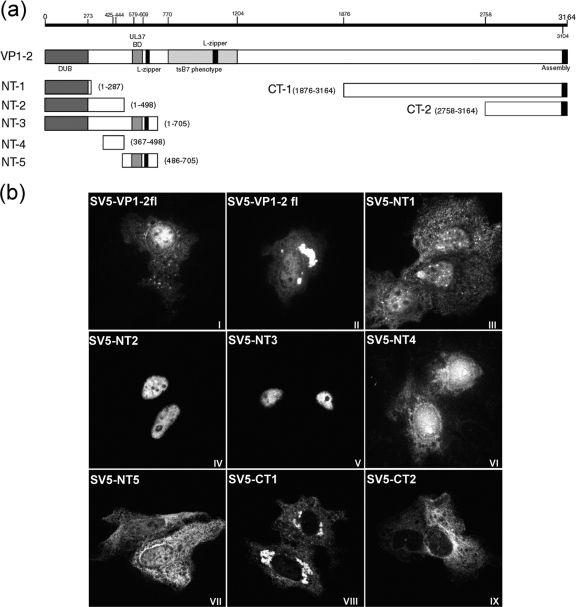
FIG. 3.
Cellular localization by immunofluorescence of transfected VP1-2 and VP1-2 fragments. (a) Schematic illustration of HSV-1 17 strain VP1-2 indicating positions of certain published features (including information on the PrV homologue), from the N-terminal end (NT) to the C-terminal end (CT): the deubiquitination enzymatic domain (DUB) (1 to 273), the putative NLS (425 to 444) region from the present work, the UL37 binding domain (UL37 BD) (579 to 609), putative leucine zipper sequences (L-zipper), a SalI fragment mapped as the tsB7 phenotype (770 to 1204), and the CT end involved in nuclear assembly (3104 to 3164). Subregions expressed from the various constructs are illustrated, including amino acid boundaries. (b) Monolayers of Vero cells were transfected with the plasmids showed in panel a. The cells were fixed with methanol 24 h after transfection and analyzed for localization using the monoclonal antibody against the SV5 N-terminal epitope tag. Representative images are shown for each fragment, including two examples of VP1-2 full length (VP1-2fl).
Full-length VP1-2, when expressed in isolation, exhibited a mixed pattern of localization, including majority cytoplasmic or diffuse cytoplasmic and nuclear (panels I and II). In many cells, pronounced accumulations of VP1-2 could also be detected. The shortest N-terminal product, NT1 (residues 1 to 287), which encompasses the previously characterized ubiquitin hydrolase domain, exhibited little specific localization with a diffuse cytoplasmic and nuclear speckled signal (panel III). Extension of the N-terminal region to include residues 1 to 498 (NT2) now resulted in a product which exhibited virtually exclusively nuclear localization (panel IV). Efficient nuclear accumulation was also observed with the 705-residue protein encoded by NT3 (panel V). In contrast, the C-terminal fragments CT1 (1,290 residues) and the shorter 406 residue protein CT2 showed exclusive cytoplasm distribution (panels VIII and IX).
While the NT1 product may be small enough to diffuse into the nucleus and thus with no specific signal be observed throughout the cell, the exclusively nuclear localization of the larger NT2 and NT3 proteins indicated that there may be a specific localization signal within the region 287 to 498. To pursue this, we constructed two additional vectors, expressing residues 367 to 498 (NT4) or 486 to 705 (NT5). Consistent with the proposal for a specific localization signal, NT4 again exhibited pronounced nuclear localization (panel VI) while NT5 exhibited little specific localization, being present in a diffuse pattern throughout cytoplasm and nucleus (panel VII). Taken together, the results indicated the presence of an NLS within the region of 367 and 498 and the possibility that the basic motif identified and conserved at residue 425 was involved.
The defining feature of an NLS is that it is able to confer nuclear import on a large protein which would otherwise not be transported to the nucleus. To further study the role of the basic motif at position 425, we generated a series of constructs in which different short overlapping sections around this position were fused in frame to the β-Gal protein (Fig. 4a). Cells, in this case COS cells, were then transfected and localization of the different β-Gal fusion constructs examined by immunofluorescence (Fig. 4b). As a control, we used the prototypical NLS from SV40 T-antigen (panel II), which clearly resulted in efficient and almost exclusively nuclear localization compared to the abundant cytoplasmic localization seen with parental β-Gal alone (panel I). Constructs containing the entire NT4 region showed prominent nuclear accumulation (panel III), as did the subregions, region 1 (panel IV), region 2 (panel V), region 3 (panel VII), and region 4 (panel VIII). Region 4 encompassed the short (18-amino-acid) conserved region, including the run of basic residues adjacent to a section enriched in proline and serine or threonine residues (PT/S section). Interestingly, while this latter part of the region was well conserved throughout the alphaherpesvirus VP1-2 proteins, it was lacking in the members of the beta- and gammaherpesvirus homologues. To examine specificity of the NLS function within the region, we mutated the lysine residue at the start of the basic run to serine in the context of either region 2 or region 4 (Fig. 4a). In both cases this single substitution completely abolished the NLS function of the corresponding region (Fig. 4b, panels VI and IX).
To refine determinants involved in the NLS function, we examined in isolation the region (region 5) immediately C-terminal to the conserved motif, observing no specific localization function (panel X), indicating that the NLS activity was contained within region 4 (panel VIII). However, region 7 (panel XII), encompassing the motif LPKRRRP, exhibited a definite localization function, though not as efficiently as region 4. Interestingly, deletion of the flanking prolines in region 4 abolished NLS function (region 6, panel XI). Taken together, the results indicate that the conserved LPKRRRP region functions as an NLS but that extension to encompass the adjacent PT/S region enhances NLS function and that the initial basic residue of the string is critical to function.
Function of the conserved NLS in the context of full-length VP1-2.
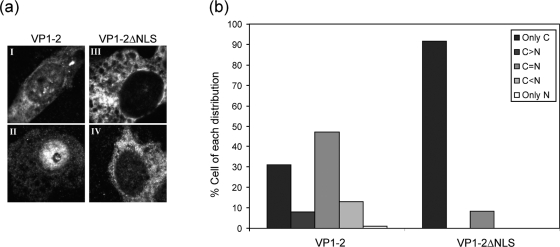
Having demonstrated that the motif was a transferable, functional NLS, we next wished to examine its role in the context of full-length VP1-2. We constructed a version of VP1-2 lacking seven residues, LPKRRRP, defined in Fig. 4 as the core NLS, and compared localization with that of the w/t protein after transient expression. As indicated in Fig. 3, full-length VP1-2 exhibited a mixed pattern of localization, including majority cytoplasmic or diffuse cytoplasmic plus nuclear, and in many cells pronounced accumulations of VP1-2 could also be detected in the nucleus (Fig. 5a, panels I and II). The distribution of VP1-2 into localization categories is shown in panel b. Deletion of the NLS had a very marked effect. The majority of cells exhibited a completely cytoplasm-restricted localization, with only a small minority of cells (10%) showing some nuclear presence together with the cytoplasmic presence. Representative fields are shown in Fig. 5, panels III and IV, and the distribution is summarized in panel b.
FIG. 5.
Effect of NLS deletion on localization of VP1-2. (a) COS cells were transfected with 1 μg of the pcDNA3-SV5-UL36fl (panels I and II) or pcDNA3-SV5-UL36ΔNLS (panels III and IV) expression vector. After 24 h, the coverslips were fixed and VP1-2 localization examined using the monoclonal antibody against the V5 tag. Panel b shows the distribution of the percentages of cells with localization patterns of VP1-2 or VP1-2ΔNLS, defined as follows: only cytoplasmic (C) or only nuclear (N); equal distribution in both compartments, C = N; cytoplasmic more abundant than nuclear, C>N; nuclear more abundant than cytoplasmic, C<N. At least 100 cells were counted. VP1-2 distribution was mixed, with a significant percentage of cells showing cytoplasmic and nuclear distribution or nuclear distribution greater than cytoplasmic. The effect of the NLS deletion was clear, with the majority of cells exhibiting cytoplasm-restricted localization.
The NLS in VP1-2 is required for functional complementation of a UL36-ve virus.
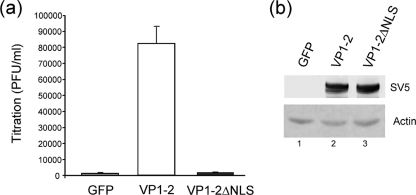
We next wished to address the physiological relevance of the NLS motif in the context of virus infection. To examine this, we established a complementation assay, using the UL36-ve virus HSV-1 KΔUL36 (5). This virus lacks the majority of the UL36 open reading frame and fails to grow on noncomplementing cell lines (5). Noncomplementing cells (COS cells) were transfected with a control plasmid or VP1-2 or VP1-2 ΔNLS expression plasmid and subsequently infected with HSV-1 KΔUL36 at a MOI of 5. Any virus not entering cells was inactivated by a low-pH wash, and the cells were incubated for a further 16 h. The cells were then harvested and virus yields determined on complementing HS30 cells. Expression of w/t VP1-2 increased virus yields by almost 50-fold, while expression of the VP1-2 ΔNLS had no measurable effect on complementation and resulted in yields not significantly above background levels (Fig. 6a). A background for this assay is likely unavoidable and was detectable due to the fact that the stock has to be grown on complementing lines resulting in a minor population of recombinant virus as originally described (5). The results were clear and striking and were not attributed to any difference in overall expression levels of the w/t versus ΔNLS versions. Expression levels were assessed and shown to be equivalent (Fig. 6b). These results strongly indicate that the motif, which we demonstrate here to be conserved, functional, and transferable, is required for a physiologically relevant role of VP1-2 during the virus life cycle.
FIG. 6.
The VP1-2 NLS is required for complementation of HSV-1ΔUL36 virus. (a) COS cells were transfected in triplicate with 1 μg of the pEGFP, pcDNA3-SV5-UL36fl, or pcDNA3-SV5-UL36ΔNLS vector. After 24 h posttransfection, the cells were infected with HSV-1 KΔUL36 at 5 PFU/cell and unabsorbed virus was inactivated by an acid wash (40 mM citric acid, 135 mM NaCl, 10 mM KCl, pH 3.0). At 16 h after infection, cells and media were harvested together, lysed by freeze/thawing three times, and the viral yield determined by plaque assay on HS30 cells. Results are plotted as virus yield in PFU/ml with the average and standard deviation for each series. (b) In parallel, cells were examined for expression levels by Western blotting of both proteins, VP1-2 and VP1-2ΔNLS, as described in Materials and Methods. Actin levels were determined as well as a loading control.
DISCUSSION
VP1-2, encoded by the UL36 gene, is conserved across the herpesvirus family and is essential in those viruses where it has been examined (1, 4, 6, 14, 16, 19). In HSV, deletion of VP1-2 results in a failure to undergo normal assembly in the cytoplasm of infected cells (4), while in PrV, deletion of UL36 appears to affect not only cytoplasmic assembly (6) but also nuclear exit, with mutants exhibiting pronounced accumulation of nuclearly restricted capsids (19). VP1-2 has been reported to be present in both the cytoplasm and nucleus of infected cells (21), while a subsequent study reported perinuclear and cytoplasmic accumulation, although with little protein in the nucleus (26). In PrV, immunoelectron microscopy has indicated that VP1-2 is tightly bound to incoming capsids, but the protein was not found on intranuclear capsids (14), although the general distribution of PrV VP1-2 by immunofluorescence has not been reported. A recent report demonstrated that VP1-2 was present on nuclear C capsids and indicated that the protein was likely recruited onto capsids within the nucleus, requiring that the protein directly or by association localized to the nucleus (2), although this is not consistent with the data of Trus et al. (31), where VP1-2 was not found to be associated with nuclear C-capsids.
In this work we examined HSV VP1-2 localization and in particular identified a conserved motif with properties of a potent NLS. We developed an antibody to VP1-2 and detected the protein in both the cytoplasms and nuclei of infected Vero cells, consistent with an earlier report (21). Within the cytoplasm, VP1-2 was frequently present in perinuclear vesicular aggregates, and while this was suggestive, it was difficult to precisely colocalize this with Golgi, since we found dispersal or loss of Golgi markers (data not shown), as previously described (35). Within the nucleus, VP1-2 was observed both in a heterogeneous diffuse pattern typical of large lobular replication compartments and in punctate foci on the periphery of these compartments. The latter foci colocalized in part with foci of VP26-GFP in infected cells. Although VP1-2 localization has been examined by immunoelectromicroscopy, there are comparatively few reports on the localization of the intact protein in relation to other capsid components. VP26 localization in punctate foci on the periphery of replication compartments has been reported in many studies, and these foci have been proposed as “assemblons” or otherwise as potential sites of assembly. Therefore, the colocalization of VP1-2 with VP26 capsid component could reflect recruitment or assembly of VP1-2 to the capsid, a conclusion consistent with the recent observation by biochemical fractionation and protein separation of a VP1-2 presence in isolated nuclear capsids (2). In this regard, we provide several lines of evidence for the presence of a potent N-terminal nuclear localization in VP1-2, including its strong sequence conservation, the nuclear accumulation of N-terminal regions accommodating the motif, its transferability to β-Gal, and the effect of its deletion on the localization patterns of intact VP1-2. Thus, the NLS may play a role in VP1-2 localization to its site of recruitment. We note that the NLS, together with the N-terminal ubiquitin hydrolase domain (DUB) and C-terminal region (see below), is extremely well conserved throughout the VP1-2 family members. Furthermore, in PrV, deletion of a 70-amino-acid region that included the conserved NLS motif completely abolished replication (16). This is consistent with results in this report which demonstrate that a refined deletion of just seven residues encompassing the core NLS completely abrogates the ability of the protein to complement growth of a VP1-2-ve mutant. Together with the results showing localization of the protein and demonstrating NLS function of the conserved motif, our results make a compelling case that this motif is an NLS playing a critical role in VP1-2 function. However, as discussed below, other possibilities remain in dissecting the complex roles of VP1-2.
The NLS motif is conserved in both position and sequence just adjacent to the N-terminal domain that has been previously reported to be a ubiquitin hydrolase domain (12). While the isolated N-terminal ubiquitin hydrolase domain exhibited a generalized distribution in both the cytoplasm and nucleus, inclusion of the conserved motif (position 425) resulted in highly efficient nuclear restricted localization. It is interesting that in the original identification of the ubiquitin hydrolase in HSV-infected cells, the activity was identified in an N-terminal product, not the full length (12). Although subsequently (34), DUB activity was found in the full-length protein (and the N-terminal product), there remains a possibility that VP1-2 is processed and that the NLS could be involved in localization of subdomains encompassing the ubiquitin hydrolase. Finally, as noted earlier, VP1-2 may have a role in the earliest phases of infection. This was proposed from the observations that a temperature-sensitive defect in the protein results in capsid accumulation at the nuclear pore (1, 14). Consistent with a role at this stage, VP1-2 has been shown to be tightly bound to the capsid (10, 13, 21, 22, 30, 38), and previous work has demonstrated an in vitro association of capsids with nuclear pores and that this requires importin β (28). It remains possible, therefore, that the NLS could be involved in VP1-2 function in early phases of nuclear pore interaction in addition to a role in import of the intact de novo protein synthesized later in infection. Although the defect in VP1-2 in tsB7 was mapped to a fragment more C-terminal to the NLS, it is possible that presentation of the N-terminal region and NLS is affected by the temperature-sensitive conformation.
It also remains possible that VP1-2 localization involves other interactions, and indeed, VP1-2 has been shown to interact with a number of other virus components, including UL37 (13, 33), VP5 (21), and in PrV, UL25 (3). In particular, UL25 has been reported to interact with the C terminus of VP1-2, and relocation of a C-terminal fragment of VP1-2 from the cytoplasm to the nucleus has been reported to be dependent upon UL25 (3). If this interaction with the C terminus relates to intact VP1-2, it is possible that both determinants are involved in VP1-2 trafficking, or if the C-terminal UL25 binding domain served as the main determinant of VP1-2 localization, it is possible that the NLS has other functions as indicated above.
Finally we note that while the NLS appears to be required for some aspect of the physiologically relevant roles of VP1-2, this does not address the question of nuclear capsid recruitment, and the role of the NLS could be in entry or some other aspect of assembly or both. The complementation assay, while confirming a critical role of the NLS, does not discriminate between these possibilities. Since any HSV UL36 deletion mutant will need to be grown on complementing lines, the mutant will have the w/t protein assembled into its virion and supporting entry. However, we have now incorporated a VP26-GFP protein into the backbone of tsB7, allowing visualization of the incoming capsid in the presence of the defective VP1-2 on the tsB7 virion. Using this reagent, we hope in future studies to establish assays to refine the importance of the NLS demonstrated in this work to one or more of the aspects of VP1-2 in the virus life cycle.
Acknowledgments
We thank Tohru Daikoku for contributions in the early phase of this work in the initial cloning of UL36, Gill Elliott for test plasmids and VP26-GFP-expressing HSV-1, and Prashant Desai for the HSV-1 KΔUL36 virus and complementing cell line.
This work was funded by Marie Cure Cancer Care. F.A. received a fellowship from the Spanish Ministerio de Educacion y Ciencia.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucks, M. A., K. J. O'Regan, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coller, K. E., J. I. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1-2 proteins. J. Virol. 8111790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 7411608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 734110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 7811879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 101044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 793200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaves, R. F., and P. O'Hare. 1990. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding protein and target TAATGARAT sequences. J. Virol. 642716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus XII. The virion polypeptides of type 1 strains. J. Virol. 14640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 7610365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19547-557. [DOI] [PubMed] [Google Scholar]
- 13.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 763065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.La Boissiere, S., A. Izeta, S. Malcomber, and P. O'Hare. 2004. Compartmentalisation of VP16 in cells infected with a recombinant herpes simplex virus expressing VP16-green fluorescent protein fusions. J. Virol. 788002-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 8012086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leuzinger, H., U. Ziegler, E. M. Schraner, C. Fraefel, D. L. Glauser, I. Heid, M. Ackermann, M. Mueller, and P. Wild. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 7913047-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 1025832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luxton, G. W., J. I. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNabb, D. S., and R. J. Courtney. 1992. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 667581-7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190221-232. [DOI] [PubMed] [Google Scholar]
- 22.Mettenleiter, T. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mettenleiter, T. C., and T. Minson. 2006. Egress of alphaherpesviruses. J. Virol. 801610-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michael, K., S. Bottcher, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2006. Pseudorabies virus particles lacking tegument proteins pUL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J. Gen. Virol. 873503-3507. [DOI] [PubMed] [Google Scholar]
- 25.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 801332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison, E. E., Y. F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 727108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naldinho-Souto, R., H. Browne, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 802582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ojala, P. M., B. Sodeik, M. W. Ebersold, U. Kutay, and A. Helenius. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 204922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rixon, F. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4135-144. [Google Scholar]
- 30.Smith, G. A., and L. W. Enquist. 2002. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18135-161. [DOI] [PubMed] [Google Scholar]
- 31.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol. Cell 26479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verhagen, J., I. Hutchinson, and G. Elliott. 2006. Nucleocytoplasmic shuttling of bovine herpesvirus 1 UL47 protein in infected cells. J. Virol. 801059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 799566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 806003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward, P. L., E. Avitabile, G. Campadelli-Fiume, and B. Roizman. 1998. Conservation of the architecture of the Golgi apparatus related to a differential organization of microtubules in polykaryocytes induced by syn- mutants of herpes simplex virus 1. Virology 241189-199. [DOI] [PubMed] [Google Scholar]
- 36.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 704623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Mueller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 791071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfstein, A., C. H. Nagel, K. Radtke, K. Dohner, V. J. Allan, and B. Sodeik. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7227-237. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, H.-Z., D. H. Chen, J. Jakana, F. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type I virions. J. Virol. 733210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]