Abstract
In this study, we report the characterization of a novel calicivirus (CV), the Tulane virus (TV), which was isolated from stool samples of captive juvenile rhesus macaques (Macaca mulatta) of the Tulane National Primate Research Center. The complete genome of TV contains 6,714 nucleotides plus a poly(A) tail and is organized into three open reading frames (ORFs) that encode the nonstructural (NS) polyprotein (ORF1); the capsid protein (ORF2), with an estimated molecular mass of 57.9 kDa; and a possible minor structural protein (ORF3), with an isoelectric point (pI) of 10.0 and a calculated molecular mass of 22.8 kDa. The NS polyprotein revealed all typical CV amino acid motifs, including GXXGXGKT (NTPase), EYXEX (Vpg), GDCG (protease), and GLPSG and YGDD (polymerase). Phylogenetic trees constructed for the NS polyprotein, NTPase, protease, polymerase, and capsid protein sequences consistently placed the TV on a branch rooted with Norovirus, but with distances equal to those between other genera. The TV can be cultured in a monkey kidney cell line (LLC-MK2) with the appearance of typical cytopathic effect. TV exhibits a typical CV morphology, with a diameter of 36 nm, and has a buoyant density of 1.37 g/ml. According to these physicochemical and genetic characteristics, TV represents a new CV genus for which we propose the name “Recovirus” (rhesus enteric CV). Although the pathogenicity of TV in rhesus macaques remains to be elucidated, the likelihood of TV causing intestinal infection and the availability of a tissue culture system make this virus a valuable surrogate for human CVs.
The family Caliciviridae consists of four genera, Norovirus (NV), Sapovirus (SV), Lagovirus, and Vesivirus (11, 12, 16). The recent genomic characterization of unique bovine enteropathogenic caliciviruses (CVs) (Newbury agent-1 and Nebraska) revealed that these viruses represent a distinct fifth genus with the proposed name Becovirus or Nabovirus (26, 27, 30). CVs cause a wide spectrum of diseases in animals, including respiratory infections, vesicular lesions, gastroenteritis, and hemorrhagic disease. NVs and SVs are important etiologic agents of acute gastroenteritis in humans and therefore are also referred as human CVs (HuCVs). Based on phylogenetic analysis, both HuCV genera are further divided into five genogroups (GI to GV) and several genetic clusters or genotypes (9, 42). Viruses genetically and antigenically closely related to HuCVs have also been isolated from animals (6, 18, 21, 36), which has raised a concern about CV gastroenteritis as a zoonotic disease and the role of animals as reservoirs for HuCVs. Since there is no effective tissue culture system or animal model available for HuCVs, animal CVs are often used as surrogates to model HuCV stability in the environment (7, 8, 37), replication (2, 3), and pathogenesis (40). Tissue culture propagation of an animal SV (porcine enteric CV [PEC]) and the murine NV (MNV-1) have previously been reported (28, 39). Recently, a gnotobiotic pig model for NVs has also been developed (4, 5).
CVs are small, nonenveloped, icosahedral viruses with a positive-sense, single-stranded, polyadenylated RNA genome (of about 7.5 to 8.5 kb). The Lagovirus, SV, and Becovirus genomes are organized into two open reading frames (ORFs); ORF1 encodes the nonstructural (NS) proteins and the capsid protein, while the NV and Vesivirus genomes contain three ORFs, where the capsid protein is encoded by ORF2, a separate ORF. The translated proteins of all CVs contain several conserved amino acid (aa) motifs that are similar to those of picornaviruses (23): 2C helicase/NTPase (GXXGXGKS/T), 3C protease (GDCG), and RNA-dependent RNA polymerase (RdRp; GLPSG and YGDD). The ORF closest to the 3′ end of the CV genome (ORF2 for Lagovirus, SV, and Becovirus and ORF3 for NV and Vesivirus) encodes a basic protein (pI, ∼10.0) that has been reported to be a minor structural protein for feline CV and the Norwalk virus (10, 34).
The only CV (Pan-1) that has been isolated from nonhuman primates belongs to the Vesivirus genus (31). Several studies have been conducted for the development of a primate animal model for studying HuCV disease. Chimpanzees inoculated with the Norwalk virus developed subclinical infections, with seroresponses and virus shedding but no vomiting or diarrhea (41). In a more recent study, Subekti et al. reported the development of clinical illness characterized by diarrhea, dehydration, vomiting, and viral shedding in newborn pigtail macaques inoculated with the Toronto virus (35). Recently, Rockx et al., evaluated the susceptibility of four different Old World monkey species to NVs (29) and found that rhesus macaques might be susceptible to NV infection, since one of the three animals inoculated with the Norwalk virus shed the virus for up to 19 days postinoculation and developed specific immunoglobulin M and immunoglobulin G responses. In the above-mentioned study, serological evidence of natural NV infection could not be demonstrated among different monkey species; however, Jiang et al. previously reported a high prevalence of anti-NV antibodies (both GI and GII) in serum samples collected from nonhuman primate species, including rhesus macaques (14).
In this study, we report the isolation and characterization of a novel, tissue culture propagable CV, the Tulane virus (TV), which was isolated from stool samples of rhesus macaques and represents a new genus within the Caliciviridae.
MATERIALS AND METHODS
Stool samples.
One hundred twenty stool samples were collected from 58 randomly selected macaques between April 2004 and January 2006, including animals with (n = 20) and without (n = 38) clinical histories of diarrhea. These stool samples were tested for the presence of CVs by reverse transcription-PCR (RT-PCR). The animals were housed in the nursery of the Tulane National Primate Research Center, under biosafety level 2 conditions in accordance with the standards of the Association for Assessment and Accreditation of Laboratory Animal Care. Investigators adhered to the Guide for the Care and Use of Laboratory Animals prepared by the National Research Council. All samples were handled in compliance with biosafety level 2 laboratory practices approved by the Institutional Biosafety Committee of Cincinnati Children's Hospital Medical Center.
Detection of CV RNA in stool specimens by RT-PCR.
Total RNA was extracted from 100 μl of 10% (wt/vol) stool specimens with the TRIzol reagent (Gibco BRL, Gaithersburg, MD) according to the manufacturer's instructions. Precipitated RNA was resolved in 20 μl of molecular-biology-grade water (Eppendorf AG, Hamburg, Germany), and 1 μl was used as the template in the RT-PCR. The remainder of the samples was stored at −70°C for repeated assays. RNA was amplified in 25-μl reaction mixtures by using the AccessQuick RT-PCR system (Promega, Madison, WI) according to the manufacturer's protocol. Primers P289H,I/P290H,I,J,K targeting conserved sequences in the RdRp region were used (9, 15). These primers are able to amplify both NVs and SVs. RT was performed at 48°C for 45 min, followed by a 3-min denaturation step at 94°C and 35 PCR cycles of denaturation for 30 s at 94°C, annealing for 1 min at 49°C, and elongation for 1 min at 72°C, followed by a final extension of 10 min at 72°C. Twelve microliters of each reaction mixture was analyzed on 1% agarose gels in the presence of ethidium bromide.
Amplification and sequence analysis of the TV genome.
The primers used in this study are listed in Table 1. Based on the sequence data obtained from the short RdRp amplicon, a TV-specific primer (P776) was designed and used with an oligo(dT) primer to obtain an approximately 3-kb amplicon, including the 3′ end of ORF1, the complete ORF2 and ORF3, and the 3′ nontranslated region of the genome. Extended amplification of the genome toward the 5′ end has been achieved by TV-specific reverse primers that were designed based on newly obtained sequences and degenerated forward primers targeting the GDCG (P774) and the GXXGXGKT amino acid motifs (P802) conserved in CV proteases and NTPases, respectively. The 5′-end sequence of the genome was determined by the 5′ rapid amplification of cDNA ends system (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations by using TV-specific primers in combination with the abridged anchor primer and abridged universal primer and viral RNA extracted from tissue-culture-adapted, CsCl-purified TV as the template. Both homopolymeric deoxycytidine-tailed and deoxyadenine-tailed cDNAs were made to determine the exact 5′ end. Additionally, the end ligation of proteinase-K-treated viral RNA with T4 RNA ligase I (New England Biolabs, Ipswich, MA) was also performed according to the method described previously by Mandl et al. (20), with slight modifications. Briefly, 1 μg extracted viral RNA was digested with proteinase K (100 μg/ml proteinase K, 30 mM Tris-Cl [pH 8.0], 30 mM EDTA, 0.5% sodium dodecyl sulfate [SDS]) at 55°C for 1 h, recovered by using an RNeasy mini column (Qiagen, Valencia, CA), and ligated with T4 RNA ligase I (New England Biolabs, Inc., Ipswich, MA) at 4°C overnight. The ligated RNA was recovered on an RNeasy mini column and used for cDNA synthesis with reverse primer P1080. First-round PCRs were performed with primers P1068 and P1080, and a second-round PCR was performed with primers P1035 and P1082, which flanked the ligated 5′-3′ end junction of the genome. The PCR yielded a smeared product, with fragments of approximately 400 to 500 bp, which were gel purified and ligated into the pGEM-T vector (Promega, Madison, WI). Clones were PCR screened with M13 R/F primers, and clones with the longest inserts were selected for sequencing.
TABLE 1.
List of primers used in the study
| Name | Position or motifa | Polarity | Sequence (5′ to 3′) |
|---|---|---|---|
| P776 | 3724-3741 | + | GCAAGGTCATCGTCAACG |
| P774b | GDCG | + | CCACACCAGGWGAYTGYGG |
| P802b | GXXGXGKT | + | GGCCMCCCKGGIWKIGGIAA |
| P1080 | 510-489 | − | GGAGTTGTTGTGTCCATCCGC |
| P1068 | 6360-6400 | + | CAAGTGGGATACAAACTATGCACTCTGCCCTTTCTGCTAAG |
| P1035 | 6568-6588 | + | TAGTGGTTCTCTGAGTAGGTT |
| P1082 | 272-242 | − | GCCCCAGGGATGGGTCAGGCCATTGAGACGG |
| P1F | 5-26 | + | GACTAGAGCTATGGATACGTCC |
| P1R | 1479-1497 | − | TTGCCATACGAGCATTCCC |
| P2F | 1214-1236 | + | TTGGCTCAACACTGTGCAAAAG |
| P2R | 2998-2979 | − | TACCAAATCTGGGTGGTCTC |
| P3F | 2397-2414 | + | CCCAGTGATGATTATTAC |
| P3R | 4358-4338 | − | TCACAAGAATCCAGAACAACC |
| P4F | 4239-4258 | + | ATTAAAGACAAGTGTGCTCG |
| P4R | 5993-5973 | − | CGCCAGCCATTATCTAAAGAC |
| P5F | 5656-5674 | + | CTTCAAAAACCACGACTAC |
| P5Rc | Poly(A) tail | − | AGCGTCGAGCGGCCGC(T)15 |
| P885 | 1214-1236 | + | TTGGCTCAACACTGTGCAAAAG |
| P886 | 1737-1716 | − | CTGATTGCATTTTCCAAACAGC |
Position nos. or motif of the 5′ and 3′ nucleotides on the TV genome.
Primers targeting conserved amino acid motifs in the protease or NTPase, respectively.
Oligo(dT) primer with a NotI site overhang.
Finally, the full TV genome was reamplified as five overlapping fragments from an RNA template extracted from a TV-positive stool sample using primer pairs P1F/P1R to P5F/P5R (Table 1). Amplicons were gel purified and sequenced directly in both directions (ABI Prism 3730 DNA analyzer; Applied Biosystems, Foster City, CA). The full genomic sequence of the wild-type TV was deposited into GenBank under the accession number EU391643.
Phylogenetic analysis.
Sequences (published in the GenBank) used in the phylogenetic analysis were of the following full-length CVs (accession nos. are shown in parentheses): for NV, Norwalk/68 (M87661), Southampton (L07418), MD145 (AY032605), and MNV-1 (DQ285629); for SV, Manchester/93 (X86560), and PEC-Cowden (AF182760); for Vesivirus, SMSV-1 (U15301), FCV-CFI68 (U13992), and FCV-Urbana (NC_001481); for Lagovirus, EBHSV-GD (NC_002615) and RHDV-FRG (M67473); and for Becovirus, BEC-NB (NC_004064) and Newbury-1 (DQ013304). Multiple alignments of nucleotides (nt) and amino acid sequences were created using the Omiga 2.0 software (Oxford Molecular Ltd, Oxford, United Kingdom). Alignments for individual NS proteins used in the analysis were created based on an initial alignment of the NS polyprotein and analysis for the presence of amino acid sequence motifs within the published proteolytic cleavage sites of other CVs (1, 19, 22, 24, 25, 32, 33). Individual NS proteins were realigned and edited to select partial alignments with relatively lower diversities. Bordering amino acid positions for the partial viral NTPase, protease, and polymerase sequences used in the final alignments are listed in Table 2. To retain the maximum number of conserved evolutionary sites, columns containing gaps were removed and the remaining consensus length alignments were used to calculate pairwise homology scores (percent) and construct phylogenetic trees. Dendrograms based on amino acid alignments were constructed by the unweighted pair group method with arithmetic mean and the neighbor-joining clustering methods of the Molecular Evolutionary Genetics Analysis (MEGA, version 3.1) software with Poisson correction distance calculations. The confidence values of the internal nodes were obtained by performing 125 and 1,025 bootstrap analyses, which yielded similar tree topologies and only slight differences (1 or 2%) in the branch bootstrap values. Pairwise distances and means within (intragenogroup) and between (intergenogroup) genogroups and clusters were also calculated.
TABLE 2.
Partial NTPase, protease, and polymerase regions used in multiple sequence alignmentsa
| Calicivirus strain | Amino acid/position no.
|
|||||
|---|---|---|---|---|---|---|
| NTPase
|
Protease
|
Polymerase
|
||||
| N terminus | C terminus | N terminus | C terminus | N terminus | C terminus | |
| Tulane | R/379 | L/531 | G/820 | Y/940 | G/977 | H/1402 |
| MD145 | R/485 | I/647 | G/1023 | Y/1151 | G/1199 | H/1649 |
| Manchester | R/470 | L/622 | G/1069 | Y/1173 | G/1213 | H/1659 |
| PEC | R/454 | L/606 | G/1063 | Y/1167 | G/1207 | H/1651 |
| FCV-Urbana | R/474 | L/638 | G/1095 | Y/1197 | G/1251 | H/1700 |
| SMSV-1 | R/580 | L/734 | G/1207 | Y/1309 | G/1364 | H/1813 |
| RHDV-FRG | R/512 | L/667 | G/1117 | L/1213 | G/1262 | H/1711 |
| BEC-NB | R/446 | L/599 | G/1010 | Y/1107 | G/1147 | H/1603 |
The N- and C-terminal amino acids and their positions of the partial NS protein sequences that were used in the multiple sequence alignments are listed. The length of the corresponding region for each strain and the consensus length of the alignments are given in Table 4.
Tissue culture propagation of TV.
A human colon carcinoma (Caco-2) and three monkey kidney (Vero, MA104, and LLC-MK2) cell lines were used in an attempt to cultivate TV. Cells were plated on six-well tissue culture plates (Corning Incorporated Life Sciences, Lowell, MA) to 75 to 80% confluence in 3 ml Dulbecco's modified Eagle's medium (MA104, Vero, and Caco-2) or M199 (LLC-MK2) medium supplemented with 10% fetal bovine serum and penicillin G (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml). Cultures were inoculated at 24 h postplating with 100 μl of sterile filtered (0.2 μm) 5% stool suspension prepared from an RT-PCR-positive stool sample in Dulbecco's modified Eagle's medium. Plates were incubated at 37°C in 5% CO2 atmosphere.
Cells and medium were harvested 5 days postinoculation by scraping, transferred to sterile 15-ml tubes (Becton Dickinson and Company, Franklin Lakes, NJ), and freeze-thawed at −20°C to release cell-associated virus. Cell debris was removed by centrifugation at 2,000 rpm (600 × g) for 5 min, and 200 μl of the cell-free medium was transferred to fresh cell cultures, which were monitored daily for cytopathic effect by light microscopy. These steps were repeated for six passages. After the 4th and 6th passages, 100 μl medium from each cell line was tested for the presence of TV by RT-PCR using TV-specific primers. Later, additional human cell lines (293T, intestine [int] 407, FHs74 int.) were also tested for their permissibility.
Plaque purification.
LLC-MK2 cell cultures grown to 80% confluence in six-well culture plates (Corning Incorporated Life Sciences, Lowell, MA) were incubated for 1 h with 1 ml of serially diluted chloroform-extracted and CsCl-purified TV obtained from the 4th passage. Following inoculation, cells were washed with medium and overlaid with 2 ml of agarose-medium containing 1 volume of 1.2% agarose in distilled water and 1 volume of 2× M199 medium supplemented with 10% fetal bovine serum. Plaque formation was evaluated daily by light microscopy. Single plaques were picked from wells with one to five isolated plaques, and agarose plugs were dissolved in 500 μl medium and used to inoculate new plates. The process was repeated six times. Viruses grown from nine final plaques were stocked at −70°C. In all subsequent experiments, plaque-purified TV M33 3/2 has been used.
Cesium chloride density gradient purification of TV.
LLC-MK2 cultures grown to 90% confluence in roller bottles (BD Biosciences, San Jose, CA) and containing 100 ml culture medium were inoculated with 3 ml of TV inoculum (105.3 50% tissue culture infective doses per milliliter). At 72 h postinoculation, cultures were freeze-thawed once and attached cells were removed by moving the ice around the flask. Cell debris was separated from culture medium in a JA-10 rotor at 8,000 rpm (10,000 × g) for 20 min in a Beckman J2-21 centrifuge. The cell pellet was resuspended in culture medium, freeze-thawed three additional times to free entrapped virus, and centrifuged at 8,000 rpm (10,000 × g) for 20 min, and the supernatant was collected and added to the culture medium. Virus particles were precipitated by mixing 1 volume of a 30% polyethylene glycol 8000-0.5 M NaCl (pH 7.4) stock solution with 4 volumes of culture medium and stirring at room temperature for 2 h and at 4°C overnight. Polyethylene glycol precipitate was collected by centrifugation at 10,000 × g for 45 min and resuspended in 1/100 of the original culture volume in phosphate-buffered saline (PBS). The resulting material was mixed with CsCl to obtain a solution with a density of 1.34 g/ml by adding 5.11 g CsCl to 10 ml suspension. Gradients were formed by centrifugation at 35,000 rpm (150,000 × g) for 27 h in a SW 41Ti rotor using a Beckman Coulter Optima L-90 K ultracentrifuge. Gradients were fractionated by bottom puncture, and the density of each fraction was determined by a traditional Abbe refractometer. Viral RNA was extracted from 1 μl of each fraction and analyzed for the presence of TV RNA by RT-PCR. Peak fractions were diluted five times in PBS, and virus particles were collected by centrifugation at 35,000 rpm (150,000 × g) for 3 h in a SW 41Ti rotor, resuspended in PBS, and stored at −70°C.
Electron microscopy (EM).
Drops of CsCl-gradient-purified TV were adsorbed onto Formvar-carbon 200-mesh copper grids (Electron Microscopy Sciences, Fort Washington, PA), stained with 1% ammonium-molybdate, and examined using a Hitachi-7600 transmission electron microscope equipped with an AMT digital camera. The mean diameter of TV particles was determined by measuring 25 virus particles at a direct magnification of ×100,000 to ×200,000.
RESULTS
CV detection in rhesus macaques.
Stool samples collected from 58 randomly selected animals with and without diarrhea were tested by RT-PCR for the presence of CVs. The degenerated primer set P289H,I/P290H,I,J,K, which is able to detect both NVs and SVs (9, 15), was used. These primers have been instrumental in the discoveries of several unique CVs in the past (9, 13, 21). Nine of the 120 samples tested positive by the RT-PCR. These samples were collected within a 2-week interval from three animals without diarrhea that were 3 to 6 weeks old. Amplicons obtained from these animals revealed identical sequences that were 6 nt shorter than the corresponding NV amplicons. A BLAST search against the GenBank nucleotide sequence database did not yield any significant sequence similarity; however, the translated amino acid sequence revealed a GLPSG motif characteristic of viral RdRps.
Genomic organization of TV.
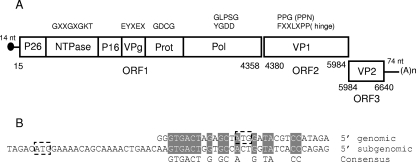
The complete genome of the TV is 6,714 nt long without the poly(A) tail, making it the shortest known CV genome. It has a ribonucleoside composition of 27.46% A, 24.58% C, 22.07% G, and 25.87% U residues and starts with three G residues that are part of a 14-nt-long 5′ untranslated region (UTR) (GGGTGACTAGAGCTATG). The genome contains three large ORFs. ORF1 spans from nt 15 to 4358, ORF2 from nt 4380 to 5984, and ORF3 from nt 5984 to 6640 (Table 3 and Fig. 1A). ORF1 and ORF2 are in frame +2 relative to the first nucleotide of the genome and separated by 21 nt, which is different from the case for NVs, where ORF1 and ORF2 are in different frames and overlap by 11 to 17 nt, but similar to the case for canine CV (GenBank accession no. NC_004542), a vesivirus, where ORF1 and ORF2 are in the same frame and are separated by 3 nt. The TV ORF3 is in frame +3 and overlaps with the last nucleotide of the ORF2 termination codon (TAATG; bold letters indicate the termination [stop] codons of ORF2; underlining the initiation [start] codons of ORF3), which is similar to NVs and different from vesiviruses and SVs (ATGA), lagovirus (ATGTCTGA), and becoviruses (TAATATG). For all three ORFs, the predicted initiation codons are in a strong consensus context for translation initiation of eukaryotic mRNA (17). The 5′ UTR did not reveal significant homologies with the 5′ region of the putative subgenomic RNA, which is different from the case for NVs; however, it aligned well to a region 21 nt downstream from the ORF2 initiation codon starting at nt 4404 (Fig. 1B). The TV genome ends with a 74-nt-long 3′ UTR and a poly(A) tail of 26 to 62 adenine residues, as determined by the sequencing of 12 clones obtained from end-ligated genomic RNA.
TABLE 3.
Comparison of the Tulane virus with representative caliciviruses of the other genera
| Calicivirus genusa | Virus strain | Accession no. | Complete genome length (nt) | UTR length
|
ORF length (nt/aa)b
|
|||
|---|---|---|---|---|---|---|---|---|
| 5′ (nt) | 3′ (nt) | 1 | 2 | 3 | ||||
| Norovirus | Norwalk | M87661 | 7,654 | 4 | 66 | 5,370/1,789 | 1,593/530 | 639/212 |
| Southampton | L07418 | 7,708 | 4 | 78 | 5,367/1,788 | 1,641/546 | 636/211 | |
| MD145 | AY032605 | 7,556 | 4 | 46 | 5,100/1,699 | 1,614/537 | 807/268 | |
| MNV-1 | DQ285629 | 7,382 | 5 | 75 | 5,058/1,685 | 1,620/539 | 627/208 | |
| Sapovirus | Manchester | X86560 | 7,431 | 12 | 82 | 6,843/2,280 | 498/165 | NA |
| PEC-Cowden | AF182760 | 7,320 | 9 | 55 | 6,765/2,254 | 495/164 | NA | |
| Vesivirus | FCV-CFI68 | U13992 | 7,677 | 19 | 43 | 5,289/1,762 | 2,007/668 | 321/106 |
| FCV-Urbana | NC_00148 | 7,683 | 19 | 46 | 5,292/1,763 | 2,007/668 | 321/106 | |
| SMSV-1 | U15301 | 8,284 | 19 | 182 | 5,640/1,879 | 2,109/702 | 333/110 | |
| Lagovirus | RHDV-FRG | M67473 | 7,437 | 9 | 59 | 7,035/2,344 | 354/117 | NA |
| EBHSV-GD | Z69620 | 7,442 | 8 | 92 | 7,005/2,334 | 345/114 | NA | |
| Becovirus | BEC-NB | AY082891 | 7,453 | 74 | 67 | 6,633/2,210 | 678/225 | NA |
| Newbury-1 | DQ013304 | 7,454 | 75 | 67 | 6,633/2,210 | 678/225 | NA | |
| Recovirus | Tulane | EU391643 | 6,714 | 14 | 74 | 4,344/1,447 | 1,605/534 | 657/218 |
Becovirus and Recovirus represent two tentative genera not yet accepted by the International Committee on Taxonomy of Viruses.
The Norovirus, Vesivirus, and Recovirus genomes are organized into three ORFs; ORF1 encodes the NS polyprotein, ORF2 encodes the capsid protein (VP1), and ORF3 encodes a minor structural protein (VP2). The Sapovirus, Lagovirus, and Becovirus genomes are organized into two ORFs; ORF1 encodes a polyprotein containing the NS proteins and the capsid protein (VP1), and ORF2 encodes the minor structural protein (VP2). NA, not applicable.
FIG. 1.
(A) Genome organization of TV. Conserved amino acid motifs are indicated. (B) Alignment of the 5′ ends of genomic and subgenomic TV RNAs. Initiation codons of the NS polyprotein and VP1 are boxed.
NS proteins encoded by ORF1.
The TV NS polyprotein is 1,447 aa long, with a calculated molecular mass of 161.9 kDa, which is the smallest among CVs. It has an organization typical for CVs, containing all the conserved amino acid motifs, including GXXGXGKT (NTPase), EYXEX (Vpg), GDCG (protease), and GLPSG and YGDD (polymerase). The TV NS polyprotein has the highest overall amino acid identity (22 to 24%) with NVs (Table 4). Based on alignments with other CVs with published polyprotein cleavage maps (1, 19, 22, 24, 25, 32, 33) and the amino acid residues at the cleavage sites (Q or E/A, G, S, T, D, and N), a putative TV NS polyprotein cleavage map, which contains the N-terminal protein (24.6 kDa), the NTPase (38 kDa), P16 (15.7 kDa), the Vpg (10.8 kDa), the protease (18 kDa), and the polymerase (52.9 kDa), was constructed (Fig. 2). The TV NTPase, protease, and polymerase have molecular masses comparable to those of NVs (less than 10% difference), while the remaining three proteins are relatively smaller (21 to 41% difference) than their NV counterparts.
TABLE 4.
Amino acid identity of the Tulane virus with representative caliciviruses of the other genera
| Calicivirus genusa | Virus strain | NS polyproteinb
|
NTPasec
|
Pro
|
Pol
|
VP1
|
VP2
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aa | % | aa | % | aa | % | aa | % | aa | % | aa | % | ||
| Norovirus | Norwalk | 1,789 | 24 | 163 | 46 | 129 | 27 | 441 | 33 | 530 | 25 | 212 | 26 |
| Southampton | 1,788 | 23 | 163 | 44 | 129 | 28 | 441 | 31 | 546 | 25 | 211 | 25 | |
| MD145 | 1,699 | 22 | 163 | 46 | 129 | 29 | 441 | 31 | 537 | 24 | 268 | 22 | |
| MNV-1 | 1,685 | 22 | 163 | 44 | 129 | 22 | 440 | 33 | 539 | 20 | 208 | 22 | |
| Sapovirus | Manchester | 1,719 | 18 | 153 | 37 | 105 | 25 | 447 | 26 | 561 | 17 | 165 | 14 |
| PEC-Cowden | 1,710 | 17 | 153 | 33 | 105 | 23 | 445 | 25 | 544 | 18 | 164 | 17 | |
| Vesivirus | FCV-CFI68 | 1,762 | 18 | 154 | 37 | 103 | 20 | 449 | 25 | 668 | 16 | 109 | 8 |
| FCV-Urbana | 1,763 | 18 | 155 | 35 | 103 | 20 | 449 | 24 | 668 | 15 | 106 | 9 | |
| SMSV-1 | 1,879 | 17 | 155 | 36 | 103 | 18 | 450 | 23 | 702 | 16 | 110 | 7 | |
| Lagovirus | RHDV-FRG | 1,765 | 19 | 156 | 33 | 97 | 20 | 450 | 27 | 579 | 17 | 117 | 11 |
| EBHSV-GD | 1,758 | 19 | 155 | 34 | 97 | 20 | 450 | 25 | 576 | 19 | 114 | 9 | |
| Becovirus | BEC-NB | 1,661 | 16 | 155 | 31 | 98 | 22 | 457 | 25 | 549 | 16 | 225 | 8 |
| Newbury-1 | 1,661 | 16 | 154 | 29 | 98 | 21 | 457 | 25 | 549 | 17 | 225 | 7 | |
| Recovirus | Tulane | 1,447 | 100 | 154 | 100 | 121 | 100 | 425 | 100 | 534 | 100 | 218 | 100 |
| Consensus length (no gap)d | 1,362 | 145 | 92 | 409 | 455 | 92 | |||||||
Becovirus and Recovirus represent two tentative genera, not yet accepted by the International Committee on Taxonomy of Viruses.
For Sapovirus, Lagovirus, and Becovirus, the length of NS polyprotein is given as the translation of ORF1, excluding the capsid protein encoding sequences.
Multiple sequence alignments for the NS polyprotein, VP1, and VP2 based on the corresponding full-length amino acid sequences. Alignments for the viral NTPase, protease, and polymerase sequences were based on partial amino acid sequences listed in Table 2.
To retain the maximum number of conserved evolutionary sites, columns containing gaps in the alignments were removed and the remaining consensus length alignments were used to calculate pairwise homology scores (percent).
FIG. 2.
Predicted NS polyprotein cleavage sites of TV based on sequence alignments with published cleavage maps of other CVs. Since the alignments revealed the highest identity between TV and NVs, only published NV cleavage maps representing GI, GII, and GV are shown. Calculated protein molecular masses (in kilodaltons) and the positions of the amino acids at the cleavage sites are shown.
Capsid (VP1) and minor structural protein (VP2).
The TV VP1 is 534 aa long, with a calculated molecular mass of 57.9 kDa, and shows the highest overall amino acid identity (20 to 25%) with NVs (Table 4). It contains the conserved PPG (PPN) motif and a FXXLXPP hinge between the S and P domains characteristic of most CV capsid proteins.
The TV VP2 is 218 aa long, with a calculated molecular mass of 22.8 kDa and an isoelectric point (pI) of 10.0, like the case for NVs. It also shows the highest amino acid identity (22 to 26%) with NVs (Table 4).
Phylogenetic analysis.
Pairwise homology scores from amino acid sequence alignments indicated that TV exhibits the highest amino acid identity with NVs in all regions analyzed (Table 4). This was confirmed by phylogenetic analyses conducted on the viral NS polyprotein, partial NTPase, protease, polymerase, and whole-capsid sequences (Fig. 3). VP2, Vpg, P16, and the N-terminal protein were not analyzed, because of the higher diversity of these proteins between the different CV genera. Phylogenetic trees obtained for each genomic region grouped viruses of the previously described genera in a manner similar to that in the trees reported by others, and TV was consistently placed on a branch rooting with NV. The mean distances between TV and the different genera also showed that TV is most closely related to NV (the mean distances between TV and NV were as follows: for ORF1 polyprotein, x̄ was 1.459 and σM was 0.041; for NTPase, x̄ was 0.791 and σM was 0.080; for protease, x̄ was 1.317 and σM was 0.162; for polymerase, x̄ was 1.124 and σM was 0.065; and for VP1, x̄ was 1.442 and σM was 0.072. x̄ indicates mean distance, and σM indicates standard error of the mean); however, these distances were longer than the distances between some of the established genera (the mean distances between SV and Vesivirus were as follows: for ORF1 polyprotein, x̄ was 1.184 and σM was 0.030; for NTPase, x̄ was 0.682 and σM was 0.068; for protease, x̄ was 1.326 and σM was 0.153; for polymerase, x̄ was 0.844 and σM was 0.047; and for VP1, x̄ was 1.322 and σM was 0.067). Also the mean distances between TV and NV were 1.66 to 3.5 times longer than the mean distances within NV (intergenogroup distance) (the mean distances within NV were as follows: for ORF1 polyprotein, x̄ was 0.550 and σM was 0.017; for NTPase, x̄ was 0.444 and σM was 0.052; for protease, x̄ was 0.367 and σM was 0.059; for polymerase, x̄ was 0.392 and σM was 0.025; and for VP1, x̄ was 0.864 and σM was 0.042). Taken together, these results indicate that TV represents a new genus within the Caliciviridae.
FIG. 3.
Unrooted phylogenetic trees based on amino acid sequence alignments of CV NTPase (A), polymerase (B), and VP1 (C). Trees were constructed by the neighbor-joining clustering method of MEGA 3.1, with Poisson distance calculations. The scale bars represent the phylogenetic distances expressed as units of amino acid substitutions per site. The confidence values of the internal nodes were obtained by performing 1,025 bootstrap analyses. Trees constructed for the NS polyprotein and protease exhibited similar topologies (data not shown).
Tissue culture propagation.
Tissue culture propagation of TV was attempted on CaCo-2, Vero, MA104, and LLC-MK2 cells from filtrates of an RT-PCR-positive stool sample. Small plaques characterized by cell rounding were observed at the 2nd passage on LLC-MK2 cells. By the 5th passage, almost 100% of the cells became rounded and detached within 72 h postinoculation. TV RNA could be detected only in the LLC-MK2 cultures by RT-PCR, but not in the other cell lines when tested at the 4th and 6th passages (Fig. 4). Attempts to propagate TV on additional human cell lines (293T, int. 407, FHs74 int.) were unsuccessful.
FIG. 4.
Detection of TV-specific RNA at the 6th passage of a TV-positive stool sample in CaCo-2, MA-104, Vero, and LLC-MK2 cells. M, molecular size marker (1-kb DNA ladder; Invitrogen, Carlsbad, CA). Mock-infected cells were passed parallel with stool-inoculated cells for each cell line. RNA was extracted from 100 μl of cell-free tissue culture medium obtained from each mock or infected (stool) cell culture. RNA extracted from 100 μl of TV-positive stool that was used to inoculate the cells originally served as a positive control (+Contr). −Contr, negative control. Primers P885 and P886 (Table 1) were used to amplify a 523-bp TV-specific product.
The TV replication was studied by infecting LLC-MK2 cells at a low (0.01) multiplicity of infection with plaque-purified TV (data not shown). Growth curves indicated a rapid replication cycle with visible cytopathic effect at about 24 h postinoculation and a peak of the virus titer at 36 to 48 h, when all cells were rounded and detached. The peak virus titer, however, even with the higher initial multiplicity of infection, reached only about 105 50% tissue culture infective doses per milliliter.
Physicochemical characterization of TV.
CsCl density gradient fractionation of plaque-purified TV revealed a clear peak, with a mean density (± standard deviation) of 1.3722 ± 0.014 g/ml containing small rounded virions with cup-like surface protrusions (Fig. 5). The mean diameter of TV particles is 35.8 ± 1.6 nm, which is similar to that of other CVs. SDS-polyacrylamide gel electrophoresis analysis of the purified TV revealed a single major protein of about 60 kDa, most likely the capsid protein (VP1) (Fig. 5).
FIG. 5.
Physicochemical properties of TV. (A) RT-PCR amplification of TV-specific RNA extracted from CsCl density gradient fractions revealed a peak with a mean density calculated as follows: x̄ = 1.37 g/ml. −Contr, negative control. (B) Virus particles with typical CV morphology and a mean diameter (calculated as x̄ = 35.8 nm) were visualized in the peak fractions. (C) A single protein band of about 60 kDa could be visualized by SDS-polyacrylamide gel electrophoresis of peak fractions.
DISCUSSION
According to the genomic and physicochemical characterizations described in this study, TV is a member of the Caliciviridae: (i) the TV genome is organized into three major ORFs, which is similar to the genomic organization of noro- and vesiviruses, (ii) all conserved amino acid sequence motifs characteristic for CVs are present in the TV NS and structural proteins and the predicted NS polyprotein cleavage map indicates the same number of cleavage products that are arranged in the same order as that described for other CVs, (iii) the buoyant density, size, and morphology of TV virions are similar to those of other CVs, (iv) TV virions consist of one major protein of about 60 kDa, and (v) phylogenetic analyses placed TV within the Caliciviridae.
The whole genome of the TV is 6,714 nt long, which is at least 600 nt shorter than the genomes of other CVs. To avoid the possibility of missing the exact 5′ end, it was determined by several methods and all of them yielded the same result. Moreover, the transfection of LLC-MK2 cells with an in vitro-transcribed TV genomic RNA yielded infectious virions (data not shown), further confirming our data.
TV represents a new CV genus that is genetically more closely related to NVs than to other CVs. Phylogenetic analysis conducted on structural and NS proteins of TV and representatives of other CV genera consistently placed TV on a separate branch rooted together with NV (Fig. 3). This placement is in accordance with the pairwise homology and phylogenetic distance calculations that also indicated the closest relationship between TV and NVs. However, the low-homology scores observed in all regions and the equivalence of phylogenetic distances between TV and NV to distances between the established CV genera clearly place TV outside the NV genus. In accordance with the proposed nomenclature of Becovirus (26), we propose the name Recovirus (rhesus enteric Calicivirus) for this new genus represented by TV.
Additional features of TV with high similarity to those of NVs have been also noted. The lengths of the TV ORF2 and ORF3 and the Mrs of the encoded structural proteins VP1 and VP2 are comparable to the corresponding values for NVs (Table 3). Also, the junction between ORF2 and ORF3, which seems to be conserved for CVs of different genera, is similar to that of NVs (TAATG). The shorter genome length of TV is clearly due to a shorter ORF1 (Table 3). The TV ORF1 is 4,344 nt long, which is more than 1,000 nt shorter than the 5,370 nt of Norwalk virus ORF1. Based on alignments of deduced NS polyprotein sequences and the predicted protein cleavage map of TV (Fig. 2), the TV NS polyprotein is cleaved into at least six functional proteins, NH2-P26.4(N-terminal)-P38(NTPase)-P16-P10.8(Vpg)-P18(Prot)-P52.9(Pol)-COOH, all of which exhibit the highest amino acid identity with the corresponding proteins of NVs. The N-terminal protein, P16, and VPg are significantly (about 20 to 40%) smaller than those of NVs, while the remaining NS proteins have Mrs comparable to those of NVs.
One significant difference between TV and HuCVs is that TV could be readily grown in vitro. Whether this result is due to the difference in NS protein function or receptor usage or to other factors is unclear and needs to be studied. It also should be noted that although TV could be grown in LLC-MK2 cells (a rhesus macaque kidney cell line), growth attempts in other cell lines failed. Thus, investigating the nature of cellular and viral elements required for TV replication may provide important clues for identifying factors required for successful in vitro growth of HuCVs. The availability of a cell culture and a reverse genetics system makes TV a valuable model to address these issues.
Since TV was isolated from stool samples of animals without symptoms of diarrhea, the clinical disease linked to TV infection and its pathogenesis and whether TV can be developed to an animal model system for CV gastroenteritis remain to be established. A recent study reported the detection of NV-like particles in nonhuman primate stool samples, including rhesus macaques, by EM (38). Whether these particles represented TV or NVs is unknown. Taken together, these findings call for extended investigations of CVs in nonhuman primates. Previous attempts at the molecular detection of CVs in primate stool samples were unsuccessful, possibly due to primer selection. The detection of TV with the primer set P289H,I/P290H,I,J,K further emphasizes the usefulness of these primers in CV detection.
Acknowledgments
This study was supported by a Cincinnati Children's Hospital Research Foundation Trustee grant awarded to T.F. and the National Institute of Allergy and Infectious diseases (R01 AI37093 and R01 AI55649), the National Institute of Child Health (PO1 HD13021), and the Department of Defense (PR033018) of the United States (to X.J.). Work with nonhuman primates was supported by a pilot study of the National Center for Research Resources (P51 RR00164) and the National Institute of Allergy and Infectious Diseases (R21 AI54146) (grants to K.S.).
We thank Xiaoyun Deng, Weiming Zhong, and Mayra A. Cantu for technical support and Irene Hofmann for helping with the EM.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Belliot, G., S. V. Sosnovtsev, T. Mitra, C. Hammer, M. Garfield, and K. Y. Green. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 7710957-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, K. O., S. S. Sosnovtsev, G. Belliot, Q. Wang, L. J. Saif, and K. Y. Green. 2005. Reverse genetics system for porcine enteric calicivirus, a prototype sapovirus in the Caliciviridae. J. Virol. 791409-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhry, Y., A. Nayak, M. E. Bordeleau, J. Tanaka, J. Pelletier, G. J. Belsham, L. O. Roberts, and I. G. Goodfellow. 2006. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 28125315-25325. [DOI] [PubMed] [Google Scholar]
- 4.Cheetham, S., M. Souza, R. McGregor, T. Meulia, Q. Wang, and L. J. Saif. 2007. Binding patterns of human norovirus-like particles to buccal and intestinal tissues of gnotobiotic pigs in relation to A/H histo-blood group antigen expression. J. Virol. 813535-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheetham, S., M. Souza, T. Meulia, S. Grimes, M. G. Han, and L. J. Saif. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 8010372-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dastjerdi, A. M., J. Green, C. I. Gallimore, D. W. Brown, and J. C. Bridger. 1999. The bovine Newbury agent-2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology 2541-5. [DOI] [PubMed] [Google Scholar]
- 7.D'Souza, D. H., A. Sair, K. Williams, E. Papafragkou, J. Jean, C. Moore, and L. Jaykus. 2006. Persistence of caliciviruses on environmental surfaces and their transfer to food. Int. J. Food Microbiol. 10884-91. [DOI] [PubMed] [Google Scholar]
- 8.Duizer, E., P. Bijkerk, B. Rockx, A. De Groot, F. Twisk, and M. Koopmans. 2004. Inactivation of caliciviruses. Appl. Environ Microbiol. 704538-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farkas, T., W. M. Zhong, Y. Jing, P. W. Huang, S. M. Espinosa, N. Martinez, A. L. Morrow, G. M. Ruiz-Palacios, L. K. Pickering, and X. Jiang. 2004. Genetic diversity among sapoviruses. Arch. Virol. 1491309-1323. [DOI] [PubMed] [Google Scholar]
- 10.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 746581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, K., R. Chanock, and A. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 12.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181S322-S330. [DOI] [PubMed] [Google Scholar]
- 13.Guo, M., J. F. Evermann, and L. J. Saif. 2001. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch. Virol. 146479-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, B., H. M. McClure, R. L. Fankhauser, S. S. Monroe, and R. I. Glass. 2004. Prevalence of rotavirus and norovirus antibodies in non-human primates. J. Med. Primatol. 3330-33. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, X., P. W. Huang, W. M. Zhong, T. Farkas, D. W. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83145-154. [DOI] [PubMed] [Google Scholar]
- 16.Koopmans, M. K., G. K. Y., T. Ando, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, A. W. Smith, M. J. Studdert, and H. J. Thiel. 2005. Caliciviridae, p. 843-851. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 17.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 26619867-19870. [PubMed] [Google Scholar]
- 18.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, B. L., G. J. Viljoen, I. N. Clarke, and P. R. Lambden. 1999. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 80291-296. [DOI] [PubMed] [Google Scholar]
- 20.Mandl, C. W., F. X. Heinz, E. Puchhammer-Stockl, and C. Kunz. 1991. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. BioTechniques 10484-486. [PubMed] [Google Scholar]
- 21.Martella, V., C. M., E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 131071-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyers, G., C. Wirblich, H. J. Thiel, and J. O. Thumfart. 2000. Rabbit hemorrhagic disease virus: genome organization and polyprotein processing of a calicivirus studied after transient expression of cDNA constructs. Virology 276349-363. [DOI] [PubMed] [Google Scholar]
- 23.Neill, J. D. 1990. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 17145-160. [DOI] [PubMed] [Google Scholar]
- 24.Oka, T., K. Katayama, S. Ogawa, G. S. Hansman, T. Kageyama, T. Miyamura, and N. Takeda. 2005. Cleavage activity of the sapovirus 3C-like protease in Escherichia coli. Arch. Virol. 1502539-2548. [DOI] [PubMed] [Google Scholar]
- 25.Oka, T., K. Katayama, S. Ogawa, G. S. Hansman, T. Kageyama, H. Ushijima, T. Miyamura, and N. Takeda. 2005. Proteolytic processing of sapovirus ORF1 polyprotein. J. Virol. 797283-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver, S. L., E. Asobayire, A. M. Dastjerdi, and J. C. Bridger. 2006. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology 350240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, S. L., A. M. Dastjerdi, S. Wong, L. El-Attar, C. Gallimore, D. W. Brown, J. Green, and J. C. Bridger. 2003. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk-like viruses) unlikely to be of risk to humans. J. Virol. 772789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parwani, A. V., W. T. Flynn, K. L. Gadfield, and L. J. Saif. 1991. Serial propagation of porcine enteric calicivirus in a continuous cell line. Effect of medium supplementation with intestinal contents or enzymes. Arch. Virol. 120115-122. [DOI] [PubMed] [Google Scholar]
- 29.Rockx, B. H., W. M. Bogers, J. L. Heeney, G. van Amerongen, and M. P. Koopmans. 2005. Experimental norovirus infections in non-human primates. J. Med. Virol. 75313-320. [DOI] [PubMed] [Google Scholar]
- 30.Smiley, J. R., K. O. Chang, J. Hayes, J. Vinje, and L. J. Saif. 2002. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 7610089-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, A. W., D. E. Skilling, M. P. Anderson, and K. Benirschke. 1985. Isolation of primate calicivirus Pan paniscus type 1 from a douc langur (Pygathrix nemaeus l.). J. Wildl. Dis. 21426-428. [DOI] [PubMed] [Google Scholar]
- 32.Sosnovtsev, S. V., G. Belliot, K. O. Chang, V. G. Prikhodko, L. B. Thackray, C. E. Wobus, S. M. Karst, H. W. Virgin, and K. Y. Green. 2006. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 807816-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sosnovtsev, S. V., M. Garfield, and K. Y. Green. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 767060-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277193-203. [DOI] [PubMed] [Google Scholar]
- 35.Subekti, D. S., P. Tjaniadi, M. Lesmana, J. McArdle, D. Iskandriati, I. N. Budiarsa, P. Walujo, I. H. Suparto, I. Winoto, J. R. Campbell, K. R. Porter, D. Sajuthi, A. A. Ansari, and B. A. Oyofo. 2002. Experimental infection of Macaca nemestrina with a Toronto Norwalk-like virus of epidemic viral gastroenteritis. J. Med. Virol. 66400-406. [DOI] [PubMed] [Google Scholar]
- 36.Sugieda, M., H. Nagaoka, Y. Kakishima, T. Ohshita, S. Nakamura, and S. Nakajima. 1998. Detection of Norwalk-like virus genes in the caecum contents of pigs. Arch. Virol. 1431215-1221. [DOI] [PubMed] [Google Scholar]
- 37.Taku, A., B. R. Gulati, P. B. Allwood, K. Palazzi, C. W. Hedberg, and S. M. Goyal. 2002. Concentration and detection of caliciviruses from food contact surfaces. J. Food Prot. 65999-1004. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Y., X. Tu, C. Humphrey, H. McClure, X. Jiang, C. Qin, R. I. Glass, and B. Jiang. 2007. Detection of viral agents in fecal specimens of monkeys with diarrhea. J. Med. Primatol. 36101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wobus, C. E., S. M. Karst, L. B. Thackray, K.-O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. Virgin IV. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wobus, C. E., L. B. Thackray, and H. W. Virgin IV. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 805104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyatt, R. G., H. B. Greenberg, D. W. Dalgard, W. P. Allen, D. L. Sly, T. S. Thornhill, R. M. Chanock, and A. Z. Kapikian. 1978. Experimental infection of chimpanzees with the Norwalk agent of epidemic viral gastroenteritis. J. Med. Virol. 289-96. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]