Abstract
The herpes simplex virus UL56 gene is conserved among most members of the Alphaherpesvirinae family and plays a critical role in viral pathogenicity in vivo. The HSV-2 UL56 protein (UL56) is a C-terminally anchored type II membrane protein that is predicted to be inserted into the virion envelope, leaving its N-terminal domain in the tegument. UL56 interacts with KIF1A and UL11. Here we report that UL56 also interacts with the ubiquitin ligase Nedd4 and increases its ubiquitination. Nedd4 was identified as a UL56-interacting protein by a yeast two-hybrid screen. UL56 bound to Nedd4 via its PY motifs. Nedd4 was phosphorylated and degraded in wild-type HSV-2-infected cells but not in cells infected with a UL56-deficient mutant. Ubiquitination assays revealed that UL56 increased ubiquitinated Nedd4, which was actively degraded in infected cells. UL56 also caused a decrease in Nedd4 protein levels and the increased ubiquitination in cotransfected cells. However, UL56 itself was not ubiquitinated, despite its interaction with Nedd4. Based on these findings, we propose that UL56 regulates Nedd4 in HSV-2-infected cells, although deletion of UL56 had no apparent effect on viral growth in vitro.
Herpes simplex virus (HSV) is a large, enveloped DNA virus with a genome possessing at least 74 different genes (19, 46). Although approximately half of the HSV genes are not essential for replication in vitro, all of these accessory gene products are predicted to play indispensable roles in viral replication and dissemination in vivo (57).
The HSV UL56 gene is an accessory gene that most members of the Alphaherpesvirinae family possess homologs for (except bovine herpes virus-1 and -5) (1, 17, 19, 22, 29, 34, 37, 46, 53, 65-70; M. Schwyzer, V. Paces, G. J. Letchworth, V. Misra, H. J. Buhk, D. E. Lowery, C. Simard, L. J. Bello, E. Thiry, and C. Vlcek, 1995, complete DNA sequence of bovine herpesvirus 1. GenBank database [http://www.ncbi.nlm.nih.gov/Genbank/index.html] accession number NC_001847) and has been shown to play an important role in HSV type 1 (HSV-1) pathogenicity in vivo. UL56-deficient HSV-1 mutants are substantially less neuroinvasive (4, 58), although little is known molecularly about how this attenuation occurs. HSV-2 UL56 is a 235-amino-acid, C-terminally anchored, type II membrane protein (39) that is predicted to be inserted into the viral envelope so that the N-terminal domain is located in the virion tegument. In this topology, UL56 is predicted to have a 216-amino-acid cytoplasmic domain. UL56 associates with the neuron-specific kinesin KIF1A (41) and the HSV-2 protein UL11 (40). KIF1A is involved in the axonal transport of synaptic vesicle precursors (51), and its association with UL56 suggests that UL56 may affect vesicular transport in infected neurons. UL11 is a tegument protein that is involved in the envelopment and egress of viral nucleocapsids (3) and has dynamic membrane-trafficking properties (45). UL56 may specifically promote UL11 function in virion envelopment and egress in infected neurons. However, the precise role and function of UL56 in viral replication and pathogenicity are still unknown.
Ubiquitin-mediated protein modifications regulate a variety of cellular processes, including protein turnover and trafficking, endocytosis, and transcription factor activation (77). Recent studies have also strongly associated ubiquitination with viral pathogenesis. The ubiquitin-proteasome system is involved in viral immune evasion, viral progeny release and (or) budding, viral transcriptional regulation, and the suppression of apoptosis (20). Ubiquitination is executed by a hierarchical cascade of three types of enzymes: ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) (55).
A major class of E3 proteins contains a domain homologous to the E6-AP carboxyl terminus (called a HECT domain). Nedd4 (neuronal precursor cell-expressed, developmentally downregulated 4) is a ubiquitin ligase with a catalytic domain of the HECT class (25). Nedd4 was originally identified as a developmentally regulated gene that was highly expressed in the mouse embryonic central nervous system (43). Further analysis revealed that Nedd4 was expressed at various levels in several embryonic and adult tissues (42). Nedd4 is the prototypical member of the Nedd4 ubiquitin ligase family and regulates diverse cellular processes including signal transduction (which is involved in cancer development [10, 49, 73, 75]), protein trafficking (52, 63), and viral budding (6, 23, 36, 60, 71, 80).
In this study, we searched for additional host cell factors that interact with UL56 to elucidate its biological role and function. We identified Nedd4 as a UL56-interacting protein. Moreover, we demonstrated that although UL56 was not ubiquitinated, it was required for enhanced degradation of Nedd4 in HSV-2-infected cells. To our knowledge, this is the first report demonstrating that an HSV-specific protein binds to and targets Nedd4.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (African green monkey kidney cells), HEp2 cells (human laryngeal carcinoma cell line), and SK-N-SH cells (human neuroblastoma cells) were obtained from the Riken BioResource Center (Ibaraki, Japan). Vero cells were maintained in Eagle's minimum essential medium (MEM) supplemented with 8% calf serum (CS), 100 U/ml penicillin, and 100 μg/ml streptomycin. HEp2 cells and SK-N-SH cells were maintained in Dulbecco's modified Eagle's medium and MEM alpha (Invitrogen, Carlsbad, CA), respectively, supplemented with 10% fetal CS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The wild-type HSV-2 strain (186) was used as the prototype strain in this study. The generation of the recombinant UL56 (ΔUL56Z), US3 (L1BR1), and US2 (YY2) deletion mutant viruses was previously described in detail (30, 40, 50). Viruses were propagated and titrated on Vero cells. Infections were routinely performed at a multiplicity of infection (MOI) of 3 PFU/cell (except where otherwise indicated).
Antibodies and reagents.
The following commercial antibodies were used: polyclonal anti-Nedd4 (Millipore, Billerica, MA) and anti-c-Myc (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies, monoclonal anti-E6AP (BD Biosciences, Franklin Lakes, NJ), anti-FLAG (M2), anti-β-actin (AC-15) (Sigma, Saint Louis, MO), and anti-VP5 (Abcam, Cambridge, United Kingdom) antibodies, horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse immunoglobulin G (IgG) (Invitrogen), and Alexa Fluor 488-conjugated goat anti-rabbit and 594-conjugated goat anti-mouse IgG (Invitrogen). Protein A affinity-purified normal rabbit IgG was purchased from Millipore. Polyclonal anti-UL56 and anti-VP16 antisera were described previously (32, 39). Anti-UL56 and preimmune sera were purified using a melon gel IgG spin purification kit (Pierce Biotechnology, Rockford, IL) for immunoprecipitation assays. Polyclonal anti-UL36 antiserum was generated using an HSV-2 UL36 (amino-terminal 483-residue)-His fusion protein to immunize rabbits. Immunization and collection of antisera were performed as previously described. The generation and evaluation of the antisera will be described in detail elsewhere. The proteasome inhibitor MG132 was purchased from Biomol International (Plymouth Meeting, PA).
Yeast two-hybrid screen.
The DNA fragment encoding a truncated UL56 protein (amino acids 1 to 210), which lacks the C-terminal putative transmembrane domain, was subcloned in frame with the Gal4 DNA binding domain of the bait plasmid, pGBT.Q. The construct was directly selected for its ability to drive tryptophane synthesis in the yeast strain PNY200. A human brain cDNA library (the prey library) was fused in frame to the Gal4 activating domain, transformed into the yeast strain BK100, and selected for based on its ability to drive leucine synthesis. PNY200 cells expressing the bait were then mated with BK100 cells expressing prey library proteins. The resulting diploid yeast cells expressed proteins that interacted with the bait protein and were selected based on their ability to synthesize tryptophane, leucine, histidine, and adenine. The library plasmids from these colonies were rescued, PCR amplified, and sequenced.
Plasmids.
Human Nedd4 cDNA (KIAA0093) in pBluescript was provided by T. Nagase (Kazusa DNA Research Institute, Chiba, Japan). The Nedd4 open reading frame (ORF) was PCR amplified and cloned into pFLAG-CMV5a (Invitrogen) to generate pFLAG-Nedd4. pcDNA-UL56 was described previously (39). The PY motifs in pcDNA-UL56 were mutated by site-directed mutagenesis using PCR and verified by sequencing. The Myc-ubiquitin expression plasmid was kindly provided by H. Kato (Kyushu University, Fukuoka, Japan) (31).
Transfection and superinfection.
Plasmid DNA was transiently transfected into Vero cells in 35-mm dishes by use of Lipofectamine 2000 (Invitrogen) and following the manufacturer's recommendations. One microgram of each plasmid was used routinely (except where otherwise indicated) and the total amounts of DNA were kept constant by adding the following empty expression plasmids: pFLAG-CMV5a, pcDNA3.1(+) (Invitrogen), and pCMV-Myc (Clontech, Mountain View, CA). In superinfection experiments, transfected cells were infected with wild-type (186) HSV-2 or ΔUL56Z 48 h posttransfection. The transfection efficiency was determined by the frequency of UL56-positive cells by use of immunofluorescent microscopy.
Extraction of cell lysates and Western blot analysis.
For blotting whole-cell lysates, cells were lysed directly with sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 6% 2-mercaptoethanol, 0.0025% bromophenol blue). Cell lysates were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Immobilon-P membrane; Millipore). Membranes were blocked for 1 h at room temperature in blocking buffer (5% skim milk, 0.1% Tween 20 in phosphate-buffered saline [PBS]). After being incubated with the appropriate primary antibodies for 1 h at room temperature, the blots were probed with horseradish peroxidase-conjugated secondary antibodies, which were detected using Westone (iNtRON Biotechnology, Gyeonggi, South Korea). When one membrane was blotted for several proteins, the membrane was incubated in Re-Blot Plus mild antibody stripping solution (Millipore) and then blocked and reprobed. The intensities of the bands were quantified by densitometry using ImageJ (NIH, Bethesda, MD).
Coimmunoprecipitation assay.
In assays with transfected cells, Vero cells in 100-mm dishes were transfected with plasmids encoding the UL56 wild-type (UL56WT) or UL56AY mutant proteins and harvested 48 h posttransfection. Cells were treated with 25 μM MG132 24 h prior to harvest. In assays with infected cells, Vero cells were infected with wild-type HSV-2 (186) and harvested 9 h postinfection (hpi). Harvested cells were lysed with 1 ml of lysis buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Nonident P-40, 1 mM EDTA, 10 mM NaF, protease inhibitor cocktail [Sigma]) supplemented with 25 μM MG132 and clarified by centrifugation at 15,000 rpm for 15 min at 4°C. The supernatants were precleared with protein A-agarose (Roche Applied Science, Mannheim, Germany) for 1 h and incubated with the appropriate antibodies (or normal rabbit IgG or preimmune serum) for 1 h at 4°C, followed by the addition of protein A-agarose for 1 h. After being washed with lysis buffer five times, the immunoprecipitated proteins were eluted in 2× SDS sample buffer and subjected to Western blot analysis.
Immunofluorescence and confocal microscopy.
Vero cells were plated on coverslips in 35-mm dishes and transfected or superinfected. Cells were fixed with 4% paraformaldehyde in PBS for 20 min and permeabilized with 0.1% Triton X-100 for 2 min at room temperature. Coverslips were incubated for 1 h at room temperature with 20% normal goat serum (Dako, Glostrup, Denmark) to block nonspecific antibody binding, anti-UL56 (1:200 dilution in PBS) and anti-FLAG (1:200) antibodies and then Alexa-conjugated anti-rabbit and anti-mouse secondary antibodies (1:300). Coverslips were mounted on glass slides by use of PermaFluor mounting medium (Thermo, Pittsburgh, PA). Confocal images were captured using a Zeiss LSM510 system (Carl Zeiss, Oberkochen, Germany).
Construction of the ΔUL56Z rescue mutant.
ΔUL56Z is a UL56-deficient derivative of wild-type HSV-2 (186), containing the lacZ gene in place of the UL56 ORF (40). To generate a rescue mutant of ΔUL56Z, a 1.1-kbp DNA fragment containing the complete UL56 ORF was PCR amplified from wild-type HSV-2 (186) genomic DNA by use of the primers ΔUL56Zrev-f (5′-TAACTCGAGGCGACGCCACAAAATC-3′; nucleotide position [NP] 116866 through 116881, GenBank accession number NC_001798 [19]) and ΔUL56Zrev-r (5′-AAGGATCCAATAAATTGCGTCTGCATG-3′; NP 117964 through 117982) with Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) and cloned into pcDNA3.1(+) to generate pcDNA-ΔUL56Zrev. Vero cells were cotransfected with this 1.1-kbp fragment and infectious ΔUL56Z DNA with the DEAE-dextran method (14). When plaques appeared, the cell monolayers were harvested and frozen and thawed once, and rescue mutants were selected as previously described (50). In brief, Vero cells were infected with progeny viruses and overlaid with 0.5% agarose in MEM with 2% CS. Plaques were simultaneously stained by adding a second overlay containing 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal; Sigma) on day 2 postinfection followed by a further incubation at 37°C for an additional 2 days. Colorless plaques were picked, amplified, and screened by PCR and Western blot analysis. Viral DNA was isolated from infected cells and PCR amplified with the primers check-f (5′-CAAAACAGACGCGGTGTGAG-3′; NP 116961 to 116980) and check-r (5′-ACGTCTGCGGTCTAGTGGTC-3′; NP 117876 to 117895). This approach distinguished the genomes of rescued viruses from those of the ΔUL56Z mutant, since the PCR products were quite different in size (935 bp versus 4 kbp). UL56 expression in infected cells was also examined by Western blot analysis. When the desired rescued viruses were identified, a single isolate was plaque purified three times, giving rise to the ΔUL56Zrev mutant virus.
Viral replication kinetics assay.
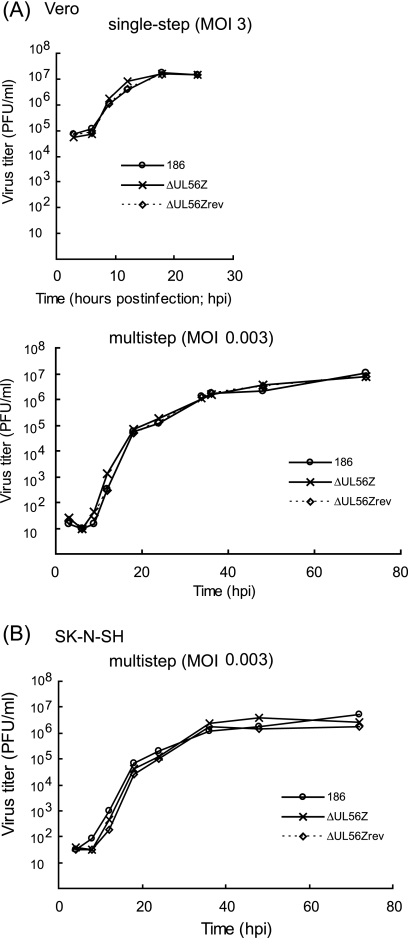
Single-step and multistep growth curve experiments were performed with Vero and SK-N-SH cells in 35-mm dishes. Cells were infected with wild-type HSV-2 (186) or the ΔUL56Z or ΔUL56Zrev mutants at an MOI of 3 or 0.003 PFU/cell. After 1 h of adsorption at 37°C, the inoculum was removed, cells were washed once with serum-free medium, and MEM containing 5% CS (for Vero) or MEM alpha containing 5% fetal CS (for SK-N-SH) was added to each culture. Cells and supernatants were harvested at the indicated times after infection. Cells were lysed by a freeze-thaw cycle, and progeny viruses were titrated on Vero cells.
Phosphatase treatment.
Vero cells infected with wild-type (186), ΔUL56Z, or ΔUL56Zrev HSV-2 were harvested 12 hpi and lysed with Nonident P-40 lysis buffer (10 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1% Nonident P-40, 1 mM EDTA) containing a protease inhibitor cocktail. Cell debris was removed by centrifugation at 12,000 rpm at 4°C for 5 min. The supernatants were mock treated or treated with 1,000 units of lambda protein phosphatase (λ-PPase; New England BioLabs, Ipswich, MA) at 30°C for 30 min. The reaction was stopped by the addition of 4× SDS buffer (1× final concentration), and the samples were subjected to Western blot analysis.
Ubiquitination assay.
In the assays with infected cells, cells were cotransfected with plasmids encoding Nedd4-FLAG and Myc-tagged ubiquitin (Myc-Ub), maintained without MG132, and then infected with wild-type (186) or ΔUL56Z HSV-2, 24 h posttransfection. Superinfected cells were maintained either without MG132 and harvested 9 hpi or treated with MG132 (25 μM) for 12 h prior to harvest (6 to 18 hpi) and harvested 18 hpi. In the assays with transfected cells, Vero cells were cotransfected with plasmids encoding Nedd4-FLAG, Myc-Ub, and UL56, treated with 25 μM MG132 for 24 h prior to harvest, and harvested 48 h posttransfection. Harvested cells were lysed with radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Nonident P-40, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 10 mM NaF, protease inhibitor cocktail) supplemented with 25 μM MG132. Immunoprecipitations were performed with anti-Nedd4 or anti-UL56 antibodies. Ubiquitinated proteins were detected using Western blot analysis with an anti-Myc antibody.
RESULTS
Several Nedd4 family ubiquitin ligases interact with UL56.
A yeast two-hybrid screen of a human brain cDNA library (using C-terminally truncated UL56 as bait) identified 10 candidate UL56-interacting genes, including the following 7 Nedd4 family genes: the Nedd4, Nedd4-2, HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 (HECW1), HECW2, itchy homolog E3 ubiquitin protein ligase (mouse) (Itch), WW domain containing E3 ubiquitin protein ligase 1 (WWP1), and WWP2 genes (data not shown).
UL56 binding to Nedd4 in transfected cells occurs via the PY motifs of UL56.
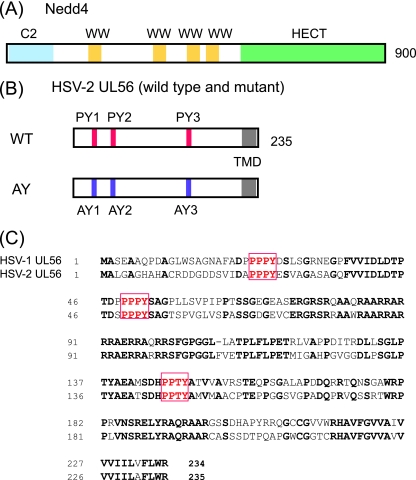
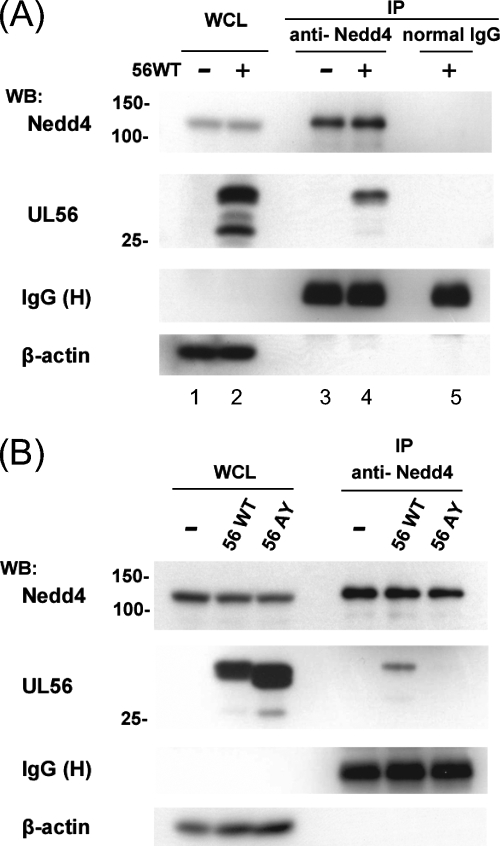
We chose to further examine Nedd4, because it is the prototypical member of the Nedd4 family of ubiquitin ligases and has been shown to be a cofactor for PPXY motif (PY motif)-dependent viral budding (6, 23, 36, 60, 71, 80). Unfortunately, other candidate Nedd4 family proteins could not be tested, either because antibodies were not available or because they were not detected in Vero cell lysates with commercially available antibodies. Nedd4 contains three types of protein domains: one amino-terminal Ca2+/lipid-binding (C2) domain (which is involved in membrane targeting), four WW domains (which bind to predominantly proline-rich motifs, including the PY motif on target proteins [11]), and a carboxyl terminal HECT domain (which coordinates with E2 and provides the catalytic E3 activity [28]) (Fig. 1A). UL56 contains three PY motifs in its N-terminal domain (two PPPY motifs and a PPTY motif), which are predicted to be located in the virion tegument or cytoplasm (Fig. 1B). These three PY motifs are conserved in HSV-1 and -2 (GenBank accession numbers NC_001806 [46] and NC_001798 [19]) (Fig. 1C), whereas VZV ORF0 (a UL56 homolog) contains only a single PPTY motif (NC_001348 [16]). To investigate whether UL56 interacts with Nedd4 in mammalian cells, we carried out coimmunoprecipitation assays with lysates obtained from Vero cells transiently expressing UL56 and immunoprecipitated with an anti-Nedd4 antibody or normal rabbit IgG (as a control). The anti-Nedd4 antibody specifically precipitated both endogenous Nedd4 and UL56 (Fig. 2A). The proteins were not detectable with control rabbit IgG (Fig. 2A, lane 5). We next tested whether the three PY motifs of UL56 were responsible for this interaction. All three proline residues in the PY motifs were mutated to alanines (UL56AY) (Fig. 1B), and UL56AY was also tested for its ability to bind to endogenous Nedd4 in the coimmunoprecipitation assay. The binding of UL56 to Nedd4 was dramatically reduced in UL56AY-expressing cells, even though UL56WT and the UL56AY mutant were expressed at comparable levels (Fig. 2B). These results demonstrate that the PY motifs of UL56 are essential for its interaction with endogenous Nedd4, although the possibility that the UL56AY mutation somehow affects the global folding of UL56 could not be ruled out.
FIG. 1.
HSV-2 UL56 contains PY motifs that interact with Nedd4. (A) Schematic representation of the Nedd4 protein. Nedd4 contains 900 amino acids that encode a Ca2+/lipid binding C2 domain (blue), three WW domains (yellow) that interact with PY motifs, and a catalytic HECT domain (green). (B) Schematic representation of UL56 and its mutant used in this study. UL56WT contains a predicted transmembrane domain (TMD) and three PY motifs (PY1, PY2, and PY3). All proline residues in the PY motifs were replaced with alanine to generate UL56AY (AY). (C) Amino acid sequence alignment of HSV-1 and -2 UL56. The HSV-1 and -2 UL56 proteins share all three PY motifs (red boxes). Identical residues are in bold.
FIG. 2.
HSV-2 UL56 interacts with Nedd4 via PY motifs in transfected cells. (A) UL56WT interacts with Nedd4. Endogenous Nedd4 was immunoprecipitated (IP) from Vero cells transiently expressing UL56WT. Western blotting (WB) with an anti-UL56 serum detected UL56 in the precipitants (second panel from the top, lane 4). Controls to detect the levels of immunoprecipitation (top, lanes 3 to 5) and expression (Nedd4, top, lanes 1 and 2; UL56, second panel from the top, lanes 1and 2) were blotted with the corresponding antibodies. (B) The UL56AY mutant does not interact with Nedd4. Immunoprecipitation and Western blot analyses were performed as described for panel A. UL56WT but not UL56AY was detected in the immunoprecipitants. β-Actin (in whole-cell lysates [WCL]) and IgG heavy chain (H) (in IP) were used as loading controls.
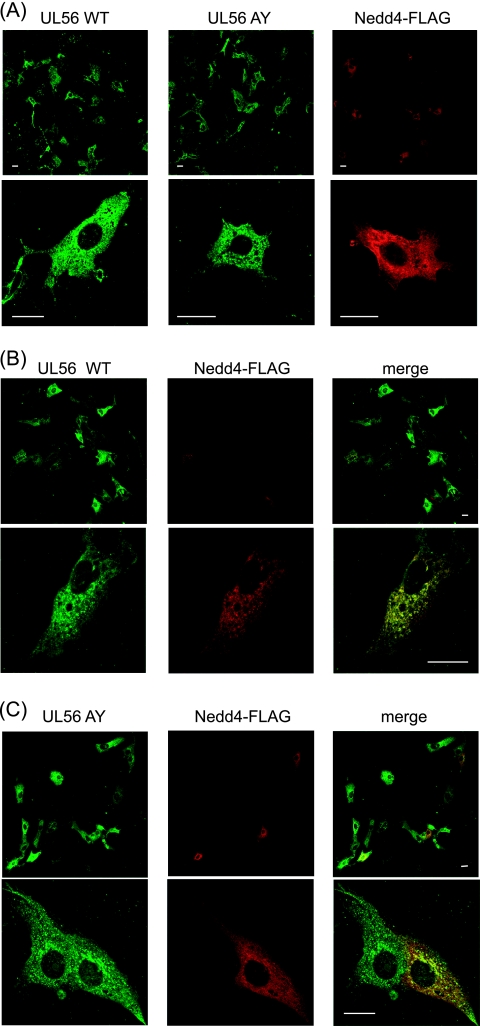
We next examined the subcellular localizations of UL56WT and -AY and FLAG-tagged Nedd4 in transiently transfected cells. Consistent with previous reports (2, 39), UL56WT was located in the perinuclear region and punctate cytoplasmic vesicles, while Nedd4-FLAG was evenly distributed throughout the cytoplasm (Fig. 3A). Coexpression of UL56 with Nedd4-FLAG markedly reduced the FLAG signal but not the UL56 signal (Fig. 3B, top), suggesting that the stability or conformation of Nedd4-FLAG was changed in the presence of UL56. However, when cells had detectable signals for both UL56WT and Nedd4, the proteins colocalized in small cytoplasmic aggregates (Fig. 3B, bottom). Similar to UL56WT, UL56AY localized to the perinuclear region and punctate cytoplasmic regions. However, in coexpressing cells, UL56AY and Nedd4-FLAG only partially colocalized with the perinuclear region (Fig. 3C). Thus, the colocalizations of UL56 and Nedd4 are consistent with the coimmunoprecipitation results and further demonstrate the importance of the PY motifs in the UL56-Nedd4 interaction.
FIG. 3.
HSV-2 UL56 and Nedd4 colocalize in cotransfected cells. (A to C) Confocal immunofluorescence analysis of the subcellular localizations of UL56WT, UL56AY, and Nedd4-FLAG. Vero cells transiently expressing Nedd4-FLAG and/or either UL56WT or UL56AY were fixed 48 h posttransfection and stained with anti-UL56 (green) and anti-FLAG (red) antibodies. (A) UL56WT and UL56AY were localized in the perinuclear region and in punctate cytoplasmic vesicles. Nedd4-FLAG was distributed throughout the cytoplasm. (B) Coexpression of UL56WT with Nedd4 reduced the FLAG signal (top). UL56 and Nedd4 colocalized in small cytoplasmic aggregates (bottom). (C) Coexpression of UL56AY with Nedd4 revealed a partial overlap in their distributions. Bars, 20 μm.
UL56 interacts with Nedd4 in infected cells.
We next tested whether UL56 and Nedd4 interacted in HSV-2-infected Vero cells. UL56 and endogenous Nedd4 were coimmunoprecipitated specifically in infected cells with the anti-Nedd4 antibody (Fig. 4A). However, two other HSV-2 proteins, which also contained PY motifs (VP5 and VP16), were not immunoprecipitated (although these proteins were readily detected in wild type-infected cell extracts). Both the PY and the P(T/S)AP motifs act as L-domains, which play a role in viral budding (5). UL36 (which contains a P(T/S)AP motif and possesses deubiquitinating enzymatic activity) and UL11 (another UL56-interacting protein) were not immunoprecipitated by the Nedd4 antibody. However, in these experiments, we found that Nedd4 was present in two forms, indicated by its different electrophoretic mobilities, and was less abundant in infected cells (Fig. 4A, top). Both forms of Nedd4 were immunoprecipitated with the anti-Nedd4 antibody.
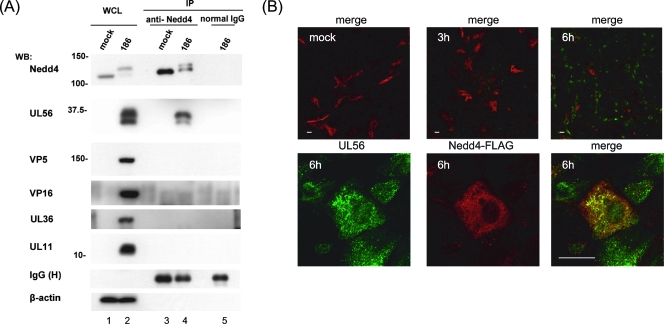
FIG. 4.
HSV-2 UL56 interacts with Nedd4 in HSV-2-infected cells. (A) Coimmunoprecipitation of UL56 with Nedd4 in HSV-2-infected cells. Lysates from Vero cells transfected with plasmids expressing Nedd4-FLAG and subsequently infected with wild-type HSV-2 were immunoprecipitated (IP) with an anti-Nedd4 antibody at 9 hpi. Western blotting (WB) with an anti-UL56 serum detected UL56 in the immunoprecipitants (second panel from the top, lane 4), whereas VP5, VP16, UL36, and UL11 were not detected by their corresponding antibodies. β-actin (in whole-cell lysates [WCL]) and IgG heavy chain (H) (in IP) were used as loading controls. (B) Colocalization of UL56 and Nedd4 in HSV-2-infected cells. Vero cells were transfected and infected as for panel A and fixed at the indicated times postinfection. Confocal immunofluorescence analysis was performed as described for Fig. 3. The detectable FLAG signal (red) was substantially reduced at 6 hpi, when UL56 (green) became detectable (top). UL56 and Nedd4-FLAG colocalized to small aggregates throughout the cytoplasm at 6 hpi. Bars, 20 μm.
We also examined the subcellular localizations of UL56 and Nedd4 in Vero cells that were transfected with a Nedd4 expression plasmid and subsequently infected with HSV-2. Nedd4-FLAG expression levels were similar for infected and mock-infected cells at 3 hpi; however, when UL56 was first detected at 6 hpi, the FLAG signal was substantially reduced (Fig. 4B, top). Moreover, when cells had detectable signals for both proteins, UL56 and Nedd4-FLAG colocalized in the cytoplasm (predominantly in the perinuclear region) (Fig. 4B, bottom). The subcellular localization of Nedd4-FLAG was not substantially changed by infection. These results indicate that UL56 also interacted with Nedd4 in infected cells and that the molecular behavior of Nedd4 was markedly affected by HSV-2 infection.
Deletion of the UL56 gene does not affect viral growth in Vero and SK-N-SH cells.
To evaluate the function of UL56, we previously generated a UL56-deficient HSV-2 mutant, the ΔUL56Z mutant (40), and then generated rescue mutants for further analysis. The plaque-purified, marker-rescued rescuant was designated the ΔUL56Zrev rescuant. We analyzed the growth properties of this mutant and rescuant to determine the contribution of the UL56 gene to HSV-2 replication in vitro. The replication patterns of the wild-type, the ΔUL56Z mutant, and the ΔUL56Zrev rescuant HSV-2 in Vero cells were virtually identical in both single-step and multistep growth analyses (Fig. 5A). Similarly, there was no significant difference between the multistep growth kinetics of the wild-type and mutant viruses in undifferentiated SK-N-SH cells (Fig. 5B). The ΔUL56Z plaques were comparable in size and morphology to those formed by the wild-type or rescuant viruses in both cell lines (data not shown). Therefore, the lack of UL56 did not cause a replication defect in these cell lines. These results agree with previous reports on HSV-1 UL56 (58).
FIG. 5.
Deletion of UL56 does not affect viral growth in Vero cells and undifferentiated SK-N-SH cells. Vero (A) and SK-N-SH (B) cells were infected with wild-type (186), ΔUL56Z, or ΔUL56Zrev HSV-2 at an MOI of 3 (single-step) or 0.003 (multistep), harvested at the indicated times postinfection, and titrated as described in Materials and Methods. The replication patterns of wild-type, ΔUL56Z, and ΔUL56Zrev viruses in Vero and SK-N-SH cells were virtually identical.
HSV-2 infection modifies Nedd4 and decreases its protein levels.
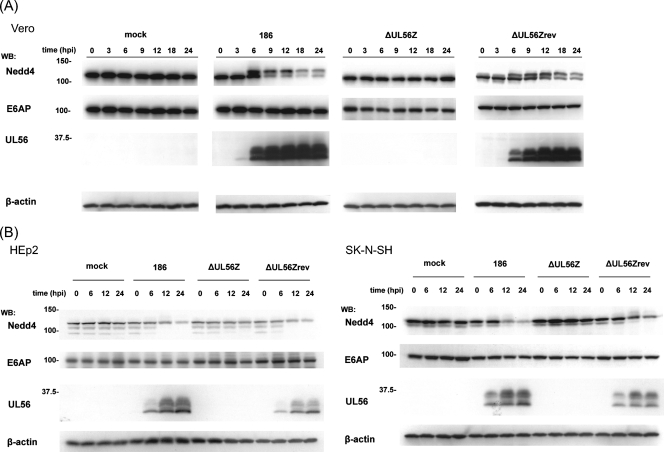
As shown in Fig. 4A, Nedd4 was detected in two forms with different electrophoretic mobilities and had decreased levels in HSV-2-infected Vero cells. We therefore investigated the kinetics of Nedd4 expression after viral infection. Early after infection with wild-type HSV-2, a faster-migrating form of Nedd4 was present (lower band, 110 kDa) (Fig. 6A). A similar band was observed for mock-infected cells. However, at 6 hpi a slower-migrating form of Nedd4 appeared (upper band, 125 kDa). The appearance of this slower-migrating form coincided with the appearance of UL56 and peaked around 9 hpi, after which both forms of Nedd4 steadily declined. In contrast, these changes in Nedd4 were not observed for mock- or ΔUL56Z-infected cells throughout a 24-h time course. E6-associated protein (E6AP), another ubiquitin ligase with a HECT domain, also did not undergo any significant changes during the course of HSV-2 infection, suggesting that HSV-2 specifically altered Nedd4. Nedd4 protein was also modified and decreased in ΔUL56Zrev-infected cells. Similar results were obtained with Vero cells infected with US2 (YY2)- or US3 (LIBR1)-deficient HSV-2 mutants (data not shown). Thus, we conclude that HSV-2 infection modifies Nedd4 and decreases its protein levels through a UL56-dependent mechanism in Vero cells.
FIG. 6.
HSV-2 infection causes a modification and decrease of Nedd4 in a UL56-dependent manner. (A) Wild-type HSV-2, but not the ΔUL56Z mutant, infection caused a modification of Nedd4 and decrease in its protein levels. Vero cells mock infected or infected with wild-type (186), ΔUL56Z, or ΔUL56Zrev HSV-2 were harvested at the indicated times postinfection. Western blots (WB) of their lysates with an anti-Nedd4 antibody showed that two forms of Nedd4 (110 kDa and 125 kDa) were present in cells infected with wild-type or ΔUL56Zrev virus, and these forms decreased as infection proceeded. E6-associated protein (E6AP), a control HECT ubiquitin ligase, showed no remarkable changes. (B) Wild-type HSV-2 infection, but not ΔUL56Z mutant infection, caused a decrease in Nedd4 protein levels in HEp2 cells and SK-N-SH cells. β-Actin was used as a loading control.
The UL56-dependent reduction in Nedd4 protein levels during HSV-2 infection was also detected in HEp2 cells and SK-N-SH cells, although the slower-migrating, modified form of Nedd4 was not as predominant (Fig. 6B).
HSV-2 infection induces Nedd4 phosphorylation and promotes proteasome-mediated degradation of Nedd4.
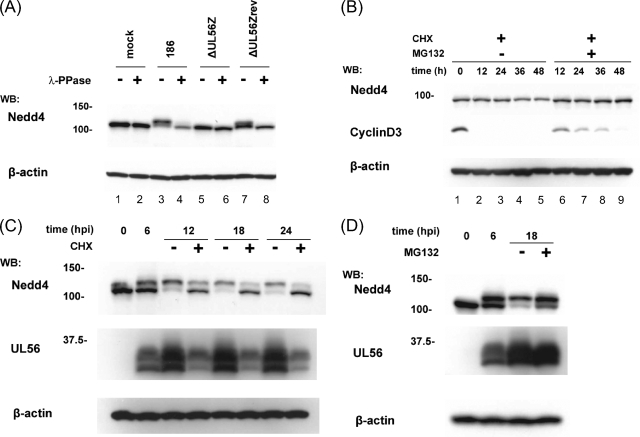
It has been shown that Nedd4 is serine/threonine phosphorylated in response to various stimuli and that its function is regulated by phosphorylation (18, 61). Therefore, we tested whether Nedd4 was phosphorylated in Vero cells during HSV-2 infection. As shown in Fig. 7A, treating cell lysates with lambda protein phosphatase converted the slower-migrating form of Nedd4 to a faster-migrating form, suggesting that Nedd4 was phosphorylated during HSV-2 infection in Vero cells.
FIG. 7.
Nedd4 is phosphorylated and actively degraded by the proteasome in HSV-2-infected cells. (A) Nedd4 is phosphorylated in HSV-2-infected cells. Lysates were extracted at 12 hpi from Vero cells that were infected with the indicated viruses, mock treated or treated with lambda protein phosphatase (λ-PPase), and subsequently analyzed by Western blotting (WB). λ-PPase treatment changed the slower-migrating form of the Nedd4 protein to a faster-migrating form in wild type (top, lanes 3 and 4)- and ΔUL56Zrev (top, lanes 7 and 8)-infected cells. (B) Nedd4 protein is more stable in noninfected cells than that in infected cells, although it is degraded by the proteasome. Uninfected Vero cells were treated with CHX (100 μg/ml) in the presence or absence of MG132 (25 μM). At the indicated times, cell lysates were analyzed by Western blotting for Nedd4 levels. Cyclin D3 was used as a control. The decrease of Nedd4 in the presence of CHX (top, lanes 4 and 5) was blocked by MG132 (top, lanes 8 and 9). (C and D) Nedd4 is degraded by the proteasome in HSV-2-infected cells. Vero cells infected with wild-type HSV-2 were mock treated or treated with CHX (100 μg/ml) (C) or MG132 (25 μM) (D) starting at 6 hpi. Cell lysates were Western blotted at the indicated times with anti-Nedd4 or anti-UL56 antibodies. β-Actin was used as a loading control. (C) CHX treatment suppressed the shift of Nedd4 to a slower-migrating form, inhibited UL56 synthesis, and promoted the decrease of the slower-migrating form of Nedd4. (D) Both forms of Nedd4 protein accumulated in cells with MG132.
We also sought to clarify the mechanism by which the levels of Nedd4 protein were decreased during HSV-2 infection. Itch and WWP1 (other Nedd4 family ubiquitin ligases) are degraded by the ubiquitin-proteasome pathway (10, 48), and Nedd4 is ubiquitinated at a steady state (78). Therefore, we investigated whether the ubiquitin-proteasome pathway degraded Nedd4 during infection.
We first evaluated the stability of Nedd4 in uninfected cells. Endogenous Nedd4 was very stable in uninfected cells in the presence of cycloheximide (CHX) (a protein synthesis inhibitor) and was maintained at constant levels for up to 24 h (Fig. 7B). Moreover, the slight reduction detected at 36 h and 48 h was completely blocked by MG132 (a proteasome inhibitor). In contrast, cyclin D3, which was used as a control (9), was rapidly degraded in the presence of CHX, and MG132 treatment delayed this degradation. These results indicate that Nedd4 is very stable but is slowly degraded by the proteasome in uninfected cells.
We next evaluated the stability of Nedd4 in infected cells. Infected cells were maintained with or without CHX (starting at 6 hpi) and harvested at various times postinfection. CHX treatment markedly reduced the slower-migrating form of Nedd4 (Fig. 7C). In the presence of CHX, the slower-migrating form of Nedd4 had almost disappeared by 24 hpi, while the faster-migrating form of Nedd4 was slightly decreased at 12 hpi and remained constant thereafter, until at least 24 hpi. From these results, it seems most likely that the slower-migrating, phosphorylated form of Nedd4 was more rapidly degraded than the faster-migrating, steady-state form in HSV-2-infected cells. However, there are two other possible explanations for the faster decrease of the phosphorylated form of Nedd4. First, because Nedd4 phosphorylation was suppressed in the presence of CHX, the phosphorylated form appeared to be more sensitive to degradation. Second, in the presence of CHX, the slower-migrating form was converted to the faster-migrating form (probably by dephosphorylation).
We further examined whether the ubiquitin-proteasome pathway was involved in the reduction of Nedd4 in infected cells. When cells were infected with wild-type virus and subsequently treated with MG132 at 6 hpi, the decrease in Nedd4 was markedly suppressed (Fig. 7D), suggesting that the ubiquitin-proteasome pathway was involved in the degradation of Nedd4. These results support the view that HSV-2 infection promotes proteasomal degradation of Nedd4 in a UL56-dependent manner.
UL56 increases ubiquitinated Nedd4 in infected cells.
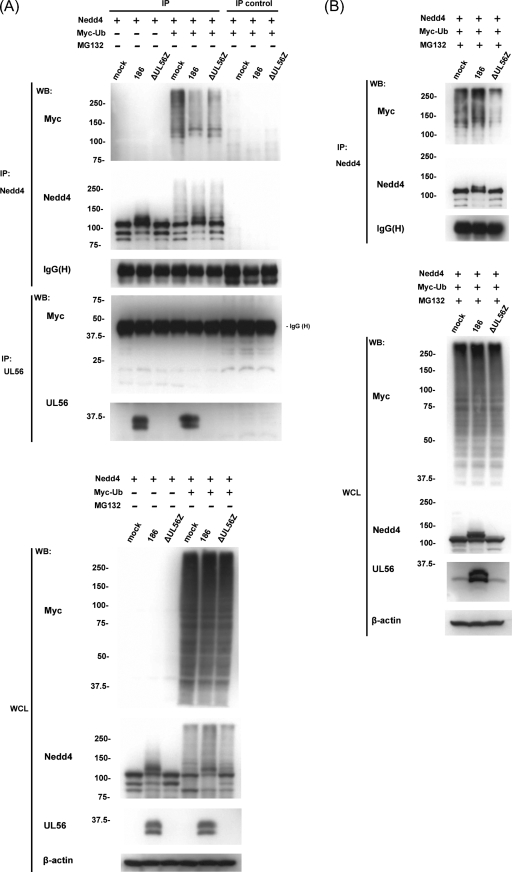
To verify this hypothesis, a ubiquitination assay was performed with superinfected cells that were transfected with plasmids expressing Nedd4-FLAG and Myc-Ub. There was no difference between the mock-infected and infected cells in their overall levels of polyubiquitination. Immunoprecipitated Nedd4 was analyzed by Western blotting with an anti-Myc antibody, which showed a high-molecular-weight smear typical of polyubiquitination. Unexpectedly, there were greater amounts of ubiquitinated Nedd4 in mock-infected cells than in cells infected with wild-type virus, and there was no significant difference between cells infected with the wild-type and ΔUL56Z viruses (Fig. 8A). We also examined the levels of Nedd4 ubiquitination in superinfected cells in the presence of MG132 (added to cultures at 6 hpi). We consistently observed that the high-molecular-weight smears detected with the anti-Myc antibody were significantly greater in wild-type virus-infected cells than in ΔUL56Z-infected cells (Fig. 8B). These results suggest that the presence of UL56 increased the ubiquitination of Nedd4 in infected cells and that ubiquitinated Nedd4 was more rapidly degraded in infected cells in the absence of MG132.
FIG. 8.
Nedd4 is polyubiquitinated at different levels in mock- and HSV-2-infected cells. Ubiquitination assays with superinfected cells in the absence (A) or presence (B) of MG132. (A) Vero cells transiently expressing Nedd4-FLAG protein and Myc-Ub were infected with wild-type (186) or ΔUL56Z HSV-2 and harvested 9 hpi. Lysates were immunoprecipitated (IP) with either anti-Nedd4 or anti-UL56 antibodies or directly subjected to Western blot (WB) analysis with anti-Myc or anti-Nedd4 antibodies. Normal rabbit IgG or preimmune serum was used as a negative control for the immunoprecipitation (IP control). Immunoprecipitated Nedd4 was Western blotted using an anti-Myc antibody, which revealed a high-molecular-weight species typical of polyubiquitination. In contrast, Western blots of immunoprecipitated UL56 showed no evidence that UL56 was ubiquitinated. The expression of Myc-Ub, Nedd4, and UL56 proteins was verified by Western blots of whole-cell lysates (WCL). (B) The experiments were performed as described for panel A, except that cells were treated with MG132 from 6 hpi and harvested 18 hpi. Polyubiquitinated Nedd4 was more abundant in wild type-infected cells than in ΔUL56Z-infected cells. The expression of the Nedd4 and UL56 proteins was verified by Western blots of WCL. β-Actin and IgG heavy chain (H) were used as loading controls. These experiments were performed three times with similar results.
UL56 decreases Nedd4 and can increase ubiquitinated Nedd4 in coexpressing cells.
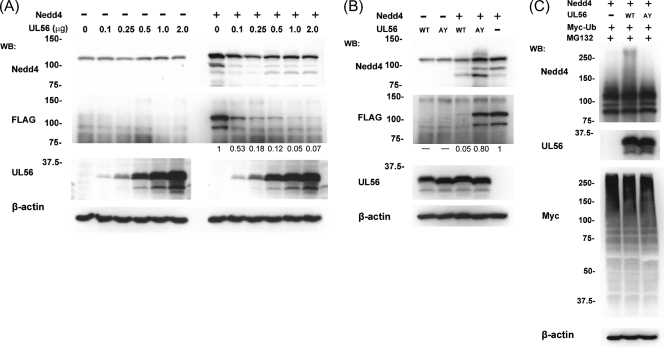
As described above, UL56 interacted with Nedd4 independent of other viral proteins. Therefore, we investigated whether UL56 affected Nedd4 ubiquitination even in the absence of other viral proteins. We first examined whether the expression of different doses of a UL56 plasmid had an effect on Nedd4. There was no significant change in endogenous Nedd4 upon expression of UL56 compared with what was seen for control transfections (Fig. 9A), which is consistent with the results in Fig. 2. However, when a constant amount of the Nedd4-FLAG expression plasmid was cotransfected with increasing amounts of the UL56 expression plasmid, Western blots with anti-FLAG antibody revealed that UL56 expression did reduce Nedd4 levels. It also showed that exogenous FLAG-tagged Nedd4 was reduced to different extents, depending on the level of UL56 expression, and was almost completely lost by the addition of 1.0 μg of UL56 expression plasmid (Fig. 9A). In contrast, when UL56AY was coexpressed with Nedd4-FLAG, there was no significant decrease in the level of Nedd4 protein (even when detected with anti-FLAG antibody) (Fig. 9B). These results indicated that UL56 expression affected the expression of exogenous Nedd4 in the absence of any other viral proteins. In these experiments, we could not rule out the possibility that endogenous Nedd4 was also affected by UL56 expression, because the transfection efficiency was 18 to 23% (data not shown). Currently, the reason why exogenous newly synthesized Nedd4 was highly sensitive to the expression of UL56 remains unknown. We next examined the effect of UL56WT and UL56AY on Nedd4 in the presence of MG132. As shown in Fig. 9C, the coexpression of UL56WT, Nedd4-FLAG, and Myc-Ub in the presence of MG132 resulted in a substantial shift to a higher-molecular-weight Nedd4 species, whereas the coexpression of UL56AY with Nedd4-FLAG and Myc-Ub did not.
FIG. 9.
UL56 affects the state of Nedd4 in cotransfected cells. (A) The overexpression of UL56 causes a decrease in the protein levels of coexpressed Nedd4. Vero cells cotransfected with a constant amount of control FLAG or Nedd4-FLAG expression plasmid and an increasing amount of UL56 expression plasmid were subjected to Western blot (WB) analysis to detect the total amounts of Nedd4 (top) and exogenous Nedd4 protein (detected with an anti-FLAG antibody; second panel from the top). The total amounts of DNA were kept constant by adding empty plasmids. The level of Nedd4 protein (especially exogenous Nedd4) decreased in a UL56 dose-dependent manner. The band intensity of FLAG was quantified and normalized to the intensity of FLAG in the absence of UL56 (values at the bottom of the FLAG panels). (B) UL56AY causes no apparent decrease in the levels of Nedd4 protein. Vero cells were cotransfected with a UL56WT or UL56AY expression plasmid or an empty expression plasmid and either a control FLAG or Nedd4-FLAG expression plasmid and analyzed as described for panel A. (C) Vero cells were cotransfected with a Nedd4-FLAG expression plasmid, a UL56WT or UL56AY expression plasmid or an empty expression plasmid, and a Myc-Ub expression plasmid in the presence of MG132. The high-molecular-weight forms of Nedd4 were the most abundant in UL56WT-expressing cells.
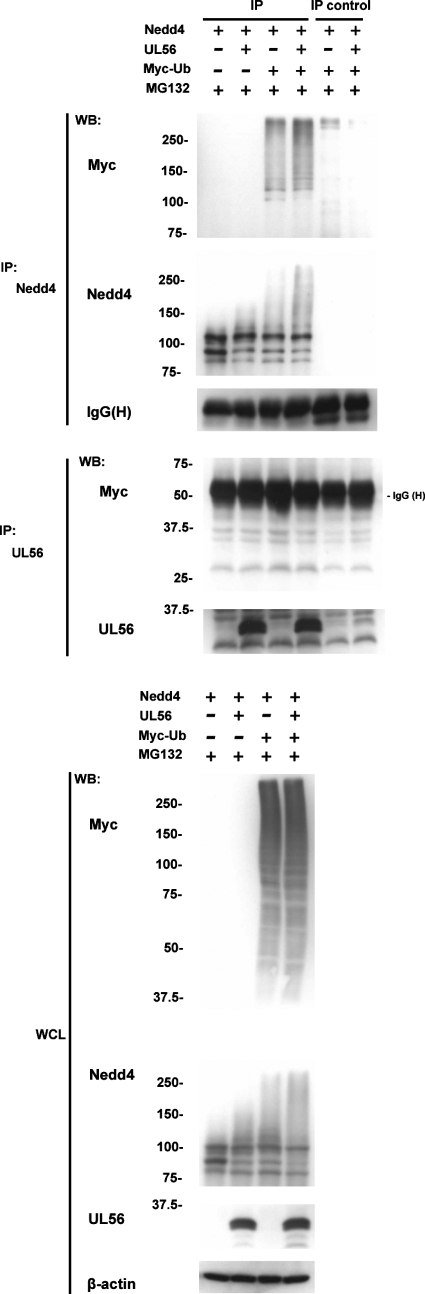
To confirm that the presence of UL56 increased the ubiquitination of Nedd4 in the absence of other viral proteins, the lysates of cotransfected cells were immunoprecipitated with an anti-Nedd4 antibody and Western blotted to detect ubiquitination with an anti-Myc antibody. These results showed that the expression of UL56 increased the ubiquitination of Nedd4 (Fig. 10).
FIG. 10.
UL56 promotes the ubiquitination of Nedd4 in cotransfected cells. Ubiquitination assays with cells overexpressing Nedd4-FLAG, UL56WT, and Myc-Ub in the presence of MG132. Cell lysates were immunoprecipitated (IP) with either anti-Nedd4 or anti-UL56 antibodies or directly subjected to Western blot (WB) analysis. Normal rabbit IgG or preimmune serum was used as a negative control for the immunoprecipitation (IP control). Immunoprecipitated Nedd4 was analyzed by Western blotting with an anti-Myc antibody, which revealed that UL56 enhanced the level of polyubiquitinated Nedd4. Western blots of immunoprecipitated UL56 showed no evidence of UL56 ubiquitination. The expression of Myc-Ub, Nedd4, and UL56 proteins was verified by Western blots of whole-cell lysates (WCL). β-Actin and IgG heavy chain (H) were used as loading controls.
UL56 is not a ubiquitination substrate.
Finally, we addressed the capacity of UL56 to serve as a substrate for Nedd4 ubiquitin ligase. Figure 8A already showed that ubiquitinated UL56 was not detected in HSV-2-infected cells. In addition, ubiquitination assays with UL56-overexpressing cells showed no evidence of UL56 ubiquitination (Fig. 10).
DISCUSSION
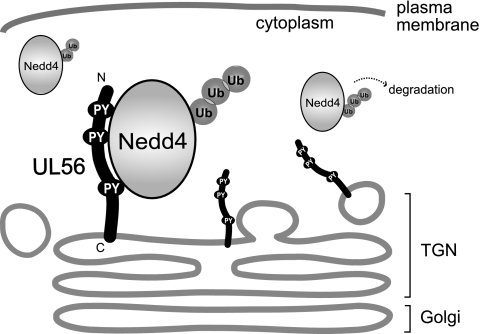
This study had four major findings. First, HSV-2 UL56 interacted with Nedd4 via its PY motifs, independent of other viral proteins. Second, HSV-2 infection induced Nedd4 phosphorylation and decreased Nedd4 protein levels in a UL56-dependent manner. Third, the expression of UL56 decreased the amount of exogenous Nedd4 (in a coexpression system), without the need for other viral proteins. This was due, at least in part, to the increased polyubiquitination of Nedd4. Fourth, UL56 was not a ubiquitin substrate. A schematic model for the interaction between UL56 and Nedd4 is shown in Fig. 11.
FIG. 11.
Model depicting the topology of HSV-2 UL56 and its interaction with Nedd4. UL56 localizes to the Golgi apparatus and cytoplasmic vesicles. UL56 possesses a hydrophobic segment near its C terminus, which anchors it to the lipid bilayer, while its N terminus resides in the cytoplasm. UL56 binds to Nedd4 via its PY motifs, which are contained in its N-terminal domain. UL56 is not ubiquitinated by Nedd4 but can increase the ubiquitination of Nedd4 and promote its degradation.
The interaction of UL56 with Nedd4 was identified using a yeast two-hybrid screen and verified by coimmunoprecipitation and immunofluorescence. The coimmunoprecipitation assay showed no evidence that Nedd4 interacted with other viral proteins containing PY motifs (VP5 and VP16), suggesting a specific interaction with UL56. However, these proteins cannot be ruled out as Nedd4-interacting proteins because Nedd4 can degrade proteins that are ubiquitination substrates. A binding assay with catalytically inactive Nedd4 would be necessary to determine whether other viral proteins that contain PY motifs interact with Nedd4. UL11, another UL56-interacting protein, was not detected in the UL56-Nedd4 complex. This result suggests that UL11 is not a member of the complex or that UL11 does not tightly interact with the complex, because the interaction between UL11 and UL56 was detected under solubilization conditions that were milder than the ones used in this study (40).
We found that HSV-2 infection caused a decrease of Nedd4 protein levels through a UL56-dependent mechanism, and this decrease was strongly inhibited by the addition of MG132, a specific inhibitor of the proteasome. We therefore investigated the involvement of the ubiquitin-proteasome pathway in the UL56-dependent degradation of Nedd4 by use of a ubiquitination assay on infected cells. However, these experiments unexpectedly revealed that ubiquitinated Nedd4 was not increased but was equally decreased in cells that were infected with wild-type and UL56-deficient HSV-2 (compared to noninfected cells). In contrast, the addition of MG132 caused a marked accumulation of Nedd4, but only in wild type-infected cells. One hypothesis explaining these results is that infection with wild-type or UL56-deficient viruses reduces a common form of ubiquitinated Nedd4, which is stable in noninfected cells, while UL56 increases another type of ubiquitinated Nedd4 that is more susceptible to proteasomal degradation. In this model, the phosphorylation of Nedd4 may be involved in its susceptibility to degradation. Substrate phosphorylation is one mechanism by which ubiquitination is regulated (55). Another explanation for the reduction in ubiquitinated Nedd4 in infected cells (without a proteasome inhibitor) is that HSV-2, like hepatitis B and C viruses, alters proteasomal activity (26, 35). The reduced viral growth in the presence of a proteasome inhibitor indicates that cellular proteasomal activity is critical for HSV replication (15). Since there was no significant difference in the overall levels of polyubiquitination between HSV-2-infected and mock-infected cells (Fig. 8A), a selective mechanism may play a role in this proteasomal degradation in infected cells.
When the ubiquitination assay was performed with transfected cells in the presence of MG132, we found that the coexpression of UL56 increased the amount of ubiquitinated Nedd4. In contrast, ubiquitinated UL56 was not detectable under the conditions of our assay, suggesting that UL56 is not a ubiquitination substrate for Nedd4 (although UL56 does interact with Nedd4 via its PY motifs). Additionally, the above observations suggest that UL56 (on its own) can increase the polyubiquitination of Nedd4. However, the mechanism regulating Nedd4 ubiquitination remains unclear. Another Nedd4 family ligase, Itch, undergoes autoubiquitination but is simultaneously deubiquitinated by deubiquitinating enzymes (DUBs) and therefore is protected from proteasomal degradation (48). Two possibilities are raised for the underlying mechanism by which UL56 increases ubiquitinated Nedd4. First, UL56 might act as an accessory or adapter protein for the ubiquitination of Nedd4. In this situation, UL56 could promote Nedd4 ubiquitination by accelerating autoubiquitination, recruiting ubiquitin ligases to Nedd4, or suppressing deubiquitination of Nedd4. HSV encodes ubiquitin-related enzymes: ubiquitin ligase (ICP0 [7, 72]) and DUB (UL36 [33]). Cellular ligases and/or DUBs could also increase Nedd4 ubiquitination and cause a decrease in the levels of Nedd4 protein in transfected cells. Second, UL56 itself may act as a ubiquitin ligase for Nedd4. However, this seems unlikely, because UL56 has no domains that are homologous to the known catalytic domains of ubiquitin ligases. Further studies will be needed to clarify the mechanisms by which UL56 increases ubiquitinated Nedd4.
Based on these findings, we propose that UL56 regulates Nedd4 during HSV-2 infection. Phosphorylation and ubiquitination of Nedd4 have profound effects on the activity of Nedd4 (18, 78). The Nedd4 family of ubiquitin ligases have been shown to regulate signal transduction during viral infection (27) and viral budding by interacting with viral proteins. However, to date there have been no reports that viral proteins regulate the Nedd4 family of ubiquitin ligases. Thus, our research represents a new interaction between the Nedd4 family of ubiquitin ligases and viral proteins. Given that alterations of Nedd4 during HSV-2 infection had no effect on viral proliferation in this study, Nedd4 may play a role only under specific conditions or may play a redundant role in HSV-2 infection. Also, Nedd4 could play a more important role during in vivo HSV infection.
Nedd4 regulates viral budding of Ebola virus (23, 80), human T-cell leukemia virus type 1 (6, 60), and Rous sarcoma virus (36, 71). In addition to these viruses, many enveloped RNA and DNA viruses exploit the multivesicular body (MVB) machinery for budding, such as retroviruses (21, 56, 74, 79, 81), rhabdoviruses (12), other filoviruses (38), arenaviruses (54, 64), paramyxoviruses (59, 62), hepatitis B virus (44, 76), HSV (8, 13), and probably orthomyxoviruses and human herpes virus type 6 (47). The cellular MVB machinery requires the coordinated action of endosomal sorting complex required for transport I (ESCRT-I), ESCRT-II, and ESCRT-III, and several Nedd4 family E3s, which act as functional links between ESCRT and the viral L-domains (including the PY motifs). Crump et al. reported that Vps4, which was essential for ESCRT function, was required for HSV-1 cytoplasmic envelopment, suggesting that HSV also exploits the MVB machinery (13). Seven L-domain containing proteins are present in both HSV-1 and -2: the PY motif-containing proteins ICP0, VP5, VP16, and UL56 and the P(T/S)AP motif-containing proteins UL29, UL36 and glycoprotein E. The roles of these putative L-domains have not been investigated to date. The PY motifs of UL56 could be linked to ESCRT by Nedd4 and function as L-domains. The functional redundancy of these L-domain-containing proteins could explain why the deletion of UL56 did not inhibit HSV replication in Vero cells and undifferentiated SK-N-SH cells.
Alternatively, like the cellular Nedd4-interacting proteins N4WBP5 (24), N4WBP5A (63), and LAPTM5 (52), UL56 could be involved in protein sorting or membrane trafficking. The association of UL56 with the neuron-specific kinesin KIF1A leads us to speculate that UL56 has a role in neurons.
In conclusion, this study showed that UL56 interacts with Nedd4 and can promote Nedd4 degradation through the ubiquitin-proteasome pathway during HSV-2 infection, suggesting that UL56 is a regulator of Nedd4. Nedd4 has been suggested to play crucial roles in the replication of various RNA and DNA viruses. The function, mechanism, and significance of regulating Nedd4 in HSV-2 infection remain unclear and warrant further investigation.
Acknowledgments
We thank Hiroki Kato for providing the Myc-ubiquitin expression plasmid and Takahiro Nagase for providing the human Nedd4 cDNA (KIAA0093). We are also grateful to Maki Kamakura, Chenghong Luo, and Yohei Yamauchi for their technical suggestions and discussions and to Kazuko Nagamoto for her technical assistance.
This study was supported by a Grant-in-Aid for Scientific Research in Priority Areas and a 21st Century Center of Excellence (COE) grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anan, T., Y. Nagata, H. Koga, Y. Honda, N. Yabuki, C. Miyamoto, A. Kuwano, I. Matsuda, F. Endo, H. Saya, and M. Nakao. 1998. Human ubiquitin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells 3751-763. [DOI] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 665168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz, C., M. Moyal, A. Rosen-Wolff, G. Darai, and Y. Becker. 1994. Herpes simplex virus type 1 (HSV-1) UL56 gene is involved in viral intraperitoneal pathogenicity to immunocompetent mice. Arch. Virol. 13473-83. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 34455-63. [DOI] [PubMed] [Google Scholar]
- 6.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 7711882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calistri, A., P. Sette, C. Salata, E. Cancellotti, C. Forghieri, A. Comin, H. Gottlinger, G. Campadelli-Fiume, G. Palu, and C. Parolin. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 8111468-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casanovas, O., M. Jaumot, A. B. Paules, N. Agell, and O. Bachs. 2004. P38SAPK2 phosphorylates cyclin D3 at Thr-283 and targets it for proteasomal degradation. Oncogene 237537-7544. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C., and L. E. Matesic. 2007. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 26587-604. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H. I., and M. Sudol. 1995. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA 927819-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 733359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 817380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen, B. R. 1987. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 152684-704. [DOI] [PubMed] [Google Scholar]
- 15.Dai-Ju, J. Q., L. Li, A. Johnson, and R. M. Sandri-Goldin. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 803567-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 671759-1816. [DOI] [PubMed] [Google Scholar]
- 17.Delhon, G., M. P. Moraes, Z. Lu, C. L. Afonso, E. F. Flores, R. Weiblen, G. F. Kutish, and D. L. Rock. 2003. Genome of bovine herpesvirus 5. J. Virol. 7710339-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinudom, A., A. B. Fotia, R. J. Lefkowitz, J. A. Young, S. Kumar, and D. I. Cook. 2004. The kinase Grk2 regulates Nedd4/Nedd4-2-dependent control of epithelial Na+ channels. Proc. Natl. Acad. Sci. USA 10111886-11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 722010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, G., and H. Luo. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 845-14. [DOI] [PubMed] [Google Scholar]
- 21.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 10755-65. [DOI] [PubMed] [Google Scholar]
- 22.Gray, W. L., B. Starnes, M. W. White, and R. Mahalingam. 2001. The DNA sequence of the simian varicella virus genome. Virology 284123-130. [DOI] [PubMed] [Google Scholar]
- 23.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 9713871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey, K. F., L. M. Shearwin-Whyatt, A. Fotia, R. G. Parton, and S. Kumar. 2002. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J. Biol. Chem. 2779307-9317. [DOI] [PubMed] [Google Scholar]
- 25.Hatakeyama, S., J. P. Jensen, and A. M. Weissman. 1997. Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J. Biol. Chem. 27215085-15092. [DOI] [PubMed] [Google Scholar]
- 26.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 737231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda, A., R. G. Caldwell, R. Longnecker, and M. Ikeda. 2003. Itchy, a Nedd4 ubiquitin ligase, downregulates latent membrane protein 2A activity in B-cell signaling. J. Virol. 775529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingham, R. J., G. Gish, and T. Pawson. 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 231972-1984. [DOI] [PubMed] [Google Scholar]
- 29.Izumiya, Y., H. K. Jang, M. Ono, and T. Mikami. 2001. A complete genomic DNA sequence of Marek's disease virus type 2, strain HPRS24. Curr. Top. Microbiol. Immunol. 255191-221. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, Y. M., H. Yamada, F. Goshima, T. Daikoku, S. Oshima, K. Wada, and Y. Nishiyama. 1998. Characterization of the herpes simplex virus type 2 (HSV-2) US2 gene product and a US2-deficient HSV-2 mutant. J. Gen. Virol. 792777-2784. [DOI] [PubMed] [Google Scholar]
- 31.Kato, H., K. Sakaki, and K. Mihara. 2006. Ubiquitin-proteasome-dependent degradation of mammalian ER stearoyl-CoA desaturase. J. Cell Sci. 1192342-2353. [DOI] [PubMed] [Google Scholar]
- 32.Kato, K., T. Daikoku, F. Goshima, H. Kume, K. Yamaki, and Y. Nishiyama. 2000. Synthesis, subcellular localization and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch. Virol. 1452149-2162. [DOI] [PubMed] [Google Scholar]
- 33.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19547-557. [DOI] [PubMed] [Google Scholar]
- 34.Kemble, G. W., P. Annunziato, O. Lungu, R. E. Winter, T. A. Cha, S. J. Silverstein, and R. R. Spaete. 2000. Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells. J. Virol. 7411311-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khu, Y. L., Y. J. Tan, S. G. Lim, W. Hong, and P. Y. Goh. 2004. Hepatitis C virus non-structural protein NS3 interacts with LMP7, a component of the immunoproteasome, and affects its proteasome activity. Biochem. J. 384401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 9811199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolesnikova, L., B. Berghofer, S. Bamberg, and S. Becker. 2004. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J. Virol. 7812277-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koshizuka, T., F. Goshima, H. Takakuwa, N. Nozawa, T. Daikoku, O. Koiwai, and Y. Nishiyama. 2002. Identification and characterization of the UL56 gene product of herpes simplex virus type 2. J. Virol. 766718-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koshizuka, T., Y. Kawaguchi, F. Goshima, I. Mori, and Y. Nishiyama. 2006. Association of two membrane proteins encoded by herpes simplex virus type 2, UL11 and UL56. Virus Genes 32153-163. [DOI] [PubMed] [Google Scholar]
- 41.Koshizuka, T., Y. Kawaguchi, and Y. Nishiyama. 2005. Herpes simplex virus type 2 membrane protein UL56 associates with the kinesin motor protein KIF1A. J. Gen. Virol. 86527-533. [DOI] [PubMed] [Google Scholar]
- 42.Kumar, S., K. F. Harvey, M. Kinoshita, N. G. Copeland, M. Noda, and N. A. Jenkins. 1997. cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics 40435-443. [DOI] [PubMed] [Google Scholar]
- 43.Kumar, S., Y. Tomooka, and M. Noda. 1992. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 1851155-1161. [DOI] [PubMed] [Google Scholar]
- 44.Lambert, C., T. Doring, and R. Prange. 2007. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J. Virol. 819050-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 7512209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 691531-1574. [DOI] [PubMed] [Google Scholar]
- 47.Mori, Y., M. Koike, E. Moriishi, A. Kawabata, H. Tang, Y. Uchiyama, and K. Yamanishi. 2007. HHV-6 infection induces multivesicular body (MVB) formation in T cells and the virus egress is gated through MVB, abstr. 6.27. Abstr. 32nd Annu. Int. Herpesvirus Workshop, Asheville, NC.
- 48.Mouchantaf, R., B. A. Azakir, P. S. McPherson, S. M. Millard, S. A. Wood, and A. Angers. 2006. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 28138738-38747. [DOI] [PubMed] [Google Scholar]
- 49.Murdaca, J., C. Treins, M. N. Monthouel-Kartmann, R. Pontier-Bres, S. Kumar, E. Van Obberghen, and S. Giorgetti-Peraldi. 2004. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J. Biol. Chem. 27926754-26761. [DOI] [PubMed] [Google Scholar]
- 50.Nishiyama, Y., Y. Yamada, R. Kurachi, and T. Daikoku. 1992. Construction of a US3 lacZ insertion mutant of herpes simplex virus type 2 and characterization of its phenotype in vitro and in vivo. Virology 190256-268. [DOI] [PubMed] [Google Scholar]
- 51.Okada, Y., H. Yamazaki, Y. Sekine-Aizawa, and N. Hirokawa. 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81769-780. [DOI] [PubMed] [Google Scholar]
- 52.Pak, Y., W. K. Glowacka, M. C. Bruce, N. Pham, and D. Rotin. 2006. Transport of LAPTM5 to lysosomes requires association with the ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J. Cell Biol. 175631-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perelygina, L., L. Zhu, H. Zurkuhlen, R. Mills, M. Borodovsky, and J. K. Hilliard. 2003. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine herpesvirus 1) from a rhesus monkey. J. Virol. 776167-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 10012978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70503-533. [DOI] [PubMed] [Google Scholar]
- 56.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 7210218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roizman, B., D. M. Knipe, and R. J. Whitely. 2007. Herpes simplex viruses, p. 2502-2601. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkin, Philadelphia, PA.
- 58.Rosen-Wolff, A., and G. Darai. 1991. Identification and mapping of the UL56 gene transcript of herpes simplex virus type 1. Virus Res. 19115-126. [DOI] [PubMed] [Google Scholar]
- 59.Sakaguchi, T., A. Kato, F. Sugahara, Y. Shimazu, M. Inoue, K. Kiyotani, Y. Nagai, and T. Yoshida. 2005. AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J. Virol. 798933-8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakurai, A., J. Yasuda, H. Takano, Y. Tanaka, M. Hatakeyama, and H. Shida. 2004. Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect. 6150-156. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez-Perez, A., S. Kumar, and D. I. Cook. 2007. GRK2 interacts with and phosphorylates Nedd4 and Nedd4-2. Biochem. Biophys. Res. Commun. 359611-615. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt, A. P., G. P. Leser, E. Morita, W. I. Sundquist, and R. A. Lamb. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 792988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shearwin-Whyatt, L. M., D. L. Brown, F. G. Wylie, J. L. Stow, and S. Kumar. 2004. N4WBP5A (Ndfip2), a Nedd4-interacting protein, localizes to multivesicular bodies and the Golgi, and has a potential role in protein trafficking. J. Cell Sci. 1173679-3689. [DOI] [PubMed] [Google Scholar]
- 64.Strecker, T., R. Eichler, J. Meulen, W. Weissenhorn, H. Dieter Klenk, W. Garten, and O. Lenz. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J. Virol. 7710700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189304-316. [DOI] [PubMed] [Google Scholar]
- 66.Telford, E. A., M. S. Watson, J. Perry, A. A. Cullinane, and A. J. Davison. 1998. The DNA sequence of equine herpesvirus-4. J. Gen. Virol. 791197-1203. [DOI] [PubMed] [Google Scholar]
- 67.Thureen, D. R., and C. L. Keeler, Jr. 2006. Psittacid herpesvirus 1 and infectious laryngotracheitis virus: comparative genome sequence analysis of two avian alphaherpesviruses. J. Virol. 807863-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 747980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tyler, S. D., G. A. Peters, and A. Severini. 2005. Complete genome sequence of cercopithecine herpesvirus 2 (SA8) and comparison with other simplexviruses. Virology 331429-440. [DOI] [PubMed] [Google Scholar]
- 70.Tyler, S. D., and A. Severini. 2006. The complete genome sequence of herpesvirus papio 2 (Cercopithecine herpesvirus 16) shows evidence of recombination events among various progenitor herpesviruses. J. Virol. 801214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vana, M. L., Y. Tang, A. Chen, G. Medina, C. Carter, and J. Leis. 2004. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J. Virol. 7813943-13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 988815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vecchione, A., A. Marchese, P. Henry, D. Rotin, and A. Morrione. 2003. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol. Cell. Biol. 233363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, H., K. M. Norris, and L. M. Mansky. 2002. Analysis of bovine leukemia virus gag membrane targeting and late domain function. J. Virol. 768485-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, X., L. C. Trotman, T. Koppie, A. Alimonti, Z. Chen, Z. Gao, J. Wang, H. Erdjument-Bromage, P. Tempst, C. Cordon-Cardo, P. P. Pandolfi, and X. Jiang. 2007. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watanabe, T., E. M. Sorensen, A. Naito, M. Schott, S. Kim, and P. Ahlquist. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. USA 10410205-10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2169-178. [DOI] [PubMed] [Google Scholar]
- 78.Woelk, T., B. Oldrini, E. Maspero, S. Confalonieri, E. Cavallaro, P. P. Di Fiore, and S. Polo. 2006. Molecular mechanisms of coupled monoubiquitination. Nat. Cell Biol. 81246-1254. [DOI] [PubMed] [Google Scholar]
- 79.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 724095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 779987-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 747250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]