Abstract
The HI loop is a prominent domain on the adeno-associated virus (AAV) capsid surface that extends from each viral protein (VP) subunit overlapping the neighboring fivefold VP. Despite the highly conserved nature of the residues at the fivefold pore, the HI loops surrounding this critical region vary significantly in amino acid sequence between the AAV serotypes. In order to understand the role of this unique capsid domain, we ablated side chain interactions between the HI loop and the underlying EF loop in the neighboring VP subunit by generating a collection of deletion, insertion, and substitution mutants. A mutant lacking the HI loop was unable to assemble particles, while a substitution mutant (10 glycine residues) assembled particles but was unable to package viral genomes. Substitution mutants carrying corresponding regions from AAV1, AAV4, AAV5, and AAV8 yielded (i) particles with titers and infectivity identical to those of AAV2 (AAV2 HI1 and HI8), (ii) particles with a decreased virus titer (1 log) but normal infectivity (HI4), and (iii) particles that synthesized VPs but were unable to assemble into intact capsids (HI5). AAV5 HI is shorter than all other HI loops by one amino acid. Replacing the missing residue (threonine) in AAV2 HI5 resulted in a moderate particle assembly rescue. In addition, we replaced the HI loop with peptides varying in length and amino acid sequence. This region tolerated seven-amino-acid peptide substitutions unless they spanned a conserved phenylalanine at amino acid position 661. Mutation of this highly conserved phenylalanine to a glycine resulted in a modest decrease in virus titer but a substantial decrease (1 log order) in infectivity. Subsequently, confocal studies revealed that AAV2 F661G is incapable of efficiently completing a key step in the infectious pathway nuclear entry, hinting at a possible perturbation of VP1 phospholipase activity. Molecular modeling studies with the F661G mutant suggest that disruption of interactions between F661 and an underlying P373 residue in the EF loop of the neighboring subunit might adversely affect incorporation of the VP1 subunit at the fivefold axis. Western blot analysis confirmed inefficient incorporation of VP1, as well as a proteolytically processed VP1 subunit that could account for the markedly reduced infectivity. In summary, our studies show that the HI loop, while flexible in amino acid sequence, is critical for AAV capsid assembly, proper VP1 subunit incorporation, and viral genome packaging, all of which implies a potential role for this unique surface domain in viral infectivity.
Adeno-associated virus (AAV), a 26-nm nonpathogenic human parvovirus, is distinct from most viruses due to the dependence on a helper virus for productive infection (adenovirus or herpes simplex virus) (6). In light of the rapidly growing applications of AAV as a gene therapy vector (51, 57), several efforts to understand events in the infectious pathway including host cell recognition (3, 11, 23, 33, 49), intracellular trafficking (5, 10), and uncoating (43) in the absence of a helper, are currently under way. Within this context, a thorough understanding of the structural correlates of the AAV capsid and how they contribute to steps during viral transduction is necessary (55). The determination of crystal structures of several AAV serotypes (32, 48, 60) and related parvoviruses (2, 22) over the past few years has been critical in this regard.
With respect to AAV, the capsid is encoded by three overlapping viral proteins (VPs), VP1, VP2, and VP3 (36), which are incorporated into a 60-subunit capsid in a 1:1:10 ratio. VP1 has a unique N terminus containing a phospholipase (PLA2) domain (15) and nuclear localization sequences (18, 42) thought to be necessary for endosomal escape (12) and possibly nuclear entry (47). VP2 also has an extended N terminus (compared to VP3) that remains internal to the capsid similar to VP1 until exposed to experimental conditions involving low pH or heat (26). Although this protein has been suggested to be nonessential for viral assembly and infectivity (50), its exact role remains unknown (18). VP3 is the primary capsid protein (contained within VP1 and VP2) that constitutes the surface topology of the AAV capsid, which in turn dictates antigenicity (20, 27) and tropism (3, 4, 31). Based on crystal structures of AAV, the VP amino acids involved in forming the icosahedral fivefold (Fig. 1B), threefold (4), and twofold symmetry interfaces have been visualized. The threefold axis has the largest amount of buried surface area and the highest contact energy, being the most interdigitated region of the capsid (60). The surface loops at the threefold axis of symmetry are thought to be involved in host cell receptor binding (4, 23) and have been the target of several mutagenesis studies (27, 31, 39, 55, 56). In addition, recent data have shown that a single amino acid change (K531E) located at the base of the threefold loops has the ability to alter the phenotypes of multiple AAV serotypes (56), suggesting an incomplete understanding of this critical region. The twofold axis of symmetry has the weakest amino acid interactions and the lowest contact energy, while the fivefold symmetry axis is thought to have intermediate interactions (60). As mentioned above, there have been many studies on the threefold axis of symmetry; however, the role of surface loops that form the interactions at the fivefold and twofold symmetry axes remains to be determined.
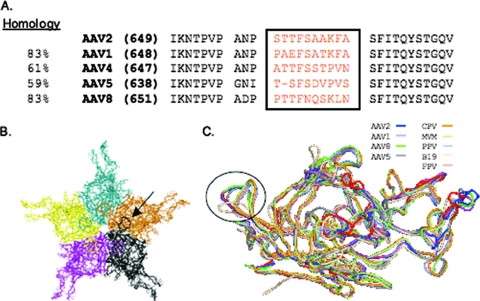
FIG. 1.
HI loop comparison of various AAV serotypes and autonomous parvoviruses. (A) Comparison of amino acid sequence homologies among representative AAV serotypes. Boxed are the 10 most variable amino acids. (B) An arrow indicates the HI loop on an AAV2 pentamer, which extends from one VP subunit and overlaps the neighboring VP. (C) Circled in black are the HI loops on a structural superimposition of the VP monomer subunits of multiple AAV serotypes, i.e., AAV2 (59) (blue), AAV4 (32) (red), AAV5 (48) (gray), AAV1 (28) (purple), and AAV8 (30) (green), and the autonomous parvoviruses canine parvovirus (CPV; 45) (orange), feline panleukopenia virus (FPV; 1) (salmon), human parvovirus B19 (22) (wheat), monoclinic canine parvovirus (MVM; 45) (lime), and porcine parvovirus (PPV; 40) (slate).
The pentameric assembly of VP3 subunits results in the formation of 12 pores at the fivefold axis of symmetry (Fig. 1B) which have been the focus of several recent investigations. Mutagenesis of residues that constitute the pore has suggested a critical role in assembly and packaging (8, 17, 55). Therefore, it is likely that the fivefold pore is necessary for DNA packaging, including Rep protein binding, capsid assembly, and VP1 N terminus exposure. Surrounding this pore at the fivefold axis of symmetry is a prominent region of the AAV capsid—the HI loop located between β strands βH and βI, which spans residues 653 to 669 (VP1 numbering) and extends to overlap each adjacent subunit (Fig. 1). Recent data have shown that the HI loop conformationally changes upon capsid interaction with the primary receptor heparan sulfate proteoglycan (M. Agbandje-McKenna unpublished data), alluding to an important capsid conformational change for subsequent stages of the AAV life cycle. In this study, we have carried out a thorough characterization of the HI loop through deletion-and-substitution mutagenesis, as well as a battery of biochemical assays to assess the role of this surface feature in the AAV life cycle. Our results help demonstrate the plasticity of the HI loop and imply a potential role in viral genome packaging. Simultaneously, we identified a critical residue within the HI loop that dictates proper incorporation of VP1 in the viral capsid.
MATERIALS AND METHODS
Generation of mutants.
All constructs were generated in the pXR2 (34) backbone by using primers and restriction sites for PCR or oligonucleotide insertion, respectively. PCR was used to generate AAV2 polyglycine and AAV2 HI−/− mutants. PCR was performed with the Expand Long Template PCR kit from Roche. All other mutants generated were the result of enzyme digests and oligonucleotide insert ligations. Restriction sites were placed downstream and upstream of the HI loop, i.e., Sbf1 and Afe1 (pXSA), respectively. The HI loops from AAV4 and AAV5 were amplified with these restriction sites on the 5′ and 3′ ends, digested, and inserted into the digested pXSA backbone. pXR1 and pXR8 were digested with Sbf1 and Afe1, removing the HI loop, which was then ligated into pXSA. Restriction enzyme sites were generated at amino acid positions 648 (Age1) and 666 (Nhe1), surrounding the HI loop, in order to insert oligonucleotides into this region. Oligonucleotides were ordered with corresponding restriction sites at the 5′ and 3′ ends, digested, and ligated into the digested backbone. All oligonucleotides were synthesized by Integrated DNA Technologies (www.idtdna.com). Site-directed mutagenesis was also used to generate point mutations within the pXR2 backbone within the HI loop with the Stratagene QuikChange site-directed mutagenesis kit. The primers generated are listed in Table 1.
TABLE 1.
Primers used for AAV2 HI loop mutant capsid generation
| Primer | Forward | Reverse |
|---|---|---|
| AAV2 HI−/− | 5′ TCC TTC ATC ACA CAG TAC TCC ACG GGA CAG G 3′ | 5′ AGG ATT CGC AGG TAC CGG GGT GTT CTT GAT GAG 3′ |
| AAV2 poly-glycine | 5′ GGA GGA GGA GGA GGA GGA GGA GGA GGA GGA TCC TTC ATC ACA CAG TAC TCC ACG GGA CAG G 3′ | TCC TCC TCC TCC TCC TCC TCC TCC TCC TCC AGG ATT CGC AGG TAC CGG GGT GTT CTT GAT GAG 3′ |
| AAV2 poly-glycine F661 | 5′ GCG AAT CCT GGA GGA GGA TTC GGA GGA GGA GGA GGA 3′ | NA |
| AAV2 poly-glycine K665 | 5′ GGA GGA GGA GGA GGA AAG GGA GGA TCC TTC ATC 3′ | NA |
| AAV2 poly-glycine F666 | 5′ GGA GGA GGA GGA GGA TTT GGA TCC TTC ATC ACA CAG 3′ | NA |
| pXSA for HI loop serotype swaps | (Sbf1) 5′ AGA GAT GTG TAC CTG CAG GGG CCC ATC TGG 3′ | NA |
| (Afe1) 5′ AAG GAA AAC AGC AAG CGC TGG AAT CCC GAA 3′ | ||
| AAV5 HI loop | 5′ GAT CCT GCA GGG ACC CAT CTG GGC CAA GAT C 3′ | 5′ GCT TGG AGT TTT CCT TCT TGA GCT CCC AC 3′ |
| AAV4 HI loop | 5′ GAT CCT GCA GGG TCC CAT TTG GGC CAA GAT T 3′ | 5′ GTT TGG ACC GCT CCT TCT GGA TCT CCC 3′ |
| AAV2 HI5 TTSF | 5′ CCC GGA AAT ATC ACC ACC AGC TTC TCG GAC GTG 3′ | NA |
| pAge1 Nhel | (Age1) 5′ CTC ATC AAG AAC ACA CCG GTA CCT GCG 3′ | NA |
| (Nhe1) 5′ GCG GCA AAG TTT GCT AGC TTC ATC ACA CAG TAC TCC 3′ | ||
| AAV2 SIGYPLP | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT AGC ATT GGT TAT CCT CTT CCT AAG TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA CTT AGG AAG AGG ATA ACC AAT GCT AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 VNTANST | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT GTT AAT ACT GCT AAT AGC ACT AAG TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA CTT AGT GCT ATT AGC AGT ATT AAC AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 QPEHSST | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT CAA CCT GAA CAT AGC AGC ACT AAG TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA CTT AGT GCT GCT ATG TTC AGG TTG AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 RGD 658 | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT CGA GGA GAC TTC AGT GCG GCA AAG TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA CTT TGC CGC ACT GAA GTC TCC TCG AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 RGD 660 | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT TCG ACC CGA GGA GAC GCG GCA AAG TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA CTT TGC CGC GTC TCC TCG GGT CGA AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 RGD 662 | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT TCG ACC ACC TTC CGA GGA GAC AAG TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA CTT GTC TCC TCG GAA GGT GGT CGA AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 RGD 663 | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT TCG ACC ACC TTC AGT CGA GGA GAC TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA GTC TCC TCG ACT GAA GGT GGT CGA AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 SGRGDS | 5′ AAG AAC ACA CCG GTA CCT GCG AAT CCT TCG GGA CGA GGA GAC TCG GCG AAG TTT GCT AGC TTC ATC 3′ | 5′ GAT GAA GCT AGC AAA CTT CGC CGA GTC TCC TCG TCC CGA AGG ATT CGC AGG TAC CGG TGT GTT CTT 3′ |
| AAV2 F661G | 5′ GCG AAT CCT TCG ACC ACC GGC AGT GCG GCA AAG TTT GCT TCC 3′ | NA |
Virus production.
Virus was produced by the triple-transfection method developed in our laboratory as described by Xiao and Samulski in 1998 (58). Cells were transfected with pXR2 containing the capsid mutations, the pXX6-80 helper plasmid, and pTR-CMV-Luciferase containing the luciferase reporter transgene flanked by terminal repeats. Cells were harvested at 60 h posttransfection and purified by cesium chloride gradient density centrifugation for 5 h at 65,000 rpm or overnight at 55,000 rpm. Gradients were fractionated, and the virus was dialyzed against 1× phosphate-buffered saline (PBS) supplemented with calcium and magnesium. Viral titers were determined in triplicate by treating 2 μl of the virus fractions with DNase, digesting the capsid with proteinase K, and loading the viral genomic DNA onto a Hybond-XL membrane (Amersham). The viral DNA was detected with a 32P-labeled probe complementary to the luciferase transgene. To carry out the experiments described here, we generated more than 35 capsid mutants, in excess of 70 vector preparations. Mutants that were important for the main conclusion of this report were generated multiple times. For example, we generated AAV2 HI−/− more than five times, AAV2 HI4 two times, AAV2 HI5 more than five times, and AAV2 F661G two times. Each mutant virus preparation was made in conjunction with AAV2 for a transfection control and titer comparison. Representative titers and phenotypes are documented in Results.
Western dot blotting, heat treatment, and Western blotting.
Production of empty and full capsids was determined posttransfection by loading 2 μl of the virus fractions onto a nitrocellulose membrane in a dot blotting apparatus. Membranes were blocked in 10% milk in PBS for 30 min at room temperature (RT) and incubated with primary antibody A20 (dilution, 1:20) (52) in 2% milk for 1 h at RT. Membranes were washed five times with 1× PBS and incubated with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Pierce; dilution, 1:5,000) for 30 min. The membranes were washed as described above, and capsid production was visualized with the SuperSignal West Femto Maximum Sensitivity Substrate chemiluminescence kit from Pierce. To examine VP1 exposure, capsids were heat treated over a range of temperatures (see Results) and blotted onto a nitrocellulose membrane through a dot blotting apparatus. The membrane was incubated as described above, except that the A1 (1:20) and B1 (1:20) (52) primary antibodies were used to detect VP1 exposure and capsid VP dissociation upon heat treatment, respectively. For Western blotting (immunoblotting), approximately 1010 dialyzed vector genome (VG)-containing particles were mixed with NuPAGE LDS sample buffer (Invitrogen), run on a NuPAGE gel (Invitrogen), transferred to a nitrocellulose membrane (Invitrogen), and blotted as described above. Other antibodies used to analyze the VP1 unique region during Western blotting were against amino acids 15 to 29 (1:1,000) and amino acids 60 to 74 (1:1,000) (Pacific Immunology; J. C. Grieger and R. J. Samulski, unpublished data). Exposure times ranged from 10 s to 1 min.
Viral transduction assay.
Viral transduction was analyzed by quantifying the luciferase transgene expression in 293 cell lysate at no more than 24 h postinfection. 293 cells (2 × 105) were transduced with 3,000 VGs/cell and lysed with 1× passive lysis buffer provided by Promega. Relative light units were analyzed with a Victor2 luminometer (Perkin-Elmer) postaddition of the d-luciferin substrate (NanoLight) to the cell lysates.
Electron microscopy (EM).
Ten microliters of purified and dialyzed full and empty virus particles in 1× PBS with Ca2+ and Mg2+ were pipetted onto a glow-discharged copper grid. The grid was washed twice with water and then stained with 2% uranyl acetate. EM images were taken with a LEO EM 910 transmission electron microscope at various magnifications at the University of North Carolina microscopy laboratories.
Heparin binding assay.
We incubated 1010 VG-containing particles of virus with preequilibrated heparin type III-S agarose beads (Sigma). The flowthrough was collected, and the beads were washed two times with 1× PBS. The washes were collected, and the beads were washed with increasing salt concentrations of 0.2 to 0.6 M PBS. A loading control, the flowthrough, washes, and elutions were blotted onto a nitrocellulose membrane with a dot blotting apparatus. The membrane was blocked and incubated with antibody as described in Materials and Methods. In this case, A20 was used as the primary antibody in order to detect intact capsids and determine the affinity of virus mutants for heparin beads.
Confocal microscopy.
Coverslips were plated with 50,000 HeLa cells/slip in a 24-well plate. Each well was infected with 30,000 VGs/cell. At 12 h postinfection, cells were fixed with 2% paraformaldehyde and washed with 1× PBS. Cells were permeabilized with 0.1% Triton X-100 at RT for 5 min and washed with 1× PBS and 1× immunofluorescence wash buffer (IFWB; distilled H2O, 20 mM Tris [pH 7.5], 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 5 mg/ml bovine serum albumin, 0.05% Tween). Cells were incubated in primary antibody A20 (1:10) in IFWB for 1 h at 37°C. Cells were washed with 1× PBS and incubated with a 488-nm fluorophore-conjugated secondary antibody (1:1,250) in IFWB for 1 h at 37°C (Abcam). Coverslips were mounted onto slides with ProLong Gold Antifade with 4′,6′-diamidino-2-phenylindole (DAPI) mounting medium. Slides were viewed on a Leica microscope in the Michael Hooker Microscopy Facility at the University of North Carolina, Chapel Hill.
Molecular modeling studies.
Homologous models of HI loop mutants were generated with VIPER (35) and/or Swiss-Model (http://swissmodel.expasy.org) in order to visualize the effects of mutagenesis on the virus capsid. The available structure of AAV2 (PDB accession no. 1LP3) was supplied as a template for the mutant models generated in the AAV2 background. Once the Swiss-Model PDBs were generated, the program O (21a) was used to generate symmetry-related models of the monomer subunits by icosahedral matrix multiplication. Structure visualization of mutant models and the structures of other AAV serotypes whose HI loops were substituted into AAV2, e.g., AAV4 (PDB accession no. 2G8G) (16), AAV8 (PDB accession no. 2QA0) (30), AAV1, and AAV5 (unpublished), was performed with winCOOT (http://www.chem.gla.ac.uk/∼bernhard/coot/wincoot.html), and images were rendered with MacPyMOL (http://pymol.sourceforge.net).
RESULTS
With respect to structure and topology, the HI loop is highly conserved between the AAV serotypes and autonomous parvoviruses (Fig. 1C); however, the amino acid sequence varies significantly. In order to determine the role of the HI loop as it pertains to the AAV capsid structure and life cycle, the AAV2 HI loop sequence was mutated, swapped between serotypes, or substituted and the resulting viruses were assayed for viral assembly, encapsidation of the viral DNA, binding to heparan sulfate, and the ability to successfully infect target cells. Mutagenesis was performed on the 10 varying amino acids of the HI loop, starting with serine 658 and ending with alanine 667 in the AAV2 capsid (Fig. 1A).
Deletion and glycine substitution mutants.
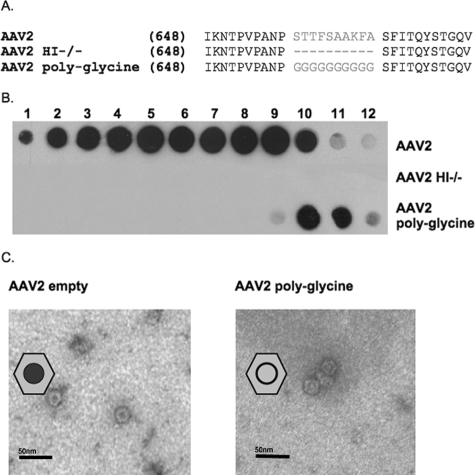
First, mutants were generated in which the AAV2 HI loop amino acids were either deleted (AAV2 HI−/−) or replaced with a polyglycine peptide (AAV2 polyglycine). In the substitution mutant, the 10 most variable HI loop amino acids were replaced with a chain of glycine residues in order to generate a flexible loop structure devoid of all amino acid side chain contacts between this loop and the EF loop (located between β strands βE and βF) in the underlying subunit (Fig. 2A). Upon removal of the HI loop, AAV2 HI−/− capsid VPs were expressed (data not shown) but were unable to assemble into particles, as determined by A20 Western dot blot analysis (Fig. 2B) and DNA dot blotting (Table 2). Interestingly, as shown in Fig. 2B, the AAV2 polyglycine mutant formed AAV particles based on Western dot blot analysis (see fractions 10 and 11 of the cesium chloride gradient); however, these particles were devoid of DNA (Table 2). The AAV2 polyglycine empty particles appear to be similar to AAV2 empty particles when analyzed by EM (Fig. 2C), with the exception of a ring-like staining pattern more apparent with AAV2 polyglycine particles. Although no other gross structural changes were evident by EM, potential conformational changes to the capsid were suggested by the ring-like staining; therefore, further biochemical analyses such as heparin binding affinity chromatography were carried out. The AAV2 polyglycine mutant had an affinity for heparin sulfate comparable to that of wild-type (WT) AAV2 particles, as determined through a heparin binding assay, eluting from the heparin column mostly at 0.4 M salt (Table 2). From these data, three important conclusions can be drawn. The presence of the HI loop is necessary for capsid assembly, and specific amino acid side chain interactions within this loop appear to be necessary for packaging the viral genomic DNA. In addition, residues forming the HI loop and adjacent capsid regions do not play a role in heparan sulfate receptor attachment.
FIG. 2.
AAV2 HI loop deletion and glycine substitution characterizations. (A) AAV2 HI loop sequence alignment showing amino acids 658 to 667, which were removed (AAV2 HI−/−) or replaced with glycine residues (AAV2 polyglycine). (B) Western dot blot analysis of CsCl gradient fractions from AAV2, AAV2 HI−/−, and AAV2 polyglycine preparations with antibody A20. Gradient fractions were collected and blotted onto a nitrocellulose membrane. The membrane was incubated with primary antibody A20 (1:20) and incubated with a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:5,000). (C) Electron micrograph images of empty AAV2 and AAV2 polyglycine particles. Gradient-purified, 1× PBS-Ca2+-Mg2+-dialyzed AAV2 (left panel) and AAV2 polyglycine (right panel) empty particles were incubated on glow-discharged copper grids negatively stained with 2% uranyl acetate.
TABLE 2.
Phenotype comparison of AAV2 HI loop capsid mutants
| Mutant | Sequence | Assembly (Western dot blotting) | Packaging (DNA dot blotting) | Heparin binding | Infectivitya (luciferase assay) | Alternative VP1 (Western blotting) |
|---|---|---|---|---|---|---|
| AAV2 HI−/− | (655) ANP----------SFI | −e | − | NAf | NA | NA |
| AAV2 poly-glycine | (655) ANP GGGGGGGGGG SFI | +c | − | + | NA | − |
| AAV2 G661F | (655) ANP GGGFGGGGGG SFI | + | − | NDg | NA | ND |
| AAV2 G665K | (655) ANP GGGGGGGKGG SFI | + | − | ND | NA | ND |
| AAV2 G666F | (655) ANP GGGGGGGGFG SFI | + | − | ND | NA | ND |
| AAV2 HI1 | (655) ANP PAEFSATKFA SFI | + | + | + | No change | − |
| AAV2 HI8 | (655) ANP PTTFNSQKLN SFI | + | + | + | No change | − |
| AAV2 HI4 | (655) ANP ATTFSSTPVN SFI | + | − | + | No change | − |
| AAV2 HI5 | (655) GNI T-SFSDVPVS SFI | − | − | NA | NA | NA |
| AAV2 RGD 658 | (655) ANP RGDFSAAKFA SFI | + | + | ND | No change | − |
| AAV2 RGD 660 | (655) ANP STRGDAAKFA SFI | + | ±d | ND | 4.7 | + |
| AAV2 RGD 662 | (655) ANP STTFRGDKFA SFI | + | + | ND | No change | − |
| AAV2 RGD 663 | (655) ANP STTFSRGDFA SFI | + | ± | ND | No changeb | − |
| AAV2 VNTANST | (655) ANP VNTANSTKFA SFI | + | ± | + | 27 | + |
| AAV2 QPEHSST | (655) ANP QPEHSSTKFA SFI | + | + | + | 2 | + |
| AAV2 SIGYPLP | (655) ANP SIGYPLPKFA SFI | + | ± | ND | 10.4 | + |
| AAV2 SGRGDS | (655) ANP SGRGDSAKFA SFI | + | ± | ND | 4.5 | + |
| AAV2 F661G | (655) ANP STTGSAAKFA SFI | + | ± | + | 13.55 | + |
Relative (n-fold) decrease in infectivity.
In SK-OV3 cells.
+, similar to WT.
±, packaging, approximately fivefold lower in titer than WT.
−, packaging, 1 log lower in titer than WT or unable to package the viral genome.
NA, not applicable.
ND, not determined.
HI loop domain swapping.
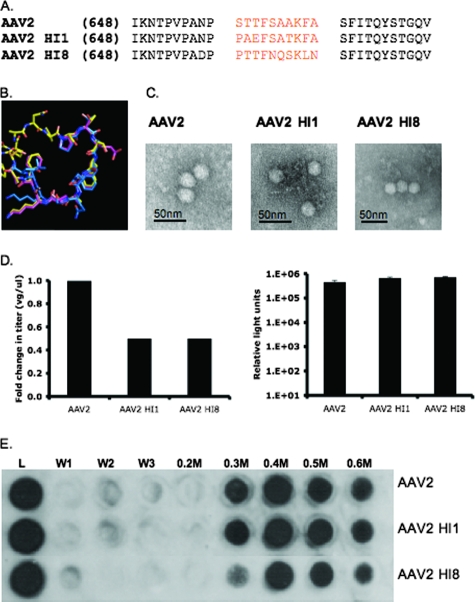
Based on the observations from the glycine residue replacement studies, we carried out HI loop swaps from representative serotypes in the hope of obtaining more information on critical amino acids that have evolved for the HI loops of specific serotypes. The serotypes chosen for this study were AAV1, AAV4, AAV5, and AAV8, which are 83%, 61%, 59%, and 83% identical to the overall amino acid sequence of the AAV2 capsid, respectively (Fig. 1A), and represent the range of sequence homology between AAV2 and the other serotypes characterized to date (13). The AAV1 and -8 HI loops are similar in conformation (Fig. 3B) but vary significantly in amino acid sequence (Fig. 3A). AAV2 with the loops from AAV1 (AAV2 HI1) and AAV8 (AAV2 HI8) generated titers only twofold lower than those of WT AAV2, at 3 × 109 VGs/μl, 3 × 109 VGs/μl, and 6 × 109 VGs/μl, respectively (Fig. 3D), and display heparan sulfate elution profiles similar to that of WT AAV2 (e.g., mostly at 0.4 M PBS; Fig. 3E). As determined through EM analysis, the full particles obtained for the mutant seem to be similar to WT AAV2 full particles in gross conformation (Fig. 3C). In addition, as shown in Fig. 3D, AAV2 HI1, AAV2 HI8, and AAV2 all transduced 293 cells (infected with 3,000 VGs/cell) with similar efficiencies, as determined through a luciferase assay.
FIG. 3.
AAV2 HI1 and AAV2 HI8 substitution mutant characterization. (A) Sequence alignment of AAV2, AAV1, and AAV8 HI loop amino acids. (B) Structural alignment of AAV2, AAV1, and AAV8 HI loops (yellow, pink, and blue). (C) Electron micrograph images of AAV2, AAV2 HI1, and AAV2 HI8 1× PBS-Ca2+-Mg2+-dialyzed, CsCl-purified (previously mentioned), DNA-containing particles. (D) Radioactive DNA dot blotting shown as relative change in titer (left side) and luciferase assay on infected 293 cells with 3,000 VGs/cell (right side; n = 3; standard deviation = black bars). (E) Heparin binding profiles of AAV2, AAV2 HI1, and AAV2 HI8. A 500-μl volume of heparin type IIIS-conjugated agarose beads (Sigma) was incubated with 1010 VG-containing particles (L = load) for 10 min at RT. The washes (1× PBS) and elutions (0.2 to 0.6 M PBS) were collected, and capsids were detected via Western dot blot analysis with monoclonal antibody A20 (1:20).
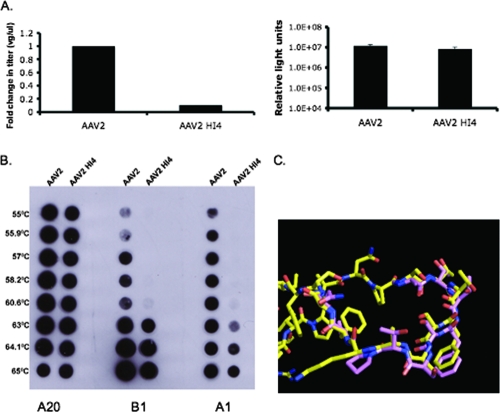
At the other end of the phenotype spectrum, swapping of the HI loops from the less homologous serotypes AAV4 and AAV5 did not produce mutant viruses that were similar to WT AAV2. As shown in Fig. 4A, AAV2 HI4, based on dot blot analysis, produced a virus titer 1 log lower than that of WT AAV2 (1.05 × 109 and 1.13 × 108 VGs/μl, respectively). However, when 293 cells were infected with the same number of VGs per cell, AAV2 HI4 and WT AAV2 infected cells with similar efficiencies (Fig. 4A). Interestingly, when WT AAV2 and AAV2 HI4 full particles were heat treated at 1° increments from 55 to 65°C, AAV2 capsid dissociation was initiated at temperatures as low as 55°C, with complete dissociation at 63°C, whereas AAV2 HI4 did not dissociate and expose VP1 until 63°C (Fig. 4B). This observation suggests that the AAV2 HI4 mutant capsid is more stable than that of WT AAV2, although we are still unsure if there is a correlation between capsid stability and titer.
FIG. 4.
AAV2 HI4 substitution mutant characterization. (A) AAV2 HI4 titer and transduction. AAV2 and AAV2 HI4 titers were quantified via radioactive DNA dot blotting against the luciferase transgene, shown as relative change in titer (left side). AAV2 HI4 transduction was quantified via luciferase assay of 293 cells infected with 3,000 VGs/cell (right side; n = 3; standard deviation = black bars). (B) AAV2 and AAV2 HI4 viral DNA-containing particles (6 × 108) were heat treated at the temperatures shown on the left, transferred to a nitrocellulose membrane, and blotted with the antibodies shown at the bottom at a ratio of 1:20. (C) Structural alignment of the AAV2 (yellow) and AAV4 (pink) HI loops from VP subunits.
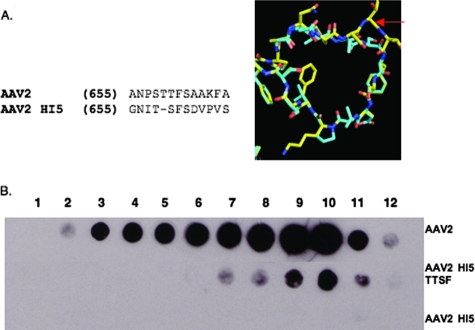
The AAV5 capsid VP subunit, with 59% homology in amino acid sequence to AAV2, contains an HI loop that is one amino acid shorter than those observed in AAV2 and the other AAV serotypes characterized to date. The AAV5 HI loop structurally lacks a threonine at the amino acid position equivalent to residue 659 in AAV2. Substitution of this HI loop into AAV2 (AAV2 HI5) resulted in a change in amino acid sequence beginning at position 655 instead of 658 due to the low homology (Fig. 5A). Although AAV2 HI5 can express VP1, -2, and -3 when the cell lysate is subjected to Western blot analysis (data not shown), the subunit proteins are unable to assemble into intact capsid particles, as determined by A20 Western dot blotting (Fig. 5B). In order to determine if the length of the HI loop contributes to the inability of AAV2 HI5 to assemble particles, a threonine was inserted into AAV2 HI5 at position 659 (AAV2 HI5 TTSF). The threonine insertion does minimally restore capsid assembly but not packaging based on Western dot blot (Fig. 5B) and DNA dot blot (Table 2) analyses, respectively.
FIG. 5.
AAV2 HI5 substitution mutant characterization. (A) Sequence alignment of AAV2 and AAV5 HI loop residues (left side) and structural alignment of AAV2 (yellow) and AAV5 (cyan) HI loops (right side). The amino acid position deleted in AAV5 relative to AAV2 in this region is depicted as a dash in the alignment, and the threonine residue inserted at this position in AAV2 is indicated by a red arrow on the structural superimposition. (B) Western dot blot analysis of AAV2, AAV2 HI5, and AAV2 HI5 TTSF (threonine inserted at amino acid position 659) CsCl gradient fractions blotted with antibody A20 (1:20).
The HI loop swap phenotypes show that specific amino acid side chain interactions of the HI loop can affect particle stability, as seen with the AAV2 HI4 mutant, and the length of the HI loop appears to be crucial for maintaining proper capsid assembly, as in AAV2 HI5; however, DNA packaging ability seems to be more stringently controlled by the loop sequence, as concluded from the AAV2 polyglycine mutant. In addition, the data show that this capsid VP region can tolerate amino acid differences in assembling an AAV capsid, as seen in AAV2 HI1 and HI8, consistent with the observation of the HI loop in all of the parvovirus structures determined to date (Fig. 1A), despite the lack of sequence similarity.
Site-directed mutagenesis.
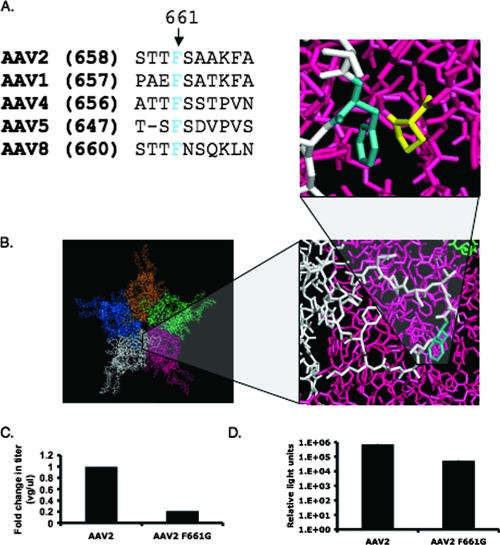
To determine which amino acids within the HI loop are critical for viral genome packaging into the assembled capsids, a series of site-directed mutants were generated in this region based on sequence conservation between the AAV serotypes and observed interactions with the underlying amino acids, since replacing the AAV2 HI loop with glycines appears to ablate DNA packaging capabilities. As shown in the serotype sequence alignment in Fig. 1A, a phenylalanine at position 661 within the HI loop is conserved in the AAV serotypes aligned. In the crystal structure of AAV2, the side chain of F661 interacts with a conserved proline at position 373 in the EF loop within the underlying subunit, possibly through Pi stacking (see Fig. 7B). Such stacking interactions have been shown to play key roles in protein stability and folding (9). Also shown in the sequence alignment, residue K665 is present in most serotypes (Fig. 1A), which, based on the crystal structure, forms a salt bridge with an aspartic acid at position 368 of the underlying subunit. Another amino acid of interest is F666, which resides in a hydrophobic pocket of the underlying subunit. As shown in the sequence and structure alignment (Fig. 1), there is a hydrophobic residue such as valine or isoleucine at this position in all of the serotypes compared. In order to determine which of these three conserved residues are critical for the AAV2 capsid to fully assemble and package the viral genome, the glycine residues at positions 661, 665, and 666 in the AAV2 polyglycine mutant were individually changed back to the amino acids present in the WT AAV2 HI loop, i.e., F661, K665, and F666, respectively. Mutating the residues one at a time did not restore the ability of the AAV2 polyglycine mutant to package the viral DNA (Table 2). This suggests that cooperative interactions facilitated by individual residues are critical for maintaining viral genome packaging capabilities, as seen in the AAV2 HI loop swap mutants (Table 2). This conclusion is further substantiated by the experiments described below.
FIG. 7.
Residue F661 structure model and sequence alignment and AAV2 F661G substitution mutant characterization. (A) Sequence alignment of representative serotypes showing that F661 is conserved (cyan). (B) The HI loop located on a pentamer at the fivefold face (left) is magnified (right), showing that the HI loop from one VP (white) extends over the underlying VP (pink). The F661 side chain (cyan) near the center of the HI loop forms a hydrophobic interaction with a conserved proline residue (yellow) of the underlying adjacent VP (top right). (C) Relative change in AAV2 F661G viral titer determined by radioactive DNA dot blotting against the luciferase transgene. (D) Transduction quantified postinfection of 293 cells with AAV2 and AAV2 F661G (3,000 VGs/cell) and evaluated via luciferase assay (n = 3; standard deviation = black bars).
Peptide substitution studies.
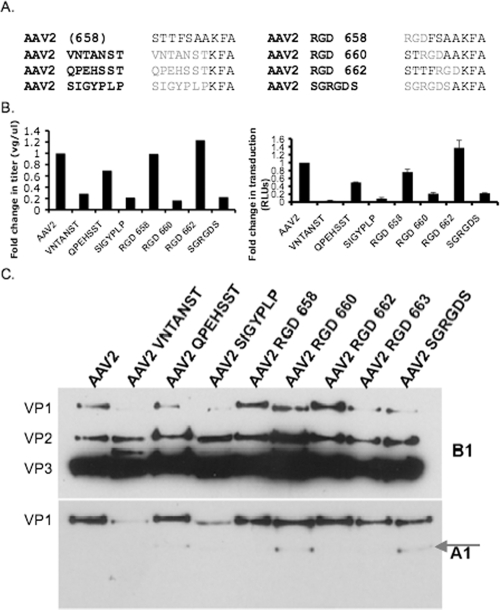
In order to determine the plasticity of the HI loop, gross mutagenesis of amino acid residues within this region was carried out on the capsid. Since the data mentioned previously suggest that a cooperative effect between amino acids is necessary for viral genome packaging, semiconserved residues K665 and F666 were left unchanged. The AAV2 HI loop was replaced with a range of peptide sequences varying in length and beginning at different amino acid positions. First, short RGD peptides were replaced in an effort to “walk through” the HI loop and characterize the effects of disparate non-AAV sequences on viral assembly and packaging. Amino acid positions 658 to 660, 660 to 662, 662 to 664, and 663 to 665 were replaced with an RGD peptide (38) (AAV2 RGD 658, AAV2 RGD 660, AAV2 RGD 662, and AAV2 RGD 663, respectively) (Fig. 6A). Most mutants were obtained at virus titers within twofold of that of WT AAV2, with the exception of mutant AAV2 RGD 660, which was obtained at a sixfold lower titer than WT AAV2, i.e., 1.48 × 108 VGs/μl compared to 8.81 × 108 VGs/μl (Fig. 6B). AAV2 RGD 658 and AAV2 RGD 662, after adjusting for VG number, had infectivities similar to that of the WT based on a luciferase assay (Table 2). However, when 293 cells were infected with AAV2 RGD 660, it was fivefold less infectious than WT AAV2 (Fig. 6B). AAV2 RGD 660 was lower in titer and infectivity than the WT and had a replacement of conserved residue F661 (see Site-directed mutagenesis) with a glycine residue, suggesting a potential role for F661 in the virus life cycle. In light of this single amino acid and its phenotypic effects, we introduced longer peptides into the AAV2 HI loop for increased amino acid variability and to gain more insight into structure-function constraints in manipulating this region.
FIG. 6.
AAV2 HI loop peptide substitution mutant characterizations. (A) Sequence alignment showing residues in the AAV2 HI loop that were replaced with the specific peptides indicated. (B) HI loop peptide substitution titer and transduction. Relative change in titer compared to AAV2 determined by radioactive DNA dot blot analysis (left side). Relative change in transduction of infected 293 cells (3,000 VGs/cell) determined by luciferase assay (right side; n = 3; standard deviation = black bars). AAV2 RGD 663 titer and SK-OV3 cell infectivity present in Table 2. (C) Western blot analysis of peptide substitution mutants incubated overnight at 4°C with the A1 (1:20) and B1 (1:20) monoclonal antibodies. The arrow indicates an additional protein band detected with the A1 antibody (bottom).
Seven-amino-acid peptides successfully used as insertions in previous capsid mutagenesis studies were chosen in order to determine if the variable region of the HI loop could tolerate these peptides as substitutions. Starting at position 658 in the AAV2 HI loop, we substituted peptides QPEHSST, VNTANST, SIGYPLP (53), and SGRGDS (25) (Fig. 6A). All of the mutants generated virus. AAV2 QPEHSST was able to make virus in titers similar to that of the WT (within 2-fold), followed by AAV2 VNTANST (3.5-fold lower than AAV2), AAV2 SIGYPLP, and AAV2 SGRGDS (4.5-fold lower than AAV2). However, AAV2 VNTANST was 27-fold less infectious than WT AAV2, while AAV2 SIGYPLP showed approximately 10-fold less infectivity (Table 1). AAV2 SGRGDS and AAV2 QPEHSST were 4.5- and 2-fold less infectious than WT AAV2, respectively (Fig. 6B). The aforementioned changes in titer and infectivity upon gross mutagenesis of the AAV2 HI loops suggest that the amino acid interactions between the HI loop and underlying subunit are crucial for maintaining AAV viability. Data in the literature suggest that the fivefold axis is necessary for viral genome packaging and VP1 externalization (8, 26, 55). We do see a viral genome packaging defect in some of the mutants mentioned above, correlating with a decrease in infectivity. To further understand the phenotypic changes observed, we carried out a battery of biochemical analyses.
Biochemical analysis.
A series of biochemical analyses such as heparin binding, heat treatment, and Western blotting were performed in order to understand why the titer and infectivity of AAV2 RGD 660, AAV2 VNTANST, AAV2 SIGYPLP, and AAV2 SGRGDS were consistently lower than those of WT AAV2; from such studies, Western blot analysis revealed an interesting capsid phenotype, as shown in Fig. 6C. We used monoclonal antibodies B1 and A1 (52), which recognize the VP3 C terminus and VP1 unique N terminus, respectively, to characterize the peptide substitution variants described above. Based on B1 staining, AAV2 VNTANST had decreased VP1 incorporation and an extra protein band between VP2 and VP3. In addition, AAV2 SIGYPLP had decreased VP1 incorporation. Interestingly, when blotted with A1 antibody against the VP1 unique region of AAV2, AAV2 QPEHSST, AAV2 RGD 660, and AAV2 SGRGDS revealed a second band at 77 kDa. Notably, AAV2 RGD 660 and the longer peptide substitutions that were detrimental to virus titer and infectivity all changed the amino acid type at the conserved phenylalanine at position 661 (Fig. 1C). Therefore, the role of this residue in AAV2 HI loop functionality was investigated further.
Analysis of the conserved F661 residue.
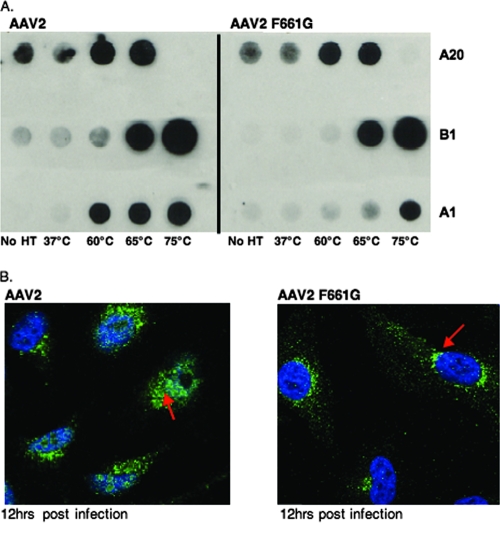
F661, which is completely conserved throughout all AAV serotypes (Fig. 7A) and interacts with a proline in the underlying VP subunit, was mutated to a glycine residue in AAV2 (AAV2 F661G). AAV2 F661G produced virus fivefold lower in titer than WT AAV2 based on dot blot analysis (8.2 × 107 and 4.1 × 108 VGs/μl, respectively; Fig. 7C). AAV2 F661G also binds heparin with an affinity similar to that of WT AAV2 based on heparin column binding and elution with increasing concentrations of salt (Table 2). Interestingly, AAV2 F661G is 1 log less infectious than AAV2 based on a luciferase assay (Fig. 7D). In addition to these data, AAV2 F661G and WT AAV2 capsids were heat treated at 37°C, 50°C, 60°C, 65°C, and 75°C. While AAV2 was able to expose the VP1 N terminus based on A1 antibody staining at 60°C, AAV2 F661G was unable to do so at this temperature and could only expose the VP1 N terminus when heated to 75°C upon capsid dissociation (Fig. 8A). To further corroborate the data showing decreased incorporation of VP1 and reduced infectivity of AAV2 F661G, we carried out intracellular trafficking studies by confocal fluorescence microscopy. Briefly, HeLa cells were infected with WT AAV2 and AAV2 F661G particles at 30,000 VGs/cell. The cells were fixed at 12 h postinfection and stained with primary antibody A20 for intact capsid detection and then a secondary goat anti-mouse 488-nm fluorophore-conjugated antibody. In addition, the nuclei were stained with DAPI. Based on this analysis, AAV2 F661G was unable to enter the nucleus efficiently and appeared to remain perinuclear, unlike WT AAV2, which trafficked into the nucleus more efficiently (Fig. 8B).
FIG. 8.
AAV2 F661G VP1 externalization and virus infectivity. (A) Heat treatment (HT) of 6 × 108 AAV2 and AAV2 F661G viral DNA-containing particles (temperatures across the bottom). Treated capsids were blotted onto a nitrocellulose membrane, and intact capsids, dissociated VPs, and externalized VP1 unique N termini were detected with antibodies A20, B1, and A1 (1:20) as indicated on the right. (B) HeLa cells were infected with 50,000 VG-containing particles of AAV2 or AAV2 F661G. Cells were fixed and permeabilized, and intact capsids were detected with primary antibody A20 (1:10) and a 488-nm fluorophore-conjugated secondary antibody (1:1,250). The nucleus was stained blue with DAPI. Red arrows indicate AAV2 (left) and AAV2 F661G (right) intact capsids.
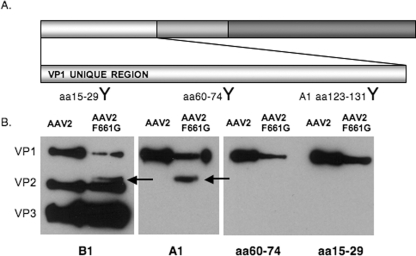
In addition, when 109 VGs of dialyzed full particles were run on a Western blot and stained with monoclonal antibodies B1 and A1, AAV2 F661G revealed a fourth-molecular-mass capsid species between VP1 and VP2 consistently running at 77 kDa. With antibodies that specifically detect amino acids 15 to 29 and 60 to 74 in the VP1 unique region (Fig. 9A), the capsid band at 77 kDa was not detected on the Western blot, further confirming an N-terminal truncation of this capsid subunit (Fig. 9B). This molecular mass species is identical to the novel protein band seen in the Western blot with the AAV2 QPEHSST, AAV2 RGD 660, and AAV2 SGRGDS capsid subunit proteins. The novel capsid subunit is approximately 100 amino acid residues shorter than VP1 (Fig. 6B), implicating potential proteolytic processing of the exposed VP1 N terminus in these HI loop variants.
FIG. 9.
AAV2 F661G VP incorporation. (A) Schematic representation of AAV2 VP3 with an enlargement of the VP1 unique region. The shaded regions represent VP1/VP2 and the VP1/VP2/VP3 common regions, respectively. Antibodies generated (Pacific Immunology) recognize specific amino acid stretches in the VP1 unique region, as indicated at the bottom (1:1,000). (B) Western blot analysis of AAV2 and AAV2 F661G capsids with antibodies B1 and A1 (1:20). An additional protein band was detected at ∼77 kDa (arrows).
Therefore, the aforementioned data demonstrate that the HI loop can tolerate most amino acid changes, and specific cooperative amino acid interactions are necessary for proper viral genome packaging. Additionally, the F661-P373 hydrophobic interaction appears crucial for proper incorporation of the VP1 subunit into the AAV2 capsid. Without the proper interactions, a distinct VP1 subunit lacking its N terminus containing the phospholipase activity is incorporated into the capsid, directly impacting virus infectivity.
DISCUSSION
With the availability of the AAV2 crystal structure (59), many aspects of the AAV life cycle, including host cell recognition (3, 11, 23, 33, 49), intracellular trafficking (5, 10), and uncoating (43), are now possible to correlate with structure. The first of such studies have centered around the threefold loops and the determination that they are key topological features in host cell recognition (4, 23). Similar structure-function studies have extended from the threefold loops to the fivefold axis and the location of the virion pore and its potential role in viral genome packaging, capsid assembly, and VP1 unique N-terminal exposure (8, 26, 55). Interestingly, the HI loop surrounds the fivefold pore and has an essential structural role in viral assembly by overlapping the neighboring VP3 subunit forming amino acid interactions with the underlying EF loop (Fig. 1). Recently, Mavis Agbandje-McKenna has observed the HI loop flipping up 90° upon AAV2-heparan sulfate proteoglycan binding, suggesting a dynamic role for this structure in the viral infectious pathway (unpublished). To better understand the role of this capsid structure, we chose to characterize in detail the HI loop as it may contribute to specific stages in the virus life cycle such as viral genome packaging, assembly, and subsequent stages during the infectious pathway. The results of this study demonstrate that the AAV2 HI loop is essential for proper capsid assembly, packaging of the viral DNA, and viral infectivity when the conserved phenylalanine at amino acid position 661 is altered.
We carried out a comprehensive amino acid deletion-and-substitution study to uncover the role of the HI loop in the AAV life cycle. From these efforts, we determined that removal of the HI loop (AAV2 HI−/−) leads to capsids that cannot assemble (Fig. 2). We assayed viral assembly primarily with the A20 monoclonal antibody (52), which detects tertiary structure of properly assembled AAV capsids. Viruses that were unable to form virions were further studied with gradients and Western blot analyses that confirmed the ability to synthesize VP subunits (data not shown). Although we relied primarily on A20 recognition to confirm the ability to form proper virion structures, additional studies such as iodixanol gradient purification of AAV2 HI−/− cell lysate, followed by EM analysis, further determined that this mutant only appears to generate monomer subunits (data not shown). In contrast, replacement of the loop with glycines (AAV2 polyglycine) generated A20-recognizable assembled capsids; however, these capsids were deficient in the ability to package the viral DNA. Even though this mutant provided sufficient structure to assemble intact AAV particles, the glycine substitutions specifically ablate amino acid side chain interactions with the EF loop of the underlying subunit, suggesting that the HI loop structure and the backbone interactions of the HI loop with the underlying subunit are sufficient to facilitate capsid formation. However, the specific amino acid interactions are required for efficient packaging of the viral DNA. Although we cannot draw from our studies the exact mechanism of the viral genome packaging deficiencies of the glycine substitution mutant, it is interesting to speculate that this phenotype can possibly be attributed to gross conformational changes in the structure of the fivefold pore since the fivefold pore has been implicated as the site of Rep protein binding, a necessary step for efficient DNA encapsidation (7, 8). As a result, it will be of interest to determine the ability of HI loop mutants to bind Rep protein in pull-down experiments as previously described (7).
In addition to the deletion-and-substitution studies described above, we decided to swap the AAV2 HI loop with those from representative serotypes. Domain swapping, specifically between virus serotypes, allows for determination of the role of that specific region of the capsid in the virus life cycle. For example, in adenovirus, the fiber necessary for host cell recognition (54), was swapped between adenovirus subgroups, resulting in alteration of intracellular trafficking of the adenovirus vectors (29). Alternatively, in the case of AAV, domains were swapped between AAV1 and -2 in order to characterize determinants necessary for AAV1 muscle tropism (19). In a similar study regarding AAV, domains from AAV2 were swapped with those from AAV8, demonstrating that loop IV of AAV8 at the threefold axis of symmetry is responsible for dictating liver tropism (37). Interestingly, in our studies, swapping the AAV2 HI loop with those of AAV1 and AAV8 did not affect titer, transduction, heparin binding, or gross conformation. However, this can be expected due to the relatively higher sequence homology between AAV2 and these serotypes. More importantly, these results suggest that the HI loop most likely does not contain determinants of tissue tropism or receptor binding. This was further confirmed via mouse intramuscular injections with AAV2 RGD 662. Briefly, bioluminescence imaging revealed similar in vivo transduction profiles for AAV2 RGD 662 and WT AAV2 1 week postadministration (data not shown).
In the case of the AAV4 and AAV5 HI loops, significant phenotypic variations were seen, possibly due to lower sequence homology with AAV2. For instance, the AAV4 HI loop is composed of a higher number of hydrophobic residues than the loop from AAV2 based on the amino acid sequence and crystal structure (16). The three-dimensional structure of the AAV2 VP3 monomer (Fig. 4C) shows that the side chains of residues 659 and 660 point away from the capsid and do not interact with the residues in the underlying subunit. On the other hand, the conserved phenylalanine at position 661 interacts with proline 373 in the EF loop of the underlying subunit, as mentioned in Results. The alanine-to-serine change at position 663 of the AAV2 HI4 mutant might contribute a hydrogen bond interaction due to a potentially accessible hydroxyl group that is not present in the AAV2 HI loop. The K665P change in AAV2 HI4 suggests a possible contribution to increased hydrophobic interactions with proline, valine, phenylalanine, and methionine residues found in the underlying subunit of AAV2. However, this assessment is based on a structure model of AAV2 HI4, and a more accurate analysis of the AAV4 HI loop amino acid contributions to AAV2 capsid stability is dependent upon knowing the crystal structure of the AAV2 HI4 mutant.
Collectively, the aforementioned amino acid changes in AAV2 HI4 could enhance HI loop-EF loop interactions and thereby could well account for increased capsid stability, as demonstrated through increased resistance to heat treatment in comparison to AAV2. In addition, such increases in capsid stability and possible gross conformational changes to the fivefold pore might account for lower packaging efficiency and titers seen with the AAV2 HI4 mutant. It is possible that the AAV capsid “breathes” or expands in volume during viral genome packaging, and if the capsid is too stable or held too tightly together, it may be more difficult for the Rep protein to package the viral genome. The idea of capsid expansion has been studied in bacteriophage, and it has been shown that during the DNA packaging process a conformational change occurs which causes an increase in capsid volume (21).
Additionally, previous data suggest that the Rep protein is bound in higher quantities to the capsids of packaging-deficient mutants (7), possibly due to “jamming” of the genome threading machinery. Such has also been noted in the case of AAV2 HI4, wherein the particle bound increased amounts of Rep protein in comparison with AAV2 (data not shown). For the AAV2 HI4 mutant, it is possible that there is increased stability of the particle based on the presence of another protein or proteins on the capsid surface.
In the case of AAV2 HI5, despite normal expression of capsid subunit proteins, no intact capsid assembly is observed. This was further confirmed via EM analysis on cesium chloride gradient fractions of the AAV2 HI5-transfected cell lysate. It was determined that the AAV2 HI5 mutant may form pentamers but does not form proper intact particles (data not shown). This phenotype is likely attributable to the fact that the AAV5 HI loop is one amino acid shorter, based on the crystal structure (48), than the WT AAV2 HI loop. In corollary, insertion of the missing threonine at position 659 minimally rescues capsid assembly. Therefore, the length of the HI loop in relation to the underlying subunit appears to be crucial for proper capsid assembly, while the loop amino acid interactions with the underlying subunit dictate genome packaging efficiency. This was further corroborated by a new collection of HI mutants that increased the length of the HI loop by 1, 3, 5, and 7 glycine residues. The fact that the AAV2 HI loop glycine extensions formed intact virus particles but were unable to package the viral genome, based on DNA dot blot (data not shown) and Western dot blot (data not shown) analyses, further supports the importance of the length of the HI loop.
Following the domain swaps, we decided to use peptide substitutions in order to mutate multiple residues of the HI loop. Many groups have successfully inserted peptides, specifically at the threefold loops, on the capsid surface as a means to retarget the virus for specific tissue types (14, 38, 39, 53). In this study, we used peptides not as insertions but as substitutions in a novel region of the capsid. Peptide substitution within the AAV2 HI loop showed that certain amino acid changes do not affect virus titer and transduction, as seen with the AAV2 RGD 658, AAV2 RGD 662, and AAV2 QPEHSST mutants. However, some peptide substitutions resulted in marked changes in phenotype that were dependent on the amino acid position substituted. For instance, replacement with peptide RGD at position 660 (AAV2 RGD 660), SGRGDS starting at position 658 (AAV2 SGRGDS), VNTANST starting at position 658 (AAV2 VNTANST), and SIGYPLP also starting at position 658 (AAV2 SIGYPLP) resulted in decreased titer and infectivity. In all of these mutants, the conserved F661 residue observed in all serotypes is replaced.
Interestingly, a number of these mutants, such as AAV2 VNTANST, SIGYPLP, RGD 660, and SGRGDS, also revealed differential banding patterns seen with B1 antibody staining of a Western blot (Fig. 6C). Specifically, there appears to be a decreased incorporation of VP1 capsid subunits in these mutant capsids. It is likely that such a phenotype, which would reduce the effectiveness of the PLA2 domain (located in the VP1 N-terminal domain) required for endosomal escape and nuclear entry of the viral capsid, could explain the decrease in transduction seen with these mutants (15, 17, 26, 42). The lower titers of the aforementioned mutants can possibly be attributed to improper capsid assembly (7, 44) and defective packaging (7). In the AAV2 VNTANST mutant, there is an additional protein band seen between VP2 and VP3 with B1 staining that has yet to be identified. The observed protein product is most likely due to proteolytic processing of VP1, which could also account for the decreased amount of VP1 present in this capsid mutant. The 77-kDa protein band in AAV2 RGD 660 and AAV2 SGRGDS shown by A1 staining further corroborates these speculations (Fig. 6C).
As mentioned above, one trait shared by these defective peptide substitution mutants is that they span the conserved phenylalanine at amino acid position 661. F661 interacts with P373 in the EF loop in the underlying subunit of all serotypes through stacking interactions (Fig. 7B). This interaction appears critical for the stability (9) of assembled capsid subunits since the HI loop is the only region at the fivefold axis of symmetry that extends from one subunit and overlaps the underlying subunit. Mutation of F661 results in a phenotype similar to that seen with peptide substitutions spanning this region. Based on data shown in Fig. 9, we hypothesize that the interaction between F661 and P373 is necessary to stabilize the capsid around the fivefold axis of symmetry, being of great importance due the fivefold pores’ contribution to viral genome packaging and infectivity (8, 55). Disruption of this interaction appears, in particular, to reduce the amount of VP1 incorporated into these mutant capsids (Fig. 9B).
Additionally, such mutagenesis could result in improper incorporation of VP1 subunits at the fivefold axis of symmetry, which would expose the critical PLA2 domain to cellular proteases during virus production. If unassembled VP1 monomers or loosely assembled particles exposing the VP1 unique N terminus are present, it is possible that they may be susceptible to cellular proteases. This may not occur as readily in the WT or other mutant viruses that are able to assemble the VP monomers efficiently in a stable configuration. Further studies will help elucidate these hypotheses.
In conjunction with this observation, a similar phenomenon may be occurring in the AAV baculovirus production system (41, 46). There appears to be inefficient incorporation of VP1 into the AAV2 capsid during production in insect cells, and this may be due to the susceptibility of VP1 to cellular protease in the nonmammalian cell environment (24). The notion that VP1 is susceptible to cellular proteases is further substantiated by the fact that when mammalian cells were transfected with VP1 constructs, specifically, VP1NLSFKN and VP1FKN, a second-molecular-mass band was detected between VP1 and VP2 in the cell lysates (17), similar to the result obtained in this study. Upon mutation of F661, this molecular mass species was not only generated but also incorporated into the intact capsid.
In addition, it is not surprising that a single amino acid on the AAV capsid, such as F661, could significantly impact the biology of the virus. A recent study in our laboratory has shown that single amino acid mutations, specifically, K531E in AAV6 and E531K in AAV1, suppress and enhance heparin binding, respectively (56). Taken together, our data support the role of the HI loop as an important AAV capsid structural element necessary for proper incorporation of VP1 into an assembled infectious particle and a functional fivefold pore that allows efficient packaging of viral genomes.
Acknowledgments
Nina DiPrimio is the recipient of pharmacology training grant 2-T32-GM007040 from the National Institutes of Health (NIGMS). R.J.S. (P01HL051818) and M.A.-M. (P01 HL59412 and P01 HL51811) also acknowledge the National Institutes of Health for research funding.
Special thanks to Ryan Nelson for assistance with heat treatment experiments and Brittney Gurda in Mavis Agbandje-McKenna's laboratory at the University of Florida for performing EM analyses on AAV2 HI5 and AAV2 HI−/− mutants.
Footnotes
Published ahead of print on 26 March 2008.
REFERENCES
- 1.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16155-171. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 61369-1381. [DOI] [PubMed] [Google Scholar]
- 3.Akache, B., D. Grimm, K. Pandey, S. R. Yant, H. Xu, and M. A. Kay. 2006. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 809831-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asokan, A., J. B. Hamra, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2006. Adeno-associated virus type 2 contains an integrin α5β1 binding domain essential for viral cell entry. J. Virol. 808961-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 742777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns, K. I., and R. M. Linden. 1995. The cryptic life style of adeno-associated virus. Bioessays 17237-245. [DOI] [PubMed] [Google Scholar]
- 7.Bleker, S., M. Pawlita, and J. A. Kleinschmidt. 2006. Impact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. J. Virol. 80810-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 792528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo, M. D., G. W. Platt, R. Bofill, and M. S. Searle. 2004. Context-dependent effects of proline residues on the stability and folding pathway of ubiquitin. Eur. J. Biochem. 2714474-4484. [DOI] [PubMed] [Google Scholar]
- 10.Ding, W., L. Zhang, Z. Yan, and J. F. Engelhardt. 2005. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 12873-880. [DOI] [PubMed] [Google Scholar]
- 11.Di Pasquale, G., B. L. Davidson, C. S. Stein, I. Martins, D. Scudiero, A. Monks, and J. A. Chiorini. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 91306-1312. [DOI] [PubMed] [Google Scholar]
- 12.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. USA 10217148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 786381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girod, A., M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deleage, and M. Hallek. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 51052-1056. [DOI] [PubMed] [Google Scholar]
- 15.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83973-978. [DOI] [PubMed] [Google Scholar]
- 16.Govindasamy, L., E. Padron, R. McKenna, N. Muzyczka, N. Kaludov, J. A. Chiorini, and M. Agbandje-McKenna. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 8011556-11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grieger, J. C., J. S. Johnson, B. Gurda-Whitaker, M. Agbandje-McKenna, and R. J. Samulski. 2007. Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J. Virol. 817833-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grieger, J. C., S. Snowdy, and R. J. Samulski. 2006. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J. Virol. 805199-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauck, B., and W. Xiao. 2003. Characterization of tissue tropism determinants of adeno-associated virus type 1. J. Virol. 772768-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzog, R. W. 2007. Immune responses to AAV capsid: are mice not humans after all? Mol. Ther. 15649-650. [DOI] [PubMed] [Google Scholar]
- 21.Jardine, P. J., and D. H. Coombs. 1998. Capsid expansion follows the initiation of DNA packaging in bacteriophage T4. J. Mol. Biol. 284661-672. [DOI] [PubMed] [Google Scholar]
- 21a.Jones, T. A., J.-Y. Zou, S. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47100-119. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann, B., A. A. Simpson, and M. G. Rossmann. 2004. The structure of human parvovirus B19. Proc. Natl. Acad. Sci. USA 10111628-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 7711072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlbrenner, E., G. Aslanidi, K. Nash, S. Shklyaev, M. Campbell-Thompson, B. J. Byrne, R. O. Snyder, N. Muzyczka, K. H. Warrington, Jr., and S. Zolotukhin. 2005. Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol. Ther. 121217-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koivunen, E., D. A. Gay, and E. Ruoslahti. 1993. Selection of peptides binding to the α5β1 integrin from phage display library. J. Biol. Chem. 26820205-20210. [PubMed] [Google Scholar]
- 26.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 795296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lochrie, M. A., G. P. Tatsuno, B. Christie, J. W. McDonnell, S. Zhou, R. Surosky, G. F. Pierce, and P. Colosi. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 80821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, E. B., B. Gurda-Whitaker, L. Govindasamy, R. McKenna, S. Zolotukhin, N. Muzyczka, and M. Agbandje-McKenna. 2006. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 621271-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazawa, N., P. L. Leopold, N. R. Hackett, B. Ferris, S. Worgall, E. Falck-Pedersen, and R. G. Crystal. 1999. Fiber swap between adenovirus subgroups B and C alters intracellular trafficking of adenovirus gene transfer vectors. J. Virol. 736056-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam, H. J., M. D. Lane, E. Padron, B. Gurda, R. McKenna, E. Kohlbrenner, G. Aslanidi, B. Byrne, N. Muzyczka, S. Zolotukhin, and M. Agbandje- McKenna. 2007. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 8112260-12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 776995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padron, E., V. Bowman, N. Kaludov, L. Govindasamy, H. Levy, P. Nick, R. McKenna, N. Muzyczka, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna. 2005. Structure of adeno-associated virus type 4. J. Virol. 795047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 571-77. [DOI] [PubMed] [Google Scholar]
- 34.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy, V. S., P. Natarajan, B. Okerberg, K. Li, K. V. Damodaran, R. T. Morton, C. L. Brooks III, and J. E. Johnson. 2001. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J. Virol. 7511943-11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose, J. A., J. V. Maizel, Jr., J. K. Inman, and A. J. Shatkin. 1971. Structural proteins of adenovirus-associated viruses. J. Virol. 8766-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen, X., T. Storm, and M. A. Kay. 2007. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol. Ther. 151955-1962. [DOI] [PubMed] [Google Scholar]
- 38.Shi, W., and J. S. Bartlett. 2003. RGD inclusion in VP3 provides adeno-associated virus type 2 (AAV2)-based vectors with a heparan sulfate-independent cell entry mechanism. Mol. Ther. 7515-525. [DOI] [PubMed] [Google Scholar]
- 39.Shi, X., G. Fang, W. Shi, and J. S. Bartlett. 2006. Insertional mutagenesis at positions 520 and 584 of adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors with eliminated heparin-binding ability and introduced novel tropism. Hum. Gene Ther. 17353-361. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 3151189-1198. [DOI] [PubMed] [Google Scholar]
- 41.Sollerbrant, K., J. Elmen, C. Wahlestedt, J. Acker, H. Leblois-Prehaud, M. Latta-Mahieu, P. Yeh, and M. Perricaudet. 2001. A novel method using baculovirus-mediated gene transfer for production of recombinant adeno-associated virus vectors. J. Gen. Virol. 822051-2060. [DOI] [PubMed] [Google Scholar]
- 42.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 8011040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, C. E., T. A. Storm, Z. Huang, and M. A. Kay. 2004. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 783110-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timpe, J., J. Bevington, J. Casper, J. D. Dignam, and J. P. Trempe. 2005. Mechanisms of adeno-associated virus genome encapsidation. Curr. Gene Ther. 5273-284. [DOI] [PubMed] [Google Scholar]
- 45.Tsao, J., M. S. Chapman, H. Wu, M. Agbandje, W. Keller, and M. G. Rossmann. 1992. Structure determination of monoclinic canine parvovirus. Acta Crystallogr. B 48(Pt. 1)75-88. [DOI] [PubMed] [Google Scholar]
- 46.Urabe, M., C. Ding, and R. M. Kotin. 2002. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 131935-1943. [DOI] [PubMed] [Google Scholar]
- 47.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 761884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters, R. W., M. Agbandje-McKenna, V. D. Bowman, T. O. Moninger, N. H. Olson, M. Seiler, J. A. Chiorini, T. S. Baker, and J. Zabner. 2004. Structure of adeno-associated virus serotype 5. J. Virol. 783361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 27620610-20616. [DOI] [PubMed] [Google Scholar]
- 50.Warrington, K. H., Jr., O. S. Gorbatyuk, J. K. Harrison, S. R. Opie, S. Zolotukhin, and N. Muzyczka. 2004. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 786595-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warrington, K. H., Jr., and R. W. Herzog. 2006. Treatment of human disease by adeno-associated viral gene transfer. Hum. Genet. 119571-603. [DOI] [PubMed] [Google Scholar]
- 52.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 749281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Work, L. M., H. Buning, E. Hunt, S. A. Nicklin, L. Denby, N. Britton, K. Leike, M. Odenthal, U. Drebber, M. Hallek, and A. H. Baker. 2006. Vascular bed-targeted in vivo gene delivery using tropism-modified adeno-associated viruses. Mol. Ther. 13683-693. [DOI] [PubMed] [Google Scholar]
- 54.Wu, E., L. Pache, D. J. Von Seggern, T. M. Mullen, Y. Mikyas, P. L. Stewart, and G. R. Nemerow. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J. Virol. 777225-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 748635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, Z., A. Asokan, J. C. Grieger, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2006. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 8011393-11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, Z., A. Asokan, and R. J. Samulski. 2006. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 14316-327. [DOI] [PubMed] [Google Scholar]
- 58.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 722224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 9910405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie, Q., T. Somasundaram, S. Bhatia, W. Bu, and M. S. Chapman. 2003. Structure determination of adeno-associated virus 2: three complete virus particles per asymmetric unit. Acta Crystallogr. D Biol. Crystallogr. 59959-970. [DOI] [PubMed] [Google Scholar]